Abstract
The p21-activated kinases (PAKs) are a family of Ser/Thr protein kinases that are represented by six genes in humans (PAK 1–6), and are found in all eukaryotes sequenced to date. Genetic and knockdown experiments in frogs, fish and mice indicate group I PAKs are widely expressed, required for multiple tissue development, and particularly important for immune and nervous system function in the adult. The group II PAKs (human PAKs 4–6) are more enigmatic, but their restriction to metazoans and presence at cell-cell junctions suggests these kinases emerged to regulate junctional signaling. Studies of protozoa and fungal PAKs show that they regulate cell shape and polarity through phosphorylation of multiple cytoskeletal proteins, including microtubule binding proteins, myosins and septins. This chapter discusses what we know about the regulation of PAKs and their physiological role in different model organisms, based primarily on gene knockout studies.
Keywords: Cdc42, Rac1, cytoskeleton, development
Background
PAKs were discovered 20 years ago by serendipity in the search for new GTPase-activating (GAPs) proteins for the small Rho-like G-proteins (p21s) Cdc42 and Rac1.1 Protein bands of ~65 kDa, when immobilized on nitrocellulose filters, were found to stabilize the GTP-bound state of Rac1. The triplet of bands of apparent 62 kDa, 65 kDa and 68 kDa were seen in brain samples, from which PAK1 (αPAK in rat) was first purified and cloned.2 Because the activity associated with the purified 68 kDa Ser/Thr kinase was directly stimulated by active Cdc42 and Rac1, this protein was coined a p21-activated kinase (αPAK). By coincidence the kinase had been worked on as a “protease-activated kinase” (PAK) for many years prior in reticulocyte lysate,3 and the kinase was eventually purified and identified as PAK2.4 The homology of PAK1 to the budding yeast sterile-20 (Ste20) in both the catalytic and the Cdc42/Rac interaction/binding (CRIB) led to the discovery that the mating pathway in yeast is directly regulated by Cdc42.5 This region of PAK that binds to Cdc42 and Rac1 is related to the sequence in ACK1, a cytoplasmic Tyr- kinase,6 and effectors such as PAKs and ACK1 stabilize the active form of Cdc42 and Rac1 from intrinsic hydrolysis of GTP.2 Mammals contain at least three highly related PAKs, which are termed PAK1, PAK2 and PAK3.2,7,8 The human PAK4 was cloned in a search for other human PAK-like kinases, and contains an N-terminal CRIB motif, but none of other features seen in human PAK1.9,10
Early studies focused on the potential role of group I PAKs (see next section) in regulating actin and microtubule networks downstream of Rac1.11-13 Although PAK1 was linked to the activities of Cdc42 and Rac1 to induce actin-rich structures in cultured cells termed filopodia and lamellipodia, effector mutants of these GTPases essentially excluded PAK1 as a key driver of these activities.11,14,15 However, PAK can act upstream of Rac1 by interacting with the Rac-GEF called PAK-interacting exchange factor (PIX), leading to enhanced neurite formation in PC12 cells.16 Direct binding of group I PAKs to these PIX proteins,17 generates a large > 1 mDa complex18 that contains PIX trimer(s) and oligomeric forms of GIT1/2,19 which are enriched at focal adhesions and at the leading edge of migrating cells.20 Activation of PAKs leads to auto-phosphorylation of Ser199/Ser204 in the PIX-binding Pro-rich motif (residues 186-203).21 This negative feedback loop prevents the accumulation of PAK at focal adhesions. PAK1 has been found to associate with the centrosome by binding PIX/GIT1 where it can regulate Aurora-A activity.22 Again the kinase appears to reversibly associate with this complex through the same negative feedback mechanisms.
The ability of PAK1-like kinases to regulate cytoskeleton dynamics is underscored by the identification of a number of important targets including myosin light-chain kinase (MLCK), cortactin, the LIMK1, stathmin.23 Unfortunately none of these targets are PAK-specific, and none offer a validated “readout” of PAK activity in vivo. The phosphorylation of Mek1 at Ser298 by group I PAKs is of significant interest to understanding PAK's role in proliferation. However, although inhibition of PAK1 kinase activity does profoundly reduce MEK1 Ser298 phosphorylation in response to epithermal growth factor (EGF), this inhibition does not prevent MEK1 activation by EGF.24 Inhibiting group 1 PAKs also reduces the Ser338 phosphorylation of c-Raf in response to both PDGF and EGF; however the reduction in Ser338 phosphorylation was not accompanied by a significant decrease in c-Raf activity as expected.24 It may be that these phosphorylation events are only relevant at low growth factor concentration or in the context of integrin-initiated signaling.
The Classification of PAKs
In addition to the conserved catalytic domain, all PAKs harbor an N-terminal regulatory domain of ~50 residues which contains a CRIB motif responsible for binding Cdc42 and Rac1-like GTPases. The vertebrate PAK1-3 are highly related to each other, while their catalytic domains are very similar to the group II kinases PAK4-6. The PAK kinase domain also resembles Mst kinases, which are essential regulators of the Hippo pathway,25 though not direct targets of small G-proteins. The PAK1-like family are classified as “group I” PAKs while the PAK4-like kinases are termed “group II” or non-conventional PAKs,26-28 primarily based on the observation that of the group II PAKs are not activated by Cdc42 binding.10 Since it turns out that PAK4 is indeed activated by Cdc42, this distinction becomes less relevant. The group II PAKs differ from the group I PAKs in their mode of kinase regulation, intracellular localization and binding partners. These PAKs are found only in metazoans and perhaps evolved along with cadherin-based adherens junctions.
The metazoan PAK1-like (i.e., group I PAKs) all contain binding sites for Nck at the N-terminus and for the PIX SH3 in the region that lies between the AID and the catalytic domain (Fig. 1). Model invertebrates also contain a third class of PAK (denoted PAK3 in C. elegans and D. melanogaster), which resemble the more diverse protist PAKs. Many Drosophila species express compact versions of D. melanogaster PAK3, comprising (for example in Drosophila grimshawi) only 401 amino acids. The N-terminal regulatory domain includes a basic, CRIB and AID domain (residues 1-90) but no other protein-interaction motifs. The remainder of the protein (residues 92-401) comprises the catalytic domain. This might represent an ideal kinase species for structural determination of a full-length PAK.
