Abstract
Background
Spread of the bla NDM-1 gene that encodes the New Delhi metallo-β-lactamase (NDM-1) in Enterobacteriaceae is a major global health problem. Plasmids carrying bla NDM-1 from two different multi-drug resistant Klebsiella pneumonia isolates collected in Singapore were completely sequenced and compared to known plasmids carrying bla NDM-1.
Methodology/Principal Findings
The two plasmids, pTR3 and pTR4, were transferred to Escherichia coli recipient strain J53 and completely sequenced by a shotgun approach using 3-kb paired-end libraries on 454. Although the K. pneumoniae strains were unrelated by molecular typing using PFGE and MLST, complete sequencing revealed that pTR3 and pTR4 are identical. The plasmid sequence is similar to the E. coli NDM-1-encoding plasmid p271A, which was isolated in Australia from a patient returning from Bangladesh. The immediate regions of the bla NDM-1 gene in pTR3/4 are identical to that of p271A, but the backbone of our plasmid is much more similar to another IncN2 plasmid reported recently, pJIE137, which contained an additional 5.2-kb CUP (conserved upstream repeat) regulon region in comparison to p271A. A 257-bp element bounded by imperfect 39-bp inverted repeats (IR) and an incomplete version of this element flanking the 3.6-kb NDM-1-encoding region were identified in these plasmids and are likely to be the vestiges of an unknown IS.
Conclusions
Although the hosts are not epidemiologically linked, we found that the plasmids bearing the bla NDM-1 gene are identical. Comparative analyses of the conserved NDM-1-encoding region among different plasmids from K. pneumoniae and E. coli suggested that the transposable elements and the two unknown IR-associated elements flanking the NDM-1-encoding region might have aided the spreading of this worrisome resistance determinant.
Introduction
The NDM-1 carbapenemase gene has become an important resistant determinant in Gram-negative bacteria [1], [2]. NDM-1 is able to hydrolyze almost all β-lactam antibiotics and when combined with other resistance mechanisms, renders the host bacterium resistant to almost all antibiotics [3], [4]. The rapid spread of these multidrug resistant strains is now a matter of global concern.
Initially, plasmids encoding bla NDM-1 were observed in Klebsiella pneumoniae and Escherichia coli [5]. These plasmids can conjugatively transfer into other species. The concern in India is the heavy contamination of this gene in seepage water with the possibility of spread in the community [6]. Travelers may be colonized with NDM-1 producing strains in the gut resulting in spread of the gene to different countries [7], [8]. The bla NDM-1 gene has been identified on different plasmids types that vary in length from ∼50 to 300 kb [9], [10]. In addition, bla NDM-1 has recently been identified in the chromosome of Acinetobacter baumannii [11]. The resistance gene was also reported recently in other bacterial species, such as Vibrio cholerae [6]. Thus, the rapid global spread of bla NDM-1 may not be explainable by a single mechanism.
In this study, the complete sequence of conjugatively transferrable plasmids encoding NDM-1 from two K. pneumoniae clinical isolates were determined to investigate the genetic basis of the resistance gene. Comparative analyses were carried out with existing sequences to investigate the molecular mechanism underlying the spread of bla NDM-1 in bacteria.
Materials and Methods
Patients’ Characteristics
Patient 1 was a 36 year old male Chinese local with lymphocytic meningitis of undetermined cause. He had no recent travel history in the last year. Multi-drug resistant K. pneumoniae 43320 was a clinical isolate from urine during his rehabilitation 3 months after admission. He had a single spike of temperature but was not septic. He recovered without specific antimicrobial treatment. Patient 2 was a 22 year old male foreigner from Vietnam admitted 2 months after Patient 1 to a different ward in the same hospital with T4 hemangioma with cord compression. Multi-drug resistant K. pneumoniae 44951 was a clinical isolate from urine 8 days after admission and 10 days from the isolate from patient 1. As this was a catheter specimen, it was considered as insignificant and no specific antimicrobial treatment was given. Although their hospital stays overlapped, there was no obvious epidemiological link between the 2 patients.
