Abstract
The combination of antibiotics is one of the strategies to combat drug-resistant bacteria, though only a handful of such combinations are in use, such as the β-lactam combinations. In the present study, the efficacy of a specific sub-inhibitory concentration of cefsulodin with other β-lactams was evaluated against a range of Gram-negative clinical isolates. This approach increased the sensitivity of the isolates, regardless of the β-lactamase production. The preferred target and mechanism of action of cefsulodin were identified in laboratory strains of Escherichia coli, by examining the effects of deleting the penicillin-binding protein (PBP) 1a and 1b encoding genes individually. Deletion of PBP1b was involved in sensitizing the bacteria to β-lactam agents, irrespective of its O-antigen status. Moreover, the use of a sub-inhibitory concentration of cefsulodin in combination with a β-lactam exerted an effect similar to that one obtained for PBP1b gene deletion. We conclude that the identified β-lactam/cefsulodin combination works by inhibiting PBP1b (at least partially) despite the involvement of β-lactamases, and therefore could be extended to a broad range of Gram-negative pathogens.
Introduction
The use of antibiotics in optimum concentration is among the most desirable criteria for any treatment strategy [1]. Reports suggest that the usage of cefsulodin, a third-generation cephalosporin, in combination with other β-lactam antibiotics, such as cefazolin and cefuroxime, produces a synergistic effect on sensitivity alteration [2]. However, the optimal concentration of cefsulodin is not known and therefore, narrowing down the concentration will facilitate treatment with comparatively lower doses of antibiotics.
Cefsulodin is thought to target penicillin-binding proteins (PBPs) 1a (encoded by mrcA) and 1b (encoded by mrcB), which are bifunctional enzymes with transglycosylase and transpeptidase activity [3]; but, it is not known which one of the two PBPs is the preferred target. Reports [4], [5] claim that PBP1b deletion alters β-lactam sensitivity; however, the reversal of sensitivity by PBP1b complementation has not been established. In addition, much of the work in this area has been conducted on E. coli K-12 laboratory strains devoid of O-antigens. As O-antigen plays a significant role in β-lactam sensitization [6], it is important to study its effect in the context of PBP1b deletion.
Some β-lactams that target a specific PBP produce an effect similar to the deletion of that PBP, for e.g. inactivation of PBP1 by cefsulodin [5], [7]. Here, we report a combination of cefsulodin with other β-lactams that acts synergistically in sensitizing laboratory and clinical isolates, regardless of the presence of β-lactamases and/or O-antigens.
Materials and Methods
The clinical isolates used for the study were collected from the Tropical School of Medicine, Kolkata, India. These isolates were identified using a combination of biochemical tests and 16S ribosomal DNA sequencing. The obtained sequences were matched with the sequences in the database (National Center for Biotechnology Information), and sequences with a more than 98% match were taken into consideration (as there were no new sequence results, they have not been deposited into Genebank). Primarily, the strains were screened for the presence of β-lactamase by assessing the ability of the cell lysates to hydrolyze nitrocefin [8], and thereafter, specifically for CTX-M β-lactamases (most widespread extended-spectrum β-lactamase) by using polymerase chain reaction with specific primer pairs [9]. The minimum inhibitory concentration (MIC) values of the antibiotics from both penicillin and cephalosporin groups were determined according to CLSI guidelines either individually or in combination [6], [7], [10]. Bacterial strains used for genetic manipulations were derived from the E. coli K-12 strains, CS109 (O-antigen negative strain) and 2443 (O-antigen positive strain). The PBP genes mrcA and mrcB were deleted by P1 transductions followed by the excision of the res–npt–res cassette by transient expression of the RP4 ParA resolvase [7], [11], [12]. Strains and plasmids used in this study are listed in Table 1.