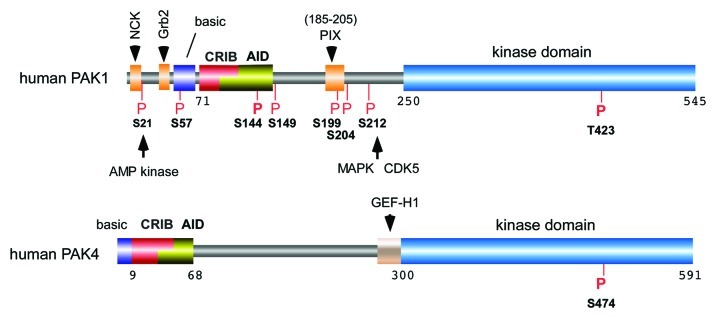
Figure 1. The domain structure of PAK1 and PAK4 highlighting the conserved features of group I kinases and the conserved sites of kinase phosphorylation. The presence of proline-rich SH3 binding sites are marked in orange. The p21-binding domain (PBD or CRIB) is indicated in red and overlaps the auto-inhibitory domain (AID) in yellow. The basic residue cluster required for phospholipid-mediated kinase activation is marked in purple. PAK1 phosphorylation sites that are conserved across other isoforms are marked in red. The activation-loop phopho-residue indicated: this is constitutively phosphorylated in the case of PAK4. Unless otherwise indicated these are auto-phosphorylation sites.
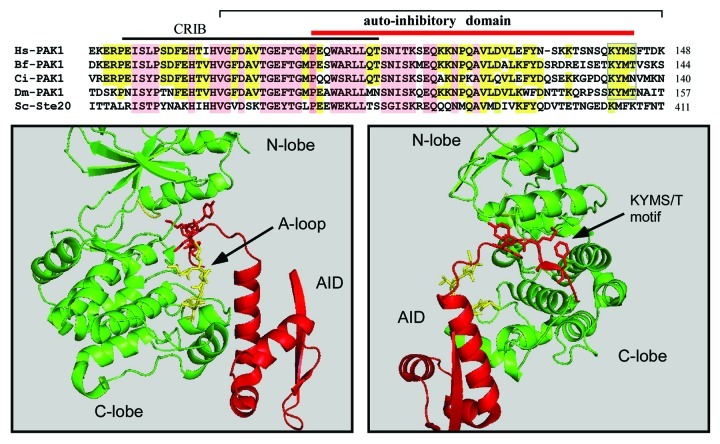
Although the protozoan PAKs are suggested to belong to the group 1 PAKs they lack the key KYMS/T (Lys-Tyr-Met-Ser/Thr) inhibitory motif (Fig. 2) in the auto-inhibitory domain (AID) which is responsible for displacing the kinase activation loop.29 The bulk of the AID is involved with contacts to the C-lobe of the kinase; however, this KYMS/T motif displaces the (activation) A-loop by interacting with the α-C helix, whose position is critical for catalytic activity. Because we do not have any structure of a protozoan-inhibited PAK, it is unclear if the auto-inhibition mechanism is identical. Certainly the core AID is well conserved, indicating that protozoan PAKs contain an AID that binds to the kinase C-lobe; however, the role of any KYMS/T like inhibitory motif is unknown. Therefore it might be prudent to regard the very diverse class of protozoan PAKs as a different class of “group III” PAKs.
Figure 2. The structure of the PAK1 AID and sequence alignment among Group I PAKs from diverse phyla. PAK1 sequences are from human (Hs, Q13153), Branchiostoma floridae (Bf, XP_002595185), Ciona intestinalis (Ci, XP_002131099), dPAK1 from Drosophila melangaster (Dm, AAC47094) and Ste20p from Saccharomyces cerevisiae (Sc, AAA35038). Completely conserved residues are in pink and partial conservation in yellow. The interaction of the KYMS box is illustrated in the figure below, which shows a complex between human PAK1 and the AID (PDB: 1F3M). The A-loop in yellow is displaced by the presence of the KYMS sequence, which occupies a position under the α-C helix. The structure was prepared using Pymol.
Group I PAKs: Conserved Motifs and Domain Structure
Group I PAKs are domain-rich proteins with a large number of conserved evolutionarily features in all metazoans (Fig. 1). Protozoan PAKs are much more diverse, and can include addition features such as the lipid binding PH domain. Three conserved Pro-rich regions in group I vertebrate kinases (residues 12-18, 40-45 and 186-203 in human PAK1) bind to NCK, Grb2 and PIX, respectively.17,30,31 The CRIB (Cdc42/Rac1 interactive binding, 75-95 in human PAK1) overlaps the AID (auto inhibitory domain, 83-149). The AID interacts with the kinase domain29 but when the kinase activated this cryptic helix can then bind fragile-X proteins32,33 and the bacterial toxin EspG,34 which maintains the kinase in an active state. The CRIB is preceded by a basic cluster, which allows the kinase to be responsive to acidic phospholipids.35 The SH3-binding motif, flanked by a poly-acidic region (9-13 acidic residues) binds to the highly conserved SH3 domain found in PIX (PAK-interacting exchange factors) proteins,17 which are found in metazoans. All PAKs contain a conserved Ser/Thr kinase domain (250-545 in human PAK1) with a single auto-phosphorylation site (T423), which is phosphorylated upon kinase activation. Mutation of certain residues in the AID lead to constitutive PAK1 activation in the absence of Cdc42.36 However, the phospho-mimetic PAK1(T423E), which has been used by a number of groups, fails to exhibit auto-phosphorylation characteristic of active kinase when expressed in mammalian cells.37 This is consistent with the observation that PAK1 (T423E) catalytic domain can be tolerated by E. coli, which are growth-sensitive to active forms of PAK1.
Mechanisms of Activation of Group I PAKs
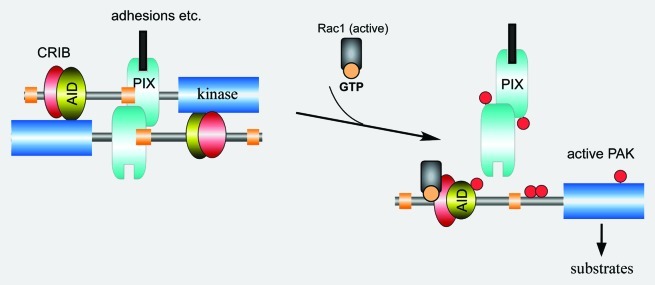
Biochemical and crystallization studies29,36 revealed that Group I PAKs are regulated via a trans auto-inhibition mechanism. PAK1 can be activated by binding to small G-proteins (Fig. 3) or sphingolipids, phosphatidyl serine or phosphatidyl inositol phosphates.38 The requirement for lipids in PAK activation is suggested by the slow kinetics of in vitro activation and the identification of multiple lipids that can activate PAK1 in the absence of Rac1 in vitro.38 Recent experiments implicate two basic regions upstream of the CRIB in lipid binding.35 The model proposed in this study is that binding of both PIP2 and Rac1 is needed for cellular PAK1 activation. The details of this cooperative activation require further study, but perhaps kinase recruitment to sites of Rac1.GTP (or other small G-proteins) can explain this behavior. The structure of the Rac3-PAK1 CRIB complex (protein database ID: 2QME) suggests a conformational change in the KID upon binding can drive its dissociation from the kinase domain, and subsequent trans-autophosphorylation at many sites, including the activation loop T42339-41 and the AID Ser144.42 A recent asymmetric dimer structure of PAK1 gives some clues as to the mode of trans-autophosphorylation of the activation loop T423 residue43 which is needed for full kinase activity. Certain proteins such as hPIP (human PAK1-interacting protein 1) and CRIPAK (Cys-rich inhibitor of PAK1) may act to prevent such PAK kinase activation.44-46 A small molecule inhibitor of PAK1 termed IPA-3 binds to the CRIB region and prevents Cdc42 interaction.47 The role of adaptor proteins Nck1 and Grb2 in recruiting PAK to phospho-Tyr containing signaling complexes such as the ligand-activated EGF receptor (ErbB1) has not yet been tested in the relevant knockout cells.