Antimicrobial Susceptibility Testing
The MICs of 15 antimicrobial agents were determined using the broth microdilution test according to the recommendations of the Clinical and Laboratory Standards Institute [12].
General characteristics of NDM-1 Carrying K. pneumoniae
The 2 carbapenem resistant K. pneumoniae were confirmed to be carrying bla NDM-1 by PCR and subsequent sequencing according to previously published primers for bla NDM-1 [13]. Plasmid conjugation was performed using E. coli J53 azide resistant strain as recipient [14]. Briefly, recipients and bla NDM-1 carrying K. pneumoniae were separately inoculated into brain heart infusion broth (Oxoid Ltd., Basingstoke, England) and incubated at 37°C for 4 h. They were then mixed at a ratio of 1∶10 (Donor:Recipient by volume) for overnight incubation at 37°C. A 0.1-ml volume of the overnight broth mixture was spread onto a MacConkey agar plate containing sodium azide (100 µ/mL) and imipenem (2 µg/mL).
Molecular Typing for NDM-1 Carrying K. pneumoniae and their Transconjugants
Multilocus sequencing typing (MLST) and Pulsed field gel electrophoresis (PFGE) were performed for both bla NDM-1 carrying K. pneumoniae strains. Sequences of seven housekeeping genes for MLST were obtained according to Diancourt et al., Sequences were compared with those on the MLST web site (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html) developed by Diancourt et al., [15] and alleles and sequence types (STs) were assigned accordingly. If there was a difference in two or more alleles the strains were considered to be unrelated.
For PFGE, DNA was prepared as described previously [16]. The restriction enzyme XbaI (New England Biolabs, Beverly, MA, USA) was used at the manufacturer’s suggested temperature. Restriction fragments were separated by PFGE in 1% agarose gel (Bio-Rad, Hercules, CA, USA) in 0.5×TBE buffer (45 mM Tris, 45 mM boric acid, 1.0 mM EDTA, pH8.0) for 22 h at 200 V at a temperature of 14°C, with ramped times of 2 to 40 s using the Bio-Rad CHEF MAPPER apparatus (Bio-Rad Laboratories, Richmond, CA, USA). Gels were then stained with ethidium bromide and photographed under ultraviolet light. The resulting genomic DNA profiles, or “fingerprints”, were interpreted according to established guidelines [17]. Plasmid replicon typing was performed for transconjugants [18].
Plasmid Sequencing
DNA sequencing of the NDM-1-carrying plasmids was performed with a whole genome shotgun approach using 3-kb paired-end libraries [19]. DNA fragments of about 3-kb in length were recovered after hydrodynamic shearing and purified using size exclusion beads (AMPure, Agencourt). The DNA fragments were subsequently linked to adaptors and circularized, then sheared again by nebulization. The resulting nucleotide fragments containing the adaptor were specifically purified, then ligated to oligomers for PCR amplification. The following emulsion-based clonal amplification (emPCR) was performed following standard 454 pyrosequencing protocols. Sequencing was performed using a 454 GS Jr (454 Life Sciences, Branford, CT, USA). The complete nucleotide sequences of plasmid pTR3 and pTR4 have been submitted to GenBank and assigned sequence accession number JQ349086 and JQ349085.
Bioinformatics Analysis
De-novo sequence assembly was performed on a computer workstation using the 454 Newbler, which automatically detects long paired-end reads (Version 2.6, 454 Life Sciences, Branford, CT, USA). The contigs were manually inspected and reassembled using the Phred/Phrap/Consed [20]. Annotation of the plasmid was manually curated after performing automatic annotation on the RAST Server [20]. Insertion sequences and transposons were further annotated using ISfinder (http://www-is.biotoul.fr) [21].