Table 1. Escherichia coli strains and plasmids.
| Strain/plasmid | Genotype/relevant features | Source |
| CS109 | W1485 rpoS rph | C. Schnaitman |
| AM1A-1 | CS109ΔmrcA | This work |
| AM1B-1 | CS109ΔmrcB | This work |
| 2443 | thr-1 leuB6 Δ(gpt-proA)66 argE3 thi-1 rfbO8 lacY1 ara-14 galK2 xyl-5 mtl-1 mgl51 rpsL31 kdgK51 supE44 | A.T. Maurelli |
| AM1OA-1 | 2443 ΔmrcA | This work |
| AG1OB-3 | 2443 ΔmrcB | This work |
| pJMSB8 | RP4::2-Tc::Mu-Km::Tn7] λ pir lysogen (AmpR) | [12] |
| pSAD588-1 | mrcB cloned in pBAD18-Cam | K.D. Young |
| pFS1A1 | mrcA cloned in pBAD18-Cam | K.D. Young |
PBP deletion was confirmed by labeling the cells with Bocillin FL (Invitrogen Inc., Carlsbad, CA, USA) and visualizing the PBPs separated in SDS-PAGE [13]. The PBP1a and 1b clones (pFS1A1 and pSAD588-1, respectively) were gifted by Professor Kevin D. Young. MIC values were determined for E. coli CS109, 2443 and their deletion mutants, before and after complementation. To check the β-lactam binding efficacy of PBP1a and 1b, competition assays between cefsulodin and Bocillin FL were performed, using mrcA and mrcB deleted strains [8]. The killing kinetics was evaluated following the methods described earlier [14].
Results and Discussion
Sub-inhibitory Concentration of Cefsulodin Combination Sensitizes Gram-negative Clinical Isolates
Clinical isolates selected for this study were identified as the members of Enterobacteriaceae group. The strains Tr1 (Salmonella enterica), Tr5 (Shigella sp.), Tr10 (Klebsiella pneumonia), Tr7 (Escherichia coli), NGM2 (Escherichia coli), NGM3 (Escherichia coli) and the type strain NCIM 2300 (Proteus mirabilis) showed significant β-lactamase activity while the strains Tr2 (Vibrio cholera), NGM6 (Escherichia coli), and the type strain NCIM 2397 (Serratia marcescens) lacked β-lactamase activity (Figure 1). Susceptibilities of these strains to various β-lactams were tested and the MICs varied from 2 mg/L to >500 mg/L (Table 2). When combined with cefsulodin, at concentrations ranging from 2 mg/L to 8 mg/L, susceptibilities of all the strains were enhanced. However, differences in susceptibilities were negligible when cefsulodin was used at concentrations higher than 4 mg/L. The combination successfully sensitized the strains 2 to 32 fold compared to their original MIC values (Table 2). Therefore, the combination of cefsulodin at a sub-inhibitory level (4 mg/L) with other β-lactam agents was effective against both β-lactamase negative and positive strains. For further experiments, the concentration of cefsulodin used in the combination was 4 mg/L.
Figure 1. β-lactamase assay for various strains used in this study.
β-lactamase activity could not be detected for Tr2, NGM6 and Serratia marcescens.
Table 2. β-lactama sensitivities of bacterial isolates in combination with Cefsulodin (4 mg/L).