Figure 3. A model for group I PAK activation. The auto-inhibited kinase is inhibited in trans as shown. The AID directly contacts the catalytic domain in order to prevent kinase auto-phosphorylation. The PAK is targeted for activation by interaction with PIX, which binds to the central proline-rich sequence. The GTP.Rac1 binds to the CRIB region and a conformational change allows autophosphorylation (red circles). Phosphorylation of Ser144 serves to disable the AID-catalytic domain interaction, while phosphorylation of Ser198/203 reduces the affinity for βPIX (which itself is modified at Ser340). Phosphorylation of the activation-loop Thr423 likely occurs in trans.
Regulation of Group II PAKs
Aside from Cdc42 no other factors are known to activate group II PAKs. The structure of the kinase domains of human PAK4-6 resemble typical Ser/Thr kinases of the Ste20 class,48,49 likely the auto-inhibited structure is quite different from PAK1.50 Certainly the CRIB of PAK4 functions as an auto-inhibitory region to allow activation by Cdc42.GTP, but the activation loop is constitutively phosphorylated.50 Generally Group II PAKs do not bind well, if at all, to Rac-family GTPases (i.e., Rac1-3 and RhoG) though the structure of a complex between Rac3.GTP and PAK4 has been solved (protein database ID: 2OV2).
It is notable that PAK4 requires Cdc42 interaction for its localization to cell-cell adherens junctions in epithelial cells45 and in the Drosophila eye.51 PAK4 likely operates downstream of Cdc42 in multiple contexts including the Met receptor.52 Human PAK6 kinase activity is reported to be upregulated by binding of androgen receptor,53,54 and the basis of this interactions requires further study. Although MKK6 can phosphorylate PAK5 and PAK655 it has not been shown that the p38 pathways is involved in kinase activation. If active Cdc42 is the key interactor that drives the association of PAK4 to cell-cell junctions10,56,57 it would be interesting to find out how this is spatially restricted. PAK4 undergoes nuclear-cytoplasmic cycling and is suggested to promote intracellular translocation and signaling of β-catenin.58 PAK4 can phosphorylate Ran on Ser135 during mitosis and both these proteins may dynamically associate with components of the microtubule spindle during mitotic progression.59 A small region upstream of the PAK4 kinase domain has been found to bind substrates such as GEF-H160 and PDZ-RhoGEF.61
The Diverse PAKs of Protozoa
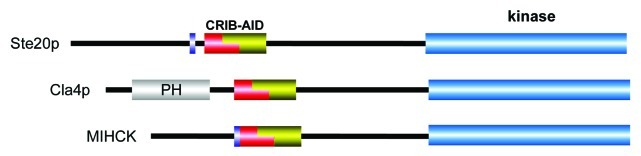
Saccharomyces cerevisiae has three conventional PAKs: Sterile 20 (Ste20), Cla4 and Skm1. These PAK members contain a typical CRIB/AID motif located N-terminal to the kinase domain (Fig. 4), but protozoa express forms of PAKs not found in metazoans that also contain a PH domain (i.e., Cla4p and Skm1p) as reviewed.62 The Ste20 and Cla4 kinases play overlapping functions,63 while Skm1 is not an essential gene.64 Loss of Ste20 results in sterility, while deletion of Cla4 leads to aberrant cytokinesis; deletion of both kinases is lethal.63 Ste20 was identified as an essential protein in the mating pathway upstream of this mitogen-activated protein kinase (MAPK) cascades.65 The mating pheromone receptors Ste2-Ste4 activate heterotrimeric G-protein signaling, which requires co-activation of the small G-protein Cdc42 via Cdc24.5,66 The Ste20 protein therefore acts upstream of the yeast MAPK pathway consists of Ste11 (MAP3K), Ste7 (MAP2K), Fus3 and Kss1 (MAPKs).67
Figure 4. Schematic diagram indicating conserved features of protozoan PAKs. All organisms seem to include Ste20p-like kinases. Both fungi and amoeba have PH containing PAKs resembling Cla4p. None of these kinases have typical protein interaction domains found in metazoan kinases.
In addition to mating, Ste20 plays an overlapping role with Cla4 in the regulation of filamentous growth and osmotolerance by activating MAP3K.68 Mating, vegetative growth and filamentous growth all require cell polarization, which is an activity of the Cdc24/Cdc42 module acting through Ste20 and Cla4. Cla4 is involved in septin ring assembly, actin polymerization and mitotic entry and exit. Cla4 kinase activity peaks at mitosis, and in this context acts late in the cell cycle.69,70 Clap4 participates in a negative feedback loop to end the polarized growth phase and thus loss of Cla4 leads to an exaggerated banana-like bud growth.63 Interestingly Skm1 is expressed in meiotic cells. In addition to Cdc42, PAK kinases in yeast are responsive to the cell cycle-dependent kinase Cdc28.71
Fission yeast such as Schizosaccharomyces pombe are genetically divergent from budding yeast strains. S. pombe contains two PAK kinases: PAK1 (Shk1 or Orb2) and PAK2, which are effectors of Cdc42,72,73 The loss of PAK1 leads to defects in actin and the microtubule cytoskeleton, loss of polarity and defects in mating.74,75 S. pombe PAK1 most closely resembles S. cerevisiae Ste20 and is an essential gene. In this organism microtubules regulate polarized vegetative growth via a landmark involving the protein Tea1 that is directly phosphorylated by PAK1.76 Consistent with a role for Tea1 as a critical downstream effector of PAK1 in this context, loss of Tea1 function rescues the PAK1 hyperactivity-induced lethal phenotype caused by loss of a PAK1 inhibitor, Skb15.76 Remarkably all phenotypes associated with Skb15 loss (i.e., cells with hyper-active PAK1), including defects in actin cytoskeletal organization, chromosome segregation and cytokinesis, are suppressed by Tea1 loss of function.76 PAK1 is also positively regulated by the SH3 domain-containing proteins Skb1 and Skb5,77 neither of which have vertebrate counterparts. These stimulate PAK1 kinase activity to regulate cell polarity during hypertonic stress.78,79 PAK2 is a PH domain containing kinase like Cla4,73 and participates in the Ras1/Cdc42-dependent regulation of morphology as well as mating responses.80
The amoebae Dictyostelium discoideum is widely used as a model to study the chemotactic directional cell movement and differentiation. The PAKs in this organism are termed DdPAKa, DdPAKb (also termed myosin I heavy chain kinase, MIHCK) and DdPAKc. The DdPAKc belongs to the Cla4 class of kinases with N-terminal PH domain, thus demonstrating that this class of kinase is not restricted to fungi and probably evolved earlier. The biochemistry of the MIHCK is well understood; the kinase is activated by Rac only in the presence of acidic phospholipids.81,82 Unlike mammalian PAK1,38 MIHCK is not activated by sphingosine or other non-negatively charged lipids.82
The kinase activity of DdPAKa is regulated by phosphoinositide 3-kinase and Akt, which phosphorylates DdPAKa at Thr-579.83 DdPAKa-null cells display defects in myosin II assembly which delay significantly the onset of chemotaxis.84 The DdPAKb (MIHCK) is activated by Rac isoforms and acidic lipids and inhibited by Ca2+-calmodulin, as also reported for Acanthamoeba MIHCK.85 In response to chemo-attractant (cAMP) stimulation,86 PAKc kinase activity is rapidly and transiently activated, with activity levels peaking at approximately 10 sec. PAKc preferentially binds the Dictyostelium RacB, which is essential for its function. Loss DdPAKb shows mild chemotaxis defects, but strains lacking both DdPAKb and DdPAKc exhibit severe loss of cell movement, suggesting that PAKc and PAKb cooperate to regulate a common chemotaxis pathway.86 Remarkably, PTEN loss in Dictyostelium, which causes elevated PIP(3) levels, is rescued (with respect to defects in cytokinesis and chemotaxis) by the removal of PAKa.87 Thus one is left with the surprising conclusion that in PTEN-null cells, excessive PIP3 levels cause cytoskeletal defects primarily through the hyper-activation of PAKa.