Results
Antimicrobial Susceptibility Testing Results for NDM-1 Carrying K. pneumoniae and their Transconjugants
Antimicrobial susceptibility testing results showed that bla NDM-1 carrying K. pneumoniae clinical isolates, 43320 and 44951, from patient 1 and 2 respectively, were resistant to all tested antibiotics (Table 1). Addition of a β-lactamase inhibitor did not increase the susceptibility to β-lactams. The E. coli transconjugants, TCJ-P1 and TCJ-P2, respectively from 43320 and 44951 had a different resistance profile when compared with the corresponding clinical isolates but were still resistant to all β-lactams except aztreonam. Reduced MICs for imipenem and meropenem were observed in both transconjugants. Both transconjugants were susceptible to non-β -lactam antibiotics including ciprofloxacin, gentamicin, tetracycline and trimethoprim/sulfamethoxazole. PFGE of 43320 and 44951 showed that they were unrelated with more than six bands difference. MLST indicated that 43320 and 44951 belonged to ST273 and ST1 respectively (data not shown). The plasmid incompatibility typing initially was positive for a product with a size consistent with the PCR product for IncN. However, subsequent sequencing of the PCR products showed unrelated sequences for a putative IS911 transposase orfA with KpLE2 phage-like element.
Table 1. Antimicrobial susceptibility test among bla NDM-1 carrying isolates and their transconjugants.
| Antibiotics | Minimal inhibitory concentration (µg/µl) | ||||
| 43320 | TCJ-P1 | 44951 | TCJ-P2 | ||
| Ampicillin | ≥32 | ≥32 | ≥32 | ≥32 | |
| piperacillin/tazobactam | ≥128 | ≥128 | ≥128 | ≥128 | |
| Cefazolin | ≥32 | ≥32 | ≥32 | ≥32 | |
| Cefpodoxime | ≥64 | ≥64 | ≥64 | ≥64 | |
| Cefoxitin | ≥128 | ≥128 | ≥128 | ≥128 | |
| Cefotaxime | ≥128 | ≥128 | ≥128 | ≥128 | |
| Cefotaxime/clavulanate | ≥128 | ≥128 | ≥128 | 64 | |
| Ceftazidime | ≥128 | ≥128 | ≥128 | ≥128 | |
| Ceftazidime/clavulanate | ≥128 | ≥128 | ≥128 | ≥128 | |
| Ceftriaxone | ≥128 | ≥128 | ≥128 | ≥128 | |
| Cefepime | ≥32 | ≥32 | ≥32 | ≥32 | |
| Aztreonam | ≥32 | ≤4 | ≥32 | ≤4 | |
| Imipenem | 16 | 8 | 16 | 4 | |
| Meropenem | ≥16 | 4 | ≥16 | 2 | |
| Ciprofloxacin | ≥4 | ≤1 | ≥4 | ≤1 | |
| Gentamicin | ≥32 | ≤4 | ≥32 | ≤4 | |
| Tetracycline | ≥32 | ≤4 | ≥32 | ≤4 | |
| †Trimethoprim/sulfamethoxazole | ≥8 | ≤1 | ≥8 | ≤1 | |
43320 and 44951, clinical isolates from patient 1 and 2 respectively; TCJ-P1 and TCJ-P2, transconjugants from 43320 and 44951 respectively.
The MIC is presented according to the concentration of trimethoprim.