| Strains | AMX | AMX+CSN | PIP | PIP+CSN | AMP | AMP+CSN | PNG | PNG+CSN | CDL | CDL+CSN | CLN | CLN+CSN | CXN | CXN+CSN | CCR | CCR+CSN | CZM | CZM+CSN | CTX | CTX+CSN | CSN |
| Proteus mirabilis NCIM 2300 | 2 | 0.5 | 4 | 0.5 | 2 | 0.5 | 16 | 2 | 8 | 1 | 8 | 0.5 | 2 | 0.25 | 2 | 0.25 | 2 | 0.25 | 1 | 0.06 | 16 |
| Serratia marcescens NCIM 2397 | 16 | 4 | 8 | 1 | 16 | 4 | 125 | 32 | 4 | 0.5 | 16 | 0.5 | 4 | 1 | 4 | 1 | 2 | 0.5 | 2 | 0.5 | 32 |
| Vibrio cholerae Tr2 | 16 | 4 | 0.25 | 0.03 | 16 | 4 | 64 | 16 | 16 | 4 | 16 | 4 | 4 | 1 | 2 | 0.5 | 1 | 0.25 | 2 | 0.25 | 32 |
| Shigella sp. Tr5 | 125 | 64 | 8 | 2 | 125 | 64 | >500 | >500 | 16 | 4 | 16 | 2 | 8 | 2 | 4 | 1 | 0.5 | 0.06 | 0.5 | 0.03 | 32 |
| Klebsiella pneumoniae Tr10 | 500 | 125 | 16 | 2 | 250 | 64 | 500 | 250 | 16 | 4 | 16 | 2 | 8 | 2 | 2 | 0.5 | 1 | 0.125 | 0.5 | 0.03 | 32 |
| Salmonella typhimurium Tr1 | 250 | 125 | 16 | 4 | 250 | 125 | 250 | 125 | 16 | 4 | 16 | 4 | 8 | 2 | 4 | 1 | 1 | 0.125 | 0.5 | 0.06 | 64 |
| Escherichia coli Tr7 | 250 | 125 | 8 | 2 | 250 | 125 | 500 | 250 | 16 | 4 | 32 | 16 | 4 | 1 | 4 | 1 | 4 | 0.25 | 0.5 | 0.06 | 32 |
| Escherichia coli NGM2 | 32 | 16 | ND | ND | ND | ND | 125 | 64 | ND | ND | 32 | 8 | 16 | 4 | 4 | 1 | 4 | 0.25 | 1 | 0.125 | 32 |
| Escherichia coli NGM3 | 16 | 8 | 16 | 1 | 8 | 2 | 250 | 125 | 125 | 16 | 16 | 4 | 16 | 4 | 4 | 1 | 4 | 0.5 | 2 | 0.25 | 32 |
| Escherichia coli NGM6 | 16 | 8 | 16 | 2 | 4 | 2 | 250 | 125 | 32 | 8 | 32 | 16 | 4 | 1 | 4 | 1 | 4 | 0.5 | 1 | 0.125 | 32 |
AMX = amoxicillin; AMP = ampicillin; PIP = piperacillin; PNG = penicillin G; CDL = cefadroxil; CLN = cefalexin; CCR = cefachlor; CXN = cefoxitin; CML = cefamandole; CZM = ceftazidime; CSN = cefsulodin; CTX = cefotaxime
O-antigen does not Impact Patterns of Antibiotic Sensitivity in PBP Mutants
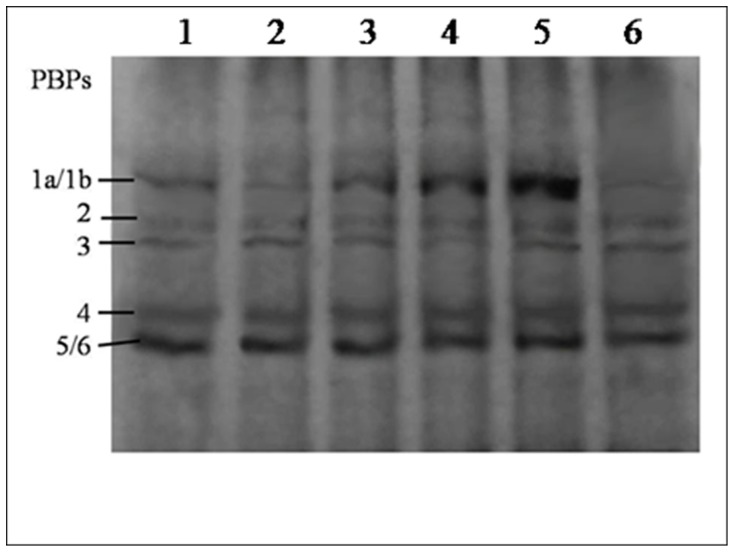
It is not known whether the preferred target of cefsulodin is PBP1a or PBP1b, so mrcA and mrcB genes were deleted separately from E. coli K12 strains that either lacked or contained O-antigen (CS109 and 2443, respectively). PBP loss was confirmed by Bocillin FL labeling (Figure 2). The effect of O-antigens on β-lactam sensitivity of the PBP mutants was tested and the patterns of alteration in β-lactam sensitivity were found identical for the strains, regardless of the presence of O-antigens (Table 3). The only difference was that the strains derived from E. coli 2443 were comparatively 2 to 4 times more sensitive to the penicillin group but not to the cephalosporin group of antibiotics [6]. As the pattern of sensitivity alterations was similar in both the mutants, unless otherwise specified, further experiments were carried out with the O-antigen positive PBP mutants.