Function of Invertebrate Group I and II PAKs
Fruit flies are one of the best studied invertebrate models. There are three PAKs in Drosophila melanogaster: dPAK1, mushroom bodies tiny (Mbt)/dPAK2 and dPAK3. The N-terminal proline-rich domain of dPAK1 is genetically linked to Dock (Nck) since photoreceptor axons under the guidance of extracellular cues require the SH2/SH3 adaptor Dock for correct targeting of these axons into the fly CNS.88 The guanine nucleotide exchange factor (GEF) Trio activates Rac, which in turn activates PAK1. Mutations in trio result in projection defects similar to those observed in both PAK and dock mutants, and Trio interacts genetically with dRac, dPAK1, and Dock.89 dPAK1 also plays role in defining the size and shape of the Drosophila embryonic salivary gland lumen by regulating the size and elongation of the cells. PAK1 mediates these effects by decreasing and increasing E-cadherin levels at the adherens junctions and basolateral membrane.90 During Drosophila oogenesis, basally localized F-actin bundles in the follicle cells covering the egg chamber are needed to drive elongation along the anterior-posterior axis. Mutations in dPAK1 disrupt the follicular epithelium that covers developing egg chambers due to failure in epithelial integrity and apical-basal polarity epithelial integrity and apical-basal polarity. Overall dPAK1 mutant egg chambers exhibit disorganized F-actin and remain spherical due to a failure to elongate. The removal of one copy of the Rho1 locus can suppress the dPAK1 phenotype, which provides strong evidence for an in vivo PAK1-Rho antagonism.91
Mushroom body tiny (Mbt) is involved in photoreceptor cell morphogenesis rather than photoreceptor axon guidance.92 The mushroom body in adult Drosophila corresponds to the hippocampus in the human brain, which is critical for many learning behaviors. Mbt-null mutants have defects in the central brain connectivity and fewer neurons. The Mbt mutant flies also have a reduced number of photoreceptor cells in the eye and disorganized adherens junctions. Similarly, active forms of Mbt disturbs the actin cytoskeleton and affects adherens junction organization.92 Cdc42 is required for recruitment of the kinase to adherens junctions.93 Both dPAK1 and Mbt have distinct functions during eye development: a genetic screen identified Twinstar/Cofilin as one target of Mbt signaling.94 However, this study failed to link these effects to the cofilin kinase dLimK, suggesting another intermediary kinase.
In both Drosophila and humans, loss of Spastin function results in reduction of synaptic connections and disabling motor defects. It is interesting to note that dPAK3 mutants (which have has mild phenotypes) have potent genetic interaction with spastin mutations. Aberrant bouton morphology, microtubule distribution and synaptic transmission caused by spastin loss of function were all restored by simultaneous dPAK3 loss.95
C. elegans expresses three PAK isoforms: CePAK1, CePAK2 and CePAK3/MAX-2.96,97 PAK-1 is a Group I kinase, PAK-2 belongs to Group II and MAX-2 is related to Drosophila dPAK3 (i.e., a peculiar form not found in vertebrates). CePAK-1 is expressed in the hypodermal cell boundaries during embryonic body elongation and co-localizes with CeRac1 and CeCdc42.96 Both CePAK1 and MAX-2 function with the Rac GTPases during axon guidance and also function redundantly in P cell migration; this pathway includes the upstream Rac activator UNC-73/Trio and Rac-like CED-10 and MIG-2.97 PAK1 scaffolding by the PIX and paxillin binding protein GIT1 are exemplified by such studies. The PAK/PIX/GIT complex can serve to allow PAKs to act independently of Rac or Cdc42 GTPases; the migration of the distal tip cells (DTCs) during morphogenesis of the C. elegans gonad requires the PAK1/CePIX/CeGIT complex, but does not require the small G-proteins.98 Only PAK1 functions in the DTC GIT/PIX/PAK pathway, while both PAK1 and MAX-2 are used redundantly for Cdc42/Rac- coupled cell migration pathways.
Of considerable interest is the discovery of a mechano-transduction pathway operating between the body-wall muscles of C. elegans and the epidermis that requires PAK1/CePIX/CeGIT. Tension exerted by adjacent muscles or externally exerted mechanical pressure maintains GIT1 at hemidesmosomes (integrin linked desmosomes) and stimulates PAK1 bound to PIX1, in this case requiring Rac.99 This pathway promotes the maturation of a hemidesmosome into a junction that can resist mechanical stress and contributes to coordinating the morphogenesis of epidermal and muscle tissues. An intermediate filament protein may be the direct kinase target of PAK1 in this context.