Sequence Annotation and Comparison of the Two blaNDM-1 Plasmids
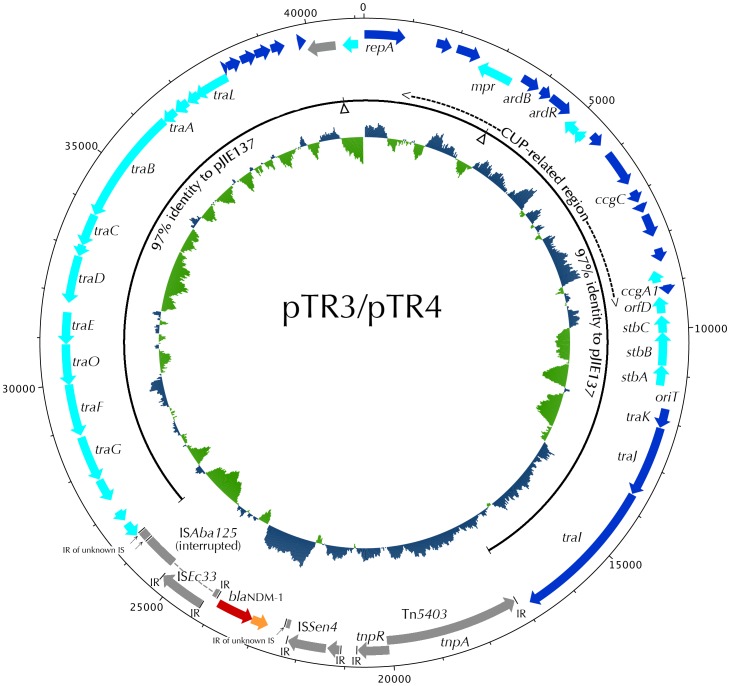
Complete sequencing was performed for the two circular bla NDM-1 plasmids, pTR3 and pTR4, respectively from 43320 and 44951. The results of the assemblies of the two plasmids based on 454 reads were almost identical, in that only seven locations of indels were found between pTR3 and pTR4. Subsequent sequence verifications by Sanger reads have shown that the two 41,188-bp plasmids are completely identical. Subsequent annotation of the plasmid, designated as pTR3/4, revealed 52 CDS (Figure 1). The nucleotide sequence of pTR3/4 is very similar to p271A, a 35,957-bp NDM-1 plasmid identified in E. coli 271 from a patient following medical transfer from a hospital in Bangladesh to Australia (GenBank: accession no. NC_015872 and [22]. Sequence comparison indicates the major difference between pTR3/4 and p271A is an additional 5.2-kb region containing hypothetical protein genes between repA and the stbABC genes in our plasmid. The genes resident in the 5.2-kb region represent the unique CUP (conserved upstream repeat)-controlled regulon of plasmid pJIE137, a 58,107-kb bla CTX-M-62-encoding plasmid from K. pneumoniae JIE137 identified in Australia [23]. Similar to p271A, and JIE137, pTR3/4 also have a backbone organization similar to the IncN plasmid R46. In addition, the conserved repA in these plasmids are unrelated to the IncN plasmids [23]. Since the 5.2-kb CUP region is missing in p271A, the backbone of pTR3/4 is more closely related to pJIE137. Comparative genomics studies revealed that, apart from a 9-kb region containing the bla NDM-1 gene in pTR3/4 (Figure 2) and two resistance regions (a class 1 integron/Tn and a complex ISEcp1-bla CTX-M-62 transposition unit) in pJIE137, the backbone sequences of pTR3/4 and pJIE137 are 97% identical (Figure 1).
Figure 1. Circular map of plasmid pTR3 and pTR4.
The open reading frames are marked along the map by arrows and significant ones are labeled. The bla NDM-1 gene (red) is located in a region with several transposon/IS-related genes (gray). The region corresponding to the IncN2 backbone of pJIE137 is indicated by a black line. Positions of the two resistance regions (a class 1 integron/Tn and a complex ISEcp1-bla CTX-M-62 transposition unit) present in pJIE137 but missing in pTR3/4 are marked by the arrowheads. The CUP-related region between repA and stbABC is missing in p271A. G+C% are shown in the inner circle.