Figure 2. Labeling of penicillin-binding proteins with fluorescent penicillin.
Total protein content (∼300 µg) were labeled with Bocillin FL (50 µM) and analyzed through 12% SDS-PAGE (100 µg/lane). Lane 1: Escherichia coli 2443; lane 2: 2443ΔmrcB; Lane 3, 4 and 5: 2443ΔmrcB/pSAD588-1 induced by 0.05%, 0.1% and 0.2% arabinose respectively; Lane 6: 2443ΔmrcA.
Table 3. β-lactama sensitivities of Escherichia coli strains and their mutants.
| MIC values (mg/L) of β-lactam antibiotics tested | ||||||||||||||
| Strains | AMX | AMP | PIP | PNG | CDL | CLN | CTN | CCR | CXN | CML | CZN | CZM | CSN | CTX |
| CS109 | 8 | 8 | 2 | 250 | 32 | 8 | 16 | 4 | 4 | 2 | 4 | 4 | 32 | 1 |
| CS109ΔmrcA | 8 | 8 | 2 | 250 | 32 | 8 | 16 | 4 | 4 | 2 | 4 | 4 | 32 | 1 |
| CS109ΔmrcB | 4 | 4 | 0.125 | 125 | 2 | 0.5 | 1 | 0.5 | 0.5 | 0.25 | 0.125 | 0.125 | 1 | 0.06 |
| CS109ΔmrcB/pSAD588-1 | 8 | 8 | 2 | 250 | 32 | 8 | 16 | 4 | 4 | 2 | 4 | 4 | 32 | 1 |
| 2443 | 4 | 2 | 1 | 125 | 16 | 8 | 16 | 4 | 4 | 2 | 4 | 4 | 32 | 1 |
| 2443ΔmrcB | 2 | 1 | 0.06 | 64 | 1 | 0.5 | 1 | 0.5 | 0.5 | 0.25 | 0.125 | 0.125 | 1 | 0.06 |
| 2443ΔmrcA | 4 | 2 | 1 | 125 | 16 | 8 | 16 | 4 | 4 | 2 | 4 | 4 | 32 | 1 |
| 2443ΔmrcB/pSAD588-1 | 4 | 2 | 1 | 125 | 16 | 8 | 16 | 4 | 4 | 2 | 4 | 4 | 32 | 1 |
| 2443ΔmrcA/pFS1A1 | 4 | 2 | 1 | 125 | 16 | 8 | 16 | 4 | 4 | 2 | 4 | 4 | 32 | 1 |
AMX = amoxicillin; AMP = ampicillin; PIP = piperacillin; PNG = penicillin G; CDL = cefadroxil; CLN = cefalexin; CTN = cefalothin; CCR = cefachlor; CXN = cefoxitin; CML = cefamandole; CZN = cefoperazone; CZM = ceftazidime; CSN = cefsulodin; CTX = cefotaxime.
Deletion of PBP1a is Unable to Sensitize E. coli Cells to β-lactams
It is believed that PBP1a and 1b compensate each other functionally for transglycosylase and transpeptidase activity; thus, in the absence of PBP1a, PBP1b can compensate its function and vice versa [11]. Therefore, to check the effect of PBP1b deletion on the sensitivity of β-lactam antibiotics, we used the CS109ΔmrcB and 2443ΔmrcB strains. These strains were sensitive to the representative antibiotics of various generations of cephalosporins, with the change in sensitivity level ranging from 16 to 32 fold, as compared to their respective parent strains (Table 3). The results indicate that in the absence of PBP1b, the intact PBP1a protein may not possess sufficient activity to compensate the physiological functions of PBP1b [15], [16]. Next, to check whether PBP1a deletion has a similar role in altering β-lactam sensitivity, the MIC values were determined for CS109ΔmrcA and 2443ΔmrcA. However, no change in β-lactam sensitivity was observed for either of the PBP1a mutants indicating that the intact PBP1b protein present in the ΔmrcA mutants is able to compensate functionally for the PBP1a deletion (Table 3).