The Physiological Roles of Vertebrate Group I PAKs
The close connection between PAK, PIX and GIT as described above, is also seen in vertebrates. Standard blast searches indicate zebrafish have genes that encode six forms of PAKs termed PAK1, PAK2a, PAK2b, PAK4, PAK5(7) and PAK6. A mutant termed redhead (rhd) represents a hypomorphic mutation in PAK2a100 in which blood leaks from vessels in the head region. Such PAK2a deficiency is the result of an autonomous endothelial cell defect in the vessels supplying the brain. In PAK2a-deficient embryos reducing PAK2b levels results in a more severe and penetrant hemorrhagic phenotype, confirming the functional overlap between the two genes. Zebrafish have three PIX genes (αPIX, βPIX and γPIX)101 that are ubiquitous expressed and abundant in the nervous system.102 The zebrafish bubblehead (bbh) mutant exhibits hydrocephalus and severe cranial hemorrhage during early embryogenesis.102 Hemorrhages are associated with poor cerebral endothelial-mesenchymal contacts and an immature vascular pattern in the head. Thus in the context of the blood-brain barrier PAK2a lies downstream of βPIX and is required for cerebro-vascular stabilization. The ability of βPIX to stabilize vessels requires GIT1 binding,103 and knockdown of GIT1 also leads to a similar hemorrhage phenotype, and is phenocopied by loss of integrin α(v) or integrin β(8). Thus in fish a PAK/βPIX/GIT complex regulates vascular stability and cerebral angiogenesis in the developing embryo.
The loss of PAK1 in fish gives a late developmental phenotype.104 The atypical Rho protein Chp (also known as RhoV), has been found to act earlier in development upstream of PAK, during the process of epiboly.101 The underlying cause is a failure to localize E-cadh and β-catenin at the adherens junctions. Loss of Chp results in delayed epiboly (and embryonic death) that can be rescued by mRNA co-injection, and closely phenocopies zebrafish E-cadh mutants. This pathway involves Chp activation of the group I PAKs, and involvement of the adaptor βPIX. These observations are consistent with studies in mammalian epithelial cells where PAK1 targeting to cell-cell junctions is required for proper contact inhibition.105
The number of PAKs expressed in Xenopus laevis is unresolved because of the lack of complete genome coverage and the tetraploid nature of this model organism. These are designated xPAK1, xPAK2, etc. in line with human numbering; however, xPAK5 is actually the ortholog of human PAK4. Active forms of xPAK1 promote oocyte arrest in the G2/prophase of the cell cycle, thereby negatively regulating oocyte maturation. This is similar to the effects of active Cdc42, which likely acts via xPAK2.106 Interestingly, xPAK2 is inactivated during maturation upon stimulation of the MPF and MAPK pathways however ectopic expression of inactive xPAK1 promotes apoptosis in the Xenopus oocytes107 indicating in this context xPAK1/2/3 support cell survival. One target of PAK1 in later stages of development is reported to be tumorhead. Phosphorylation of tumorhead enhances its binding to the apical cortex and lateral cell membrane of neural plate epithelial cells, resulting in neural plate expansion and inhibition of neuronal differentiation.108
Some indication PAK biological action is indicated by knockout phenotype in mice.26 Both PAK1-null and PAK3-null mice are viable, healthy and fertile, but the mice lacking the ubiquitous PAK2 are embryonic lethal due to development failures in multiple organs.109 PAK3 loss of function is associated with certain X-linked nonsyndromic forms of mental retardation.33 To date, five distinct point mutations have been found: three in the kinase domain (abolishing kinase activity), one in the Cdc42/Rac-binding domain and one in an intron, causing a premature stop.110,111 Behavioral and cognitive impairments can be associated with abnormal neuron plasticity, which would fit with cytoskeletal functions of PAK1/PAK3 in neurons.112 Loss of PAK1 alone gives rise to modest defects in long-term potentiation in hippocampal CA1 synapses,113 similar to the effects reported for loss of PAK3.109 However, combined PAK1- and PAK3-null mice exhibit pronounced loss of brain volume compared with that of wild-type mice, despite apparently normal brain organization.114 In these animals the morphology of neurons was much less complex, with reduced dendrite length and number of dendritic tips, showing that PAK1/PAK3 are involved in branch formation. The biochemical basis for these changes is not clear: of seven substrates tested, only cofilin was found to be significantly altered with respect to phosphorylation, which is a LIM kinase (LIMK) target. It should be noted, however, that in Drosophila (see previous section) LIMK1 is not the genetic link between Twinstar and dPAK.94
Forms of human non-syndromic X-linked mental retardation are associated with genetic mutations of βPIX,115 which is needed for PAK targeting. When expressed in neurons, the AID of PAK1 (which can inhibit PAK1-3) affects synapse morphology and consolidation of long-term memory in mice.116 Interestingly expression of this AID in the hippocampus of a mouse modeled for fragile-X syndrome (FXS) can rescue some aspects of the syndrome.33 Greater spine density and elongated spines in the cortex, which are synaptic abnormalities commonly observed in FXS, are partially restored by postnatal expression of the AID in the forebrain. The deficit in cortical long-term potentiation observed in FMR1 KO mice is fully restored by this AID.33 The connection between PAK1 and fragile-X proteins involves a direct interaction between the residue 122-135 within the AID of PAK1 and the second KH domain of FMR1.32 PAK1 binds to FMR1 only when this kinase is in an open active state and phosphorylates the fragile-X related protein (FXR1) at Ser420, which appears to be required for its biological function. While FXS arises from a loss of the FMR1 protein (in males inheriting the defective X-chromosome), a rare null mutation of FMR1 (I304N) fails to interact with PAKs,32 indicating that this interaction could be critical to the function of the fragile-X complex in the normal synaptic spine. Surprisingly the activation of both Rac1 and PAK1 is reported to be impaired at hippocampal synapses in the FMR1 knockout mice.117 Stimulation-induced activation of synaptic Rac1 which stabilizes F-actin could not be seen in the FXS mutants. Thus a Rac1/PAK activation pathway appears to be “missing” for the rapid stabilization of activity-induced actin filaments in FXS. This observation apparently contradicts the notion that PAK inhibition can ameliorate fragile X defects in the brain.33
Mammalian βPIX is critical for the correct localization of PAKs to various cellular compartments and its loss in mice is embryonic lethal (Kerstin Schilling, thesis dissertation 2008). Although βPIX is widely described as a Rac1 and Cdc42 GEF118 co-expression with PAK1 in mammalian cells does not lead to potent kinase activation, suggesting the PAK/βPIX complex does not locally activate these G-proteins, at least without other (unknown) signals. As described previously PAK1/PIX complex requires GIT1 in order to target to cell adhesions119 and the centrosome where PAK1 is activated.22 βPIX via its PDZ binding domain can interact independently of GIT1 with Scribble (Scb) at cell-cell junctions120,121 and with Shank at post-synaptic densities.122,123 Although PIX has homology to established Rac GEFs such as Vav, it has essentially no activity in vitro124 because several contact residues (for Rac1) are missing. Whether the PIX partner GIT1 or others can “activate” the GEF activity is not known. The PIX proteins all have an SH3 domain that binds to a non-canonical binding sequence conserved in group I PAKs,17 but other partners have been invoked including Cbl.125
In summary, vertebrate PAK1-3 loss of function mutants demonstrate phenotypes consistent with multiple roles for these kinases in different cell types. The underlying deficiencies in target(s) phosphorylation are not well understood, and may only be revealed through the use of unbiased large-scale phospho-proteomic analyses. It seems likely that the group I PAKs have non-overlapping functions with group II kinases, the details of which are covered in depth by Audrey Minden.126
Acknowledgments
We thank the GSK-IMCB fund for previous long-term support.