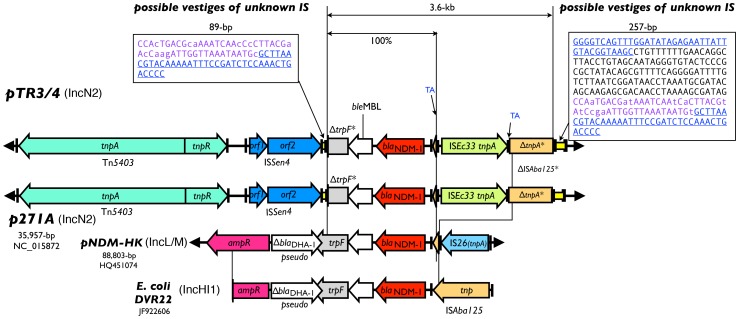
Figure 2. Schematic diagram of the NDM-1 region of pTR3 and pTR4, compared to those from the other known plasmids.
The bla NDM-1 (red), and nearby IS elements (various colors) are shown. ORFs are depicted with arrows and the IRs were depicted by short vertical bars. The regions corresponding to possible vestiges of unknown IS identified in pTR3/4 and p271A are marked by yellow rectangles. Nucleotide sequences of the two regions are shown in the boxes, of which the 39-bp putative IRs are underlined. Corresponding repeat sequences in the boxes are shown in the same color. Differences are shown in lower case.
Sequence Comparison of the Immediate Region Near blaNDM-1
The immediate flanking regions of bla NDM-1, including the ble MBL bleomycin-resistance protein gene, the trpF pseudogene, the nearby ISAba125 (interrupted), ISEc33, ISSen4 and Tn5403 are identical in pTR3/4 and p271A (Figure 2). Upstream of bla NDM-1 is a short fragment corresponding to the left extremity of an ISAba125. When compared with the E. coli DVR22 sequence from Spain [GeneBank accession no. JF922606; [24]], it is apparent that the ISAba125 was interrupted by insertion of the ISEc33, which produces a 2-bp target duplication (TA) during the event (Figure 2, marked blue adjacent to the ISEc33 IRs). When compared with the DVR22 sequence, the ISAba125 in pTR3/4 and p271A were all interrupted at the same position (…TATCÂ). A detailed analysis of the sequences adjacent to the interrupted ISAba125 revealed a 257-bp element bounded by a pair of 39-bp inverted repeats (blue and underlined in Figure 2) [23]. An 89-bp incomplete version, which consists of only the right end of the 257-bp element (11 differences in 89-bp, shown in lowercase in Figure 2), including one of the 39-bp IR, was found at the other side of the NDM-1 region. The 39-bp imperfect IR (6 differences) associated with these elements are different from the 38-bp IR of the nearby Tn5403. Compared to pNDM-HK and DVR22, the trpF pseudogenes in pTR3/4 and p271A were all truncated by this IR-associated element, of which the left extremity is further truncated by the ISSen4. We hypothesize that the 257-bp element and the 89-bp element (marked yellow and sequence shown in the boxes in Figure 2) may be the remains of an unknown IS that transposed into a progenitorial sequence similar to that of the E. coli DVR22.
Discussion
A diversity of bla NDM-1 plasmids have been observed in different published studies. Although plasmid carrying bla NDM-1 was first described in K. pneumoniae, the plasmid incompatibility type was not determined in that study [13]. Subsequent studies revealed plasmid scaffolds of IncL/M type in Hong Kong [14], IncA/C type in Japan [25], IncN2 type from Bangladesh [22], IncF, type in India [26], and recently IncP type in China [9]. In this study, two isolates carrying bla NDM-1 on plasmids similar to IncN2 were identified in two patients who were not epidemiologically linked to each other (Figure 1). These two isolates were resistant to all tested antibiotics (Table 1). Transconjugants showed resistance only to all tested β-lactams except aztreonam. Thus, chromosomal and/or other plasmid-mediated resistance to antibiotics other than β-lactams were very likely present in their parental strains. Complete sequencing showed that although the parent isolates were unrelated based on molecular typing using PFGE and MLST, the plasmid carrying the bla NDM-1 is the same.