Expression of PBP1b in Trans Restores the Lost β-lactam Resistance
From our results, we inferred a possible involvement of PBP1b in altering β-lactam sensitivity. To strengthen this hypothesis, we checked whether the expression of PBP1b in trans could reverse the augmented β-lactam sensitivity in CS109ΔmrcB and 2443ΔmrcB (Table 3). Expression of mrcB gene from plasmid pSAD588-1 (Table 1) reversed the lost β-lactam sensitivity in both the strains. However, no change in β-lactam sensitivity was observed upon expressing mrcA (from plasmid pFS1A1) in 2443ΔmrcA. Therefore, the results obtained from both the deletion and complementation experiments demonstrate that PBP1b is involved in maintaining an intrinsic β-lactam resistance, especially to cephalosporins.
Cefsulodin Binding Efficacy of PBP1a is Ten Times Higher than that of PBP1b
To understand the biochemical origins of the variation in the physiological functions of PBP1a and PBP1b, their binding efficacy for cefsulodin was determined through a competition assay between cefsulodin and Bocillin FL using the strains 2443ΔmrcA and 2443ΔmrcB. The relative efficacy of binding of cefsulodin to PBP1s was determined by their ability to inhibit the binding of Bocillin FL by 50%, also known as IC50 value (50% inhibitory concentration). Cefsulodin was found to interact specifically with PBP 1a and 1b. The band intensity of 2443ΔmrcB (where PBP1a was intact) showed 50% inhibition in presence of 10 µM cefsulodin, while 2443ΔmrcA (where PBP1b was intact) showed 50% inhibition in presence of 100 µM cefsulodin. Assessment of the IC50 of cefsulodin for the PBP1s revealed that PBP1a has an IC50 value that is approximately 10 times lower than that of PBP1b. In other words, PBP1a is 10 times more sensitive to cefsulodin than PBP1b, which resembles the result reported by Ramachandran et al [17].
Sub-inhibitory Levels of Cefsulodin in Combination with β-lactams Mimics PBP1b Deletion
The efficiency of sub-inhibitory concentrations of cefsulodin in combination with other β-lactams was tested. As described above, 4 mg/L of cefsulodin was most effective in sensitizing the clinical isolates to β-lactams. The MIC values obtained for each antibiotic tested in combination with cefsulodin (4 mg/L), resembled the pattern observed with mrcB deletion. However, the enhancements in sensitivity were in a range from 2 to 8 fold for the entire set of antibiotics tested (Table 4).
Table 4. The MIC values in combination with Cefsulodin (4 µg ml−1) against E. coli.
| E. coli strains | AMX | AMP | PIP | PNG | CDL | CLN | CTN | CXN | CCR | CFL | CZN | CZM | CTX | |
| 2443 | A | 4 | 2 | 1 | 125 | 16 | 8 | 16 | 4 | 4 | 2 | 4 | 4 | 1 |
| B | 1 | 0.5 | 0.125 | 64 | 4 | 2 | 4 | 1 | 1 | 0.5 | 0.5 | 0.5 | 0.125 | |
| CS109 | A | 8 | 8 | 2 | 250 | 32 | 8 | 16 | 4 | 4 | 2 | 4 | 4 | 1 |
| B | 4 | 4 | 0.25 | 125 | 8 | 2 | 4 | 1 | 1 | 0.5 | 0.5 | 0.5 | 0.125 |
AMX = Amoxicillin; AMP = Ampiocillin; PIP = Piperacillin; PNG = Penicillin G; CDL = Cefadroxil; CLN = Cefalexin; CTN = Cefalothin; CXN = Cefoxitin; CCR = Cefaclor; CFL = Cefamandole; CZN = Cefoperazone; CZM = Ceftazidime and CTX = Cefotaxime.