Glossary
Abbreviations:
- ACK1
activated Cdc42 associated kinase 1
- AID
autoinhibitory domain
- PBD
p21 binding domain
- PH
pleckstrin homology
- PIX
PAK-interacting exchange factor
- EGF
epithermal growth factor
- SH3
Src homology domain 3
Footnotes
Previously published online: www.landesbioscience.com/journals/cellularlogistics/article/21912
References
- 1.Manser E, Leung T, Monfries C, Teo M, Hall C, Lim L. Diversity and versatility of GTPase activating proteins for the p21rho subfamily of ras G proteins detected by a novel overlay assay. J Biol Chem. 1992;267:16025–8. [PubMed] [Google Scholar]
- 2.Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–6. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 3.Tahara SM, Traugh JA. Differential activation of two protease-activated protein kinases from reticulocytes by a Ca2+-stimulated protease and identification of phosphorylated translational components. Eur J Biochem. 1982;126:395–9. doi: 10.1111/j.1432-1033.1982.tb06793.x. [DOI] [PubMed] [Google Scholar]
- 4.Jakobi R, Chen CJ, Tuazon PT, Traugh JA. Molecular cloning and sequencing of the cytostatic G protein-activated protein kinase PAK I. J Biol Chem. 1996;271:6206–11. doi: 10.1074/jbc.271.11.6206. [DOI] [PubMed] [Google Scholar]
- 5.Zhao ZS, Leung T, Manser E, Lim L. Pheromone signalling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42p and its activator CDC24. Mol Cell Biol. 1995;15:5246–57. doi: 10.1128/mcb.15.10.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manser E, Leung T, Salihuddin H, Tan L, Lim L. A non-receptor tyrosine kinase that inhibits the GTPase activity of p21cdc42. Nature. 1993;363:364–7. doi: 10.1038/363364a0. [DOI] [PubMed] [Google Scholar]
- 7.Martin GA, Bollag G, McCormick F, Abo A. A novel serine kinase activated by rac1/CDC42Hs-dependent autophosphorylation is related to PAK65 and STE20. EMBO J. 1995;14:1970–8. doi: 10.1002/j.1460-2075.1995.tb07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bagrodia S, Taylor SJ, Creasy CL, Chernoff J, Cerione RA. Identification of a mouse p21Cdc42/Rac activated kinase. J Biol Chem. 1995;270:22731–7. doi: 10.1074/jbc.270.39.22731. [DOI] [PubMed] [Google Scholar]
- 9.Maruta H, Nheu TV, He H, Hirokawa Y. Rho family-associated kinases PAK1 and rock. Prog Cell Cycle Res. 2003;5:203–10. [PubMed] [Google Scholar]
- 10.Abo A, Qu J, Cammarano MS, Dan C, Fritsch A, Baud V, et al. PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J. 1998;17:6527–40. doi: 10.1093/emboj/17.22.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manser E, Huang HY, Loo TH, Chen XQ, Dong JM, Leung T, et al. Expression of constitutively active alpha-PAK reveals effects of the kinase on actin and focal complexes. Mol Cell Biol. 1997;17:1129–43. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daub H, Gevaert K, Vandekerckhove J, Sobel A, Hall A. Rac/Cdc42 and p65PAK regulate the microtubule-destabilizing protein stathmin through phosphorylation at serine 16. J Biol Chem. 2001;276:1677–80. doi: 10.1074/jbc.C000635200. [DOI] [PubMed] [Google Scholar]
- 13.Sells MA, Knaus UG, Bagrodia S, Ambrose DM, Bokoch GM, Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol. 1997;7:202–10. doi: 10.1016/S0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 14.Lamarche N, Tapon N, Stowers L, Burbelo PD, Aspenström P, Bridges T, et al. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell. 1996;87:519–29. doi: 10.1016/S0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- 15.Joneson T, McDonough M, Bar-Sagi D, Van Aelst L. RAC regulation of actin polymerization and proliferation by a pathway distinct from Jun kinase. Science. 1996;274:1374–6. doi: 10.1126/science.274.5291.1374. [DOI] [PubMed] [Google Scholar]
- 16.Obermeier A, Ahmed S, Manser E, Yen SC, Hall C, Lim L. PAK promotes morphological changes by acting upstream of Rac. EMBO J. 1998;17:4328–39. doi: 10.1093/emboj/17.15.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, et al. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–92. doi: 10.1016/S1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 18.Fackler OT, Lu X, Frost JA, Geyer M, Jiang B, Luo W, et al. p21-activated kinase 1 plays a critical role in cellular activation by Nef. Mol Cell Biol. 2000;20:2619–27. doi: 10.1128/MCB.20.7.2619-2627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loo TH, Ng YW, Lim L, Manser E. GIT1 activates p21-activated kinase through a mechanism independent of p21 binding. Mol Cell Biol. 2004;24:3849–59. doi: 10.1128/MCB.24.9.3849-3859.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manabe R, Kovalenko M, Webb DJ, Horwitz AR. GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. J Cell Sci. 2002;115:1497–510. doi: 10.1242/jcs.115.7.1497. [DOI] [PubMed] [Google Scholar]
- 21.Zhao ZS, Manser E, Lim L. Interaction between PAK and nck: a template for Nck targets and role of PAK autophosphorylation. Mol Cell Biol. 2000;20:3906–17. doi: 10.1128/MCB.20.11.3906-3917.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao ZS, Lim JP, Ng YW, Lim L, Manser E. The GIT-associated kinase PAK targets to the centrosome and regulates Aurora-A. Mol Cell. 2005;20:237–49. doi: 10.1016/j.molcel.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 23.Kumar R, Gururaj AE, Barnes CJ. p21-activated kinases in cancer. Nat Rev Cancer. 2006;6:459–71. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- 24.Beeser A, Jaffer ZM, Hofmann C, Chernoff J. Role of group A p21-activated kinases in activation of extracellular-regulated kinase by growth factors. J Biol Chem. 2005;280:36609–15. doi: 10.1074/jbc.M502306200. [DOI] [PubMed] [Google Scholar]
- 25.Bao Y, Hata Y, Ikeda M, Withanage K. Mammalian Hippo pathway: from development to cancer and beyond. J Biochem. 2011;149:361–79. doi: 10.1093/jb/mvr021. [DOI] [PubMed] [Google Scholar]
- 26.Arias-Romero LE, Chernoff J. A tale of two Paks. Biol Cell. 2008;100:97–108. doi: 10.1042/BC20070109. [DOI] [PubMed] [Google Scholar]
- 27.Jaffer ZM, Chernoff J. p21-activated kinases: three more join the Pak. Int J Biochem Cell Biol. 2002;34:713–7. doi: 10.1016/S1357-2725(01)00158-3. [DOI] [PubMed] [Google Scholar]