Overall, the bla NDM-1–carrying plasmid pTR3/4 is very similar to the bla NDM-1-encoding plasmid p271A from E. coli strain 271 collected in 2009 from a patient from Bangladesh [22]. A recently reported bla CTX-M-62-containing plasmid, pJIE137, also possesses a similar backbone to p271A, but carries a 5.2-kb CUP regulon region in addition [23]. These plasmids are referred to as an IncN2 subgroup which have a backbone similar to the IncN plasmid R46 but a repA gene unrelated to incN plasmids [22], [23]. Plasmid pTR3 and pTR4 also possesses the CUP regulon region and are closely related to these plasmids, especially with respect to the pJIE137 backbone. The discovery of pTR3/4 adds to the IncN2 subgroup of plasmids that cannot be classified using current PCR-based surveys. It appears that the resistance genes were acquired by this plasmid backbone and have been spreading to different locations in the world. Comparison with pJIE137 revealed that the 9,180-bp bla NDM-1-containing insert region in pTR3/4, as depicted in Figure 2, was bounded by the outermost IR of Tn5403 and the 257-bp element at the other end. It had been proposed that in p271A the formation of this insert region was probably a result of progressive insertions and deletions of transposons in the fipA gene in the pJIE137 backbone or insertion of the entire region as a hybrid transposon created elsewhere [23]. The loss of the 5.2-kb CUP regulon region in p271A, on the other hand, may be explained by recombination between CUP repeats [23], [27]. It is likely the 9,180-bp bp bla NDM-1 containing region may have been inserted and settled in the pJIE137-like backbone form pTR3/4, while subsequently loss of the CUP regulon region in pTR3/4 resulted in p271A. The unknown IR-associated elements associated with bla NDM-1 and the interrupted ISAba125 was first described in a comparative analysis between p271A and pJIE137 [23]. In our analysis, the sequence associated with the IR in the 89-bp element is 88% identical to that bounded by the IRs in the 257-bp element (11 in 89 nucleotide positions, colored purple in Figure 2). While we think these elements may be the remains of an unknown IS, it is also possible that they are from related but different IS. The similarities between these IRs and the 38-bp IR from the nearby Tn5403 (50% and 53% identity in 38 nucleotide positions) have also been reported [23]. When comparing the sequence homology to other NDM-1-encoding plasmids, the 257-bp and 89-bp elements comprised by the remains of unknown IS are very likely the factor to facilitate the transposition of bla NDM-1 from the progenitor sequence in E. coli DVR22 instead of pNDM-HK. This finding suggests that different IS elements increase the efficiency of resistance gene spreading.
In the present study, we have observed that the transmission of bla NDM-1 could be achieved by incorporation of transposable elements prior to plasmid spreading. This dual method for spreading may increase the incidence in the prevalence of bacteria carrying bla NDM-1. Since transposition could have occurred by incorporation of the resistance gene into the plasmid or chromosome, a diversity of Inc plasmid types with bla NDM-1 is to be expected and should also be identified in bacteria other than K. pneumoniae. In conclusion, we have identified a plasmid spreading in K. pneumoniae strains that are not epidemiologically linked. An unknown insertion element may be responsible for the mobilization of bla NDM-1 into an IncN2 plasmid backbone similar to pJIE137 and p271A. Comparative genomic studies on the IncN2 plasmids have revealed interesting features related to the accumulation and molecular evolution mechanisms of the plasmid scaffold.