A: The sensitivities in absence of Cefsulodin; B: The sensitivities in presence of Cefsulodin (4 µg ml−1).
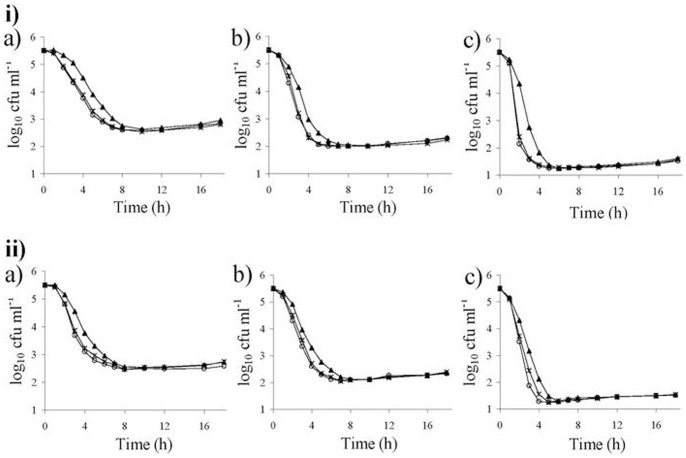
To check the effect of 4 mg/L cefsulodin in combination with other β-lactams, we evaluated the killing rate of parent and PBP1b mutant strains using 1× and 2× MICs of ampicillin and cefotaxime as representatives of penicillin and cephalosporin groups, respectively. The results were plotted as log cfu/mL versus time [14] (Figure 3). Interestingly, the killing rate of 2443ΔmrcB in the absence of cefsulodin was similar to that of the 2443 parent strain in presence of 4 mg/L cefsulodin. Therefore, β-lactam in combination with 4 mg/L cefsulodin showed an effect similar to PBP1b loss in E. coli.
Figure 3. Graphical representation of Time–kill analyses.
‘▴’ E. coli 2443, ‘o’ 2443ΔmrcB, ‘x’ 2443 plus cefsulodin (4 mg/L) in presence (a) 1×MIC, (b) 2×MIC, (c) 4×MIC of (i) ampicillin and (ii) cefadroxil.
Overall, based on the obtained results, it can be speculated that in a cell where both the PBPs are intact, cefsulodin inhibits PBP1a at a concentration 10 times lower than PBP1b. If concentration of cefsulodin is sub-optimal, majority of PBP1b remain viable for its enzymatic functions. Similar situation prevails for PBP1a deletion mutants leading to unaltered MIC values. However, if the sub-inhibitory dose of cefsulodin is sufficient to inhibit PBP1b (at least partially), the availability of functional PBP1b diminishes. In this situation, the cells become more vulnerable to β-lactams that target other essential PBPs. Therefore, a sub-inhibitory concentration of cefsulodin would be sufficient to inhibit PBP1a and at least partially inhibit PBP1b. On the other hand, when PBP1b is deleted, the intact PBP1a would be inhibited by a much lower dose of cefsulodin as compared to PBP1b, which explains the reason for enhanced β-lactam sensitivity of PBP1b deletion mutant.
Conclusion
It is inferred that PBP1b is involved in altering β-lactam sensitivity, especially for antibiotics of the cephalosporin group. We suggest that by deleting mrcB or its homologs, and/or by applying a sub-inhibitory level of cefsulodin (4 mg/L), the bacterial cells, regardless of the presence of β-lactamases, can be sensitized against conventional β-lactam agents. Further studies in this area may expand our knowledge of combinatorial therapy using cefsulodin as a key component.
Acknowledgments
We thank Professor Kevin Young for the clones of PBP1a and 1b genes, Dr. N. K. Pal and Dr. Nadeem for the clinical isolates and M. Narayani for her contribution in editing the language of the manuscript.