- 28.Zhao ZS, Manser E. PAK and other Rho-associated kinases--effectors with surprisingly diverse mechanisms of regulation. Biochem J. 2005;386:201–14. doi: 10.1042/BJ20041638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei M, Lu W, Meng W, Parrini MC, Eck MJ, Mayer BJ, et al. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell. 2000;102:387–97. doi: 10.1016/S0092-8674(00)00043-X. [DOI] [PubMed] [Google Scholar]
- 30.Bokoch GM, Wang Y, Bohl BP, Sells MA, Quilliam LA, Knaus UG. Interaction of the Nck adapter protein with p21-activated kinase (PAK1) J Biol Chem. 1996;271:25746–9. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- 31.Puto LA, Pestonjamasp K, King CC, Bokoch GM. p21-activated kinase 1 (PAK1) interacts with the Grb2 adapter protein to couple to growth factor signaling. J Biol Chem. 2003;278:9388–93. doi: 10.1074/jbc.M208414200. [DOI] [PubMed] [Google Scholar]
- 32.Say E, Tay HG, Zhao ZS, Baskaran Y, Li R, Lim L, et al. A functional requirement for PAK1 binding to the KH(2) domain of the fragile X protein-related FXR1. Mol Cell. 2010;38:236–49. doi: 10.1016/j.molcel.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi ML, Rao BS, Seo JS, Choi HS, Dolan BM, Choi SY, et al. Inhibition of p21-activated kinase rescues symptoms of fragile X syndrome in mice. Proc Natl Acad Sci U S A. 2007;104:11489–94. doi: 10.1073/pnas.0705003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selyunin AS, Sutton SE, Weigele BA, Reddick LE, Orchard RC, Bresson SM, et al. The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature. 2011;469:107–11. doi: 10.1038/nature09593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strochlic TI, Viaud J, Rennefahrt UE, Anastassiadis T, Peterson JR. Phosphoinositides are essential coactivators for p21-activated kinase 1. Mol Cell. 2010;40:493–500. doi: 10.1016/j.molcel.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao ZS, Manser E, Chen XQ, Chong C, Leung T, Lim L. A conserved negative regulatory region in alphaPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol Cell Biol. 1998;18:2153–63. doi: 10.1128/mcb.18.4.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng YW, Raghunathan D, Chan PM, Baskaran Y, Smith DJ, Lee CH, et al. Why an A-loop phospho-mimetic fails to activate PAK1: understanding an inaccessible kinase state by molecular dynamics simulations. Structure. 2010;18:879–90. doi: 10.1016/j.str.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 38.Bokoch GM, Reilly AM, Daniels RH, King CC, Olivera A, Spiegel S, et al. A GTPase-independent mechanism of p21-activated kinase activation. Regulation by sphingosine and other biologically active lipids. J Biol Chem. 1998;273:8137–44. doi: 10.1074/jbc.273.14.8137. [DOI] [PubMed] [Google Scholar]
- 39.Yu JS, Chen WJ, Ni MH, Chan WH, Yang SD. Identification of the regulatory autophosphorylation site of autophosphorylation-dependent protein kinase (auto-kinase). Evidence that auto-kinase belongs to a member of the p21-activated kinase family. Biochem J. 1998;334:121–31. doi: 10.1042/bj3340121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zenke FT, King CC, Bohl BP, Bokoch GM. Identification of a central phosphorylation site in p21-activated kinase regulating autoinhibition and kinase activity. J Biol Chem. 1999;274:32565–73. doi: 10.1074/jbc.274.46.32565. [DOI] [PubMed] [Google Scholar]
- 41.Gatti A, Huang Z, Tuazon PT, Traugh JA. Multisite autophosphorylation of p21-activated protein kinase gamma-PAK as a function of activation. J Biol Chem. 1999;274:8022–8. doi: 10.1074/jbc.274.12.8022. [DOI] [PubMed] [Google Scholar]
- 42.Chong C, Tan L, Lim L, Manser E. The mechanism of PAK activation. Autophosphorylation events in both regulatory and kinase domains control activity. J Biol Chem. 2001;276:17347–53. doi: 10.1074/jbc.M009316200. [DOI] [PubMed] [Google Scholar]
- 43.Malecka KA, Peterson JR. Face-to-face, pak-to-pak. Structure. 2011;19:1723–4. doi: 10.1016/j.str.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Xia C, Ma W, Stafford LJ, Marcus S, Xiong WC, Liu M. Regulation of the p21-activated kinase (PAK) by a human Gbeta -like WD-repeat protein, hPIP1. Proc Natl Acad Sci U S A. 2001;98:6174–9. doi: 10.1073/pnas.101137298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace SW, Durgan J, Jin D, Hall A. Cdc42 regulates apical junction formation in human bronchial epithelial cells through PAK4 and Par6B. Mol Biol Cell. 2010;21:2996–3006. doi: 10.1091/mbc.E10-05-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talukder AH, Meng Q, Kumar R. CRIPak, a novel endogenous Pak1 inhibitor. Oncogene. 2006;25:1311–9. doi: 10.1038/sj.onc.1209172. [DOI] [PubMed] [Google Scholar]
- 47.Deacon SW, Beeser A, Fukui JA, Rennefahrt UE, Myers C, Chernoff J, et al. An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chem Biol. 2008;15:322–31. doi: 10.1016/j.chembiol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eswaran J, Lee WH, Debreczeni JE, Filippakopoulos P, Turnbull A, Fedorov O, et al. Crystal Structures of the p21-activated kinases PAK4, PAK5, and PAK6 reveal catalytic domain plasticity of active group II PAKs. Structure. 2007;15:201–13. doi: 10.1016/j.str.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eswaran J, Soundararajan M, Kumar R, Knapp S. UnPAKing the class differences among p21-activated kinases. Trends Biochem Sci. 2008;33:394–403. doi: 10.1016/j.tibs.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 50.Baskaran Y, Ng YW, Selamat W, Ling FT, Manser E. Group I and II mammalian PAKs have different modes of activation by Cdc42. EMBO Rep. 2012;13:653–9. doi: 10.1038/embor.2012.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Menzel N, Melzer J, Waschke J, Lenz C, Wecklein H, Lochnit G, et al. The Drosophila p21-activated kinase Mbt modulates DE-cadherin-mediated cell adhesion by phosphorylation of Armadillo. Biochem J. 2008;416:231–41. doi: 10.1042/BJ20080465. [DOI] [PubMed] [Google Scholar]
- 52.Paliouras GN, Naujokas MA, Park M. Pak4, a novel Gab1 binding partner, modulates cell migration and invasion by the Met receptor. Mol Cell Biol. 2009;29:3018–32. doi: 10.1128/MCB.01286-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schrantz N, da Silva Correia J, Fowler B, Ge Q, Sun Z, Bokoch GM. Mechanism of p21-activated kinase 6-mediated inhibition of androgen receptor signaling. J Biol Chem. 2004;279:1922–31. doi: 10.1074/jbc.M311145200. [DOI] [PubMed] [Google Scholar]
- 54.Yang F, Li X, Sharma M, Zarnegar M, Lim B, Sun Z. Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. J Biol Chem. 2001;276:15345–53. doi: 10.1074/jbc.M010311200. [DOI] [PubMed] [Google Scholar]
- 55.Kaur R, Liu X, Gjoerup O, Zhang A, Yuan X, Balk SP, et al. Activation of p21-activated kinase 6 by MAP kinase kinase 6 and p38 MAP kinase. J Biol Chem. 2005;280:3323–30. doi: 10.1074/jbc.M406701200. [DOI] [PubMed] [Google Scholar]
- 56.Cotteret S, Jaffer ZM, Beeser A, Chernoff J. p21-Activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Mol Cell Biol. 2003;23:5526–39. doi: 10.1128/MCB.23.16.5526-5539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cotteret S, Chernoff J. Nucleocytoplasmic shuttling of Pak5 regulates its antiapoptotic properties. Mol Cell Biol. 2006;26:3215–30. doi: 10.1128/MCB.26.8.3215-3230.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Shao Y, Tong Y, Shen T, Zhang J, Li Y, et al. Nucleo-cytoplasmic shuttling of PAK4 modulates β-catenin intracellular translocation and signaling. Biochim Biophys Acta. 2012;1823:465–75. doi: 10.1016/j.bbamcr.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 59.Bompard G, Rabeharivelo G, Frank M, Cau J, Delsert C, Morin N. Subgroup II PAK-mediated phosphorylation regulates Ran activity during mitosis. J Cell Biol. 2010;190:807–22. doi: 10.1083/jcb.200912056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Callow MG, Zozulya S, Gishizky ML, Jallal B, Smeal T. PAK4 mediates morphological changes through the regulation of GEF-H1. J Cell Sci. 2005;118:1861–72. doi: 10.1242/jcs.02313. [DOI] [PubMed] [Google Scholar]
- 61.Barac A, Basile J, Vázquez-Prado J, Gao Y, Zheng Y, Gutkind JS. Direct interaction of p21-activated kinase 4 with PDZ-RhoGEF, a G protein-linked Rho guanine exchange factor. J Biol Chem. 2004;279:6182–9. doi: 10.1074/jbc.M309579200. [DOI] [PubMed] [Google Scholar]
- 62.Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cvrcková F, De Virgilio C, Manser E, Pringle JR, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–30. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- 64.Martián H, Mendoza A, Rodriáguez-Pachoán JM, Molina M, Nombela C. Characterization of SKM1, a Saccharomyces cerevisiae gene encoding a novel Ste20/PAK-like protein kinase. Mol Microbiol. 1997;23:431–44. doi: 10.1046/j.1365-2958.1997.d01-1870.x. [DOI] [PubMed] [Google Scholar]
- 65.Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–30. doi: 10.1016/S0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- 66.Bhattacharjya S, Gingras R, Xu P. An NMR-based identification of a peptide fragment from the beta-subunit of a G-protein showing specific interactions with the GBB domain of the Ste20 kinase in budding yeast. Biochem Biophys Res Commun. 2006;347:1145–50. doi: 10.1016/j.bbrc.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 67.Lee BN, Elion EA. The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc Natl Acad Sci U S A. 1999;96:12679–84. doi: 10.1073/pnas.96.22.12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Nadal E, Alepuz PM, Posas F. Dealing with osmostress through MAP kinase activation. EMBO Rep. 2002;3:735–40. doi: 10.1093/embo-reports/kvf158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benton BK, Tinkelenberg A, Gonzalez I, Cross FR. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol Cell Biol. 1997;17:5067–76. doi: 10.1128/mcb.17.9.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keniry ME, Kemp HA, Rivers DM, Sprague GF., Jr The identification of Pcl1-interacting proteins that genetically interact with Cla4 may indicate a link between G1 progression and mitotic exit. Genetics. 2004;166:1177–86. doi: 10.1534/genetics.166.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oehlen LJ, Cross FR. Potential regulation of Ste20 function by the Cln1-Cdc28 and Cln2-Cdc28 cyclin-dependent protein kinases. J Biol Chem. 1998;273:25089–97. doi: 10.1074/jbc.273.39.25089. [DOI] [PubMed] [Google Scholar]
- 72.Marcus S, Polverino A, Chang E, Robbins D, Cobb MH, Wigler MH. Shk1, a homolog of the Saccharomyces cerevisiae Ste20 and mammalian p65PAK protein kinases, is a component of a Ras/Cdc42 signaling module in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1995;92:6180–4. doi: 10.1073/pnas.92.13.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sells MA, Barratt JT, Caviston J, Ottilie S, Leberer E, Chernoff J. Characterization of Pak2p, a pleckstrin homology domain-containing, p21-activated protein kinase from fission yeast. J Biol Chem. 1998;273:18490–8. doi: 10.1074/jbc.273.29.18490. [DOI] [PubMed] [Google Scholar]
- 74.Qyang Y, Yang P, Du H, Lai H, Kim H, Marcus S. The p21-activated kinase, Shk1, is required for proper regulation of microtubule dynamics in the fission yeast, Schizosaccharomyces pombe. Mol Microbiol. 2002;44:325–34. doi: 10.1046/j.1365-2958.2002.02882.x. [DOI] [PubMed] [Google Scholar]
- 75.Ottilie S, Miller PJ, Johnson DI, Creasy CL, Sells MA, Bagrodia S, et al. Fission yeast pak1+ encodes a protein kinase that interacts with Cdc42p and is involved in the control of cell polarity and mating. EMBO J. 1995;14:5908–19. doi: 10.1002/j.1460-2075.1995.tb00278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim H, Yang P, Catanuto P, Verde F, Lai H, Du H, et al. The kelch repeat protein, Tea1, is a potential substrate target of the p21-activated kinase, Shk1, in the fission yeast, Schizosaccharomyces pombe. J Biol Chem. 2003;278:30074–82. doi: 10.1074/jbc.M302609200. [DOI] [PubMed] [Google Scholar]
- 77.Chang E, Bartholomeusz G, Pimental R, Chen J, Lai H, Wang Lh, et al. Direct binding and In vivo regulation of the fission yeast p21-activated kinase shk1 by the SH3 domain protein scd2. Mol Cell Biol. 1999;19:8066–74. doi: 10.1128/mcb.19.12.8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bao S, Qyang