Funding Statement
The work was supported by grants from National Science Council and National Health Research Institutes, Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nordmann P, Poirel L, Toleman MA, Walsh TR (2011) Does broad-spectrum β-lactam resistance due to NDM-1 herald the end of the antibiotic era for treatment of infections caused by Gram-negative bacteria? J Antimicrob Chemother 66: 689–692. [DOI] [PubMed] [Google Scholar]
- 2. Poirel L, Fortineau N, Nordmann P (2011) International transfer of NDM-1-producing Klebsiella pneumoniae from Iraq to France. Antimicrob Agents Chemother 55: 1821–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Poirel L, Lagrutta E, Taylor P, Pham J, Nordmann P (2010) Emergence of metallo-β-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob Agents Chemother 54: 4914–4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Poirel L, Ros A, Carricajo A, Berthelot P, Pozzetto B, et al. (2011) Extremely drug-resistant Citrobacter freundii isolate producing NDM-1 and other carbapenemases identified in a patient returning from India. Antimicrob Agents Chemother 55: 447–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, et al. (2010) Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10: 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Walsh TR, Weeks J, Livermore DM, Toleman MA (2011) Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11: 355–362. [DOI] [PubMed] [Google Scholar]
- 7.Struelens MJ, Monnet DL, Magiorakos AP, Santos O’Connor F, Giesecke J (2010) New Delhi metallo-β-lactamase 1-producing Enterobacteriaceae: emergence and response in Europe. Euro Surveill 15. [DOI] [PubMed]
- 8. Woodford N, Turton JF, Livermore DM (2011) Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35: 736–755. [DOI] [PubMed] [Google Scholar]
- 9. Hu H, Hu Y, Pan Y, Liang H, Wang H, et al. (2012) Novel plasmid and its variant harboring both a bla NDM-1 gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii . Antimicrob Agents Chemother 56: 1698–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Potron A, Poirel L, Nordmann P (2011) Plasmid-mediated transfer of the bla NDM-1gene in Gram-negative rods. FEMS Microbiol Lett 324: 111–116. [DOI] [PubMed] [Google Scholar]
- 11. Pfeifer Y, Witte W, Holfelder M, Busch J, Nordmann P, et al. (2011) NDM-1-producing Escherichia coli in Germany. Antimicrob Agents Chemother 55: 1318–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CLSI (2010) Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 20th informational supplement M100-S17. Wayne, PA: Clinical and Laboratory Standards Institute.
- 13. Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, et al. (2009) Characterization of a new metallo-β-lactamase gene, bla NDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53: 5046–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, et al. (2011) Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6: e17989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S (2005) Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43: 4178–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ma L, Lin CJ, Chen JH, Fung CP, Chang FY, et al. (2009) Widespread dissemination of aminoglycoside resistance genes armA and rmtB in Klebsiella pneumoniae isolates in Taiwan producing CTX-M-type extended-spectrum β-lactamases. Antimicrob Agents Chemother 53: 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, et al. (1995) Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33: 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, et al. (2005) Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63: 219–228. [DOI] [PubMed] [Google Scholar]
- 19. Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, et al. (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, et al. (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M (2006) ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34: D32–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Poirel L, Bonnin RA, Nordmann P (2011) Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob Agents Chemother 55: 4224–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Partridge SR, Paulsen IT, Iredell JR (2012) pJIE137 carrying bla CTX-M-62 is closely related to p271A carrying bla NDM-1 . Antimicrob Agents Chemother 56: 2166–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sole M, Pitart C, Roca I, Fabrega A, Salvador P, et al. (2011) First description of an Escherichia coli strain producing NDM-1 carbapenemase in Spain. Antimicrob Agents Chemother 55: 4402–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sekizuka T, Matsui M, Yamane K, Takeuchi F, Ohnishi M, et al. (2011) Complete sequencing of the bla NDM-1-positive IncA/C plasmid from Escherichia coli ST38 isolate suggests a possible origin from plant pathogens. PLoS One 6: e25334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poirel L, Dortet L, Bernabeu S, Nordmann P (2011) Genetic features of bla NDM-1-positive Enterobacteriaceae . Antimicrob Agents Chemother 55: 5403–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Delver EP, Belogurov AA (1997) Organization of the leading region of IncN plasmid pKM101 (R46): a regulation controlled by CUP sequence elements. J Mol Biol 271: 13–30. [DOI] [PubMed] [Google Scholar]