Funding Statement
The work is funded in part by two different grants, from Department of Biotechnology, Government of India [Grant #BT/215/NE/TBP/2011: http://www.dbtindia.nic.in]; and from Indian council of Medical Research [Grant #5/3/3/16/2006-ECD-1: http://www.icmr.nic.in]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Yah CS (2010) Plasmid-encoded multidrug resistance: A case study of Salmonella and Shigella from enteric diarrhea sources among humans. Biol Res 43: 141–148. [PubMed] [Google Scholar]
- 2. Kondo M, Tsuchiya K (1981) Effect of combination of cefsulodin and mecillinam. J Antibiot (Tokyo) 34: 727–738. [DOI] [PubMed] [Google Scholar]
- 3. Ghosh AS, Chowdhury C, Nelson DE (2008) Physiological functions of D-alanine carboxypeptidases in Escherichia coli . Trends Microbiol 16: 309–317. [DOI] [PubMed] [Google Scholar]
- 4. Pepper ED, Farrell MJ, Finkel SE (2006) Role of penicillin-binding protein 1b in competitive stationary-phase survival of Escherichia coli . FEMS Microbiol Lett 263: 61–67. [DOI] [PubMed] [Google Scholar]
- 5. Yousif SY, Broome-Smith JK, Spratt BG (1985) Lysis of Escherichia coli by β-Lactam Antibiotics: Deletion Analysis of the Role of Penicillin-binding Proteins 1A and 1B. J Gen Microbiol 131: 2839–2845. [DOI] [PubMed] [Google Scholar]
- 6. Sarkar SK, Ghosh AS (2008) Involvement of O8-antigen in altering β-lactam antibiotic susceptibilities in Escherichia coli . FEMS Microbiol Lett 28: 259–264. [DOI] [PubMed] [Google Scholar]
- 7. Sarkar SK, Chowdhury C, Ghosh AS (2010) Deletion of penicillin-binding protein 5 (PBP5) sensitises Escherichia coli cells to β-lactam agents. Int J Antimicrob Agents 35: 244–249. [DOI] [PubMed] [Google Scholar]
- 8. Ghosh AS, Kar AK, Kundu M (1998) Alterations in High Molecular Mass Penicillin-Binding Protein I Associated with Beta-lactam Resistance in Shigella dysenteriae . Biophys Biochem Res Commun 248: 669–672. [DOI] [PubMed] [Google Scholar]
- 9. Pitout JD (2012) Extraintestinal Pathogenic Escherichia coli: A combination of virulence with antibiotic resistance. Front Microbiol 3: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(2007) Wayne, PA. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Seventeenth informational supplement M100-S17.: CLSI.
- 11. Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD (1999) Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol 181: 3981–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nilsen T, Ghosh AS, Goldberg MB, Young KD (2004) Branching sites and morphological abnormalities behave as ectopic poles in shape-defective Escherichia coli . Mol Microbiol 52: 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chowdhury C, Nayak TR, Young KD, Ghosh AS (2010) A weak DD-carboxypeptidase activity explains the inability of PBP 6 to substitute for PBP 5 in maintaining normal cell shape in Escherichia coli . FEMS Microbiol Lett 303: 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sarkar SK, Dutta M, Chowdhury C, Kumar A, Ghosh AS (2011) PBP5, PBP6 and DacD play different roles in intrinsic β-lactam resistance of Escherichia coli. . Microbiol 157: 2702–2707. [DOI] [PubMed] [Google Scholar]
- 15. Chandrakala B, Shandil RK, Mehra U, Ravishankar S, Kaur P, et al. (2004) High-throughput screen for inhibitors of transglycosylase and/or transpeptidase activities of Escherichia coli penicillin binding protein 1b. Antimicrob Agents Chemother 48: 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jacoby GH, Young KD (1991) Cell cycle-independent lysis of Escherichia coli by cefsulodin, an inhibitor of penicillin-binding proteins 1a and 1b. J Bacteriol 173: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ramachandran V, Chandrakala B, Kumar VP, Usha V, Solapure SM, et al. (2006) Screen for inhibitors of the coupled transglycosylase-transpeptidase of peptidoglycan biosynthesis in Escherichia coli . Antimicrob Agents Chemother 50: 1425–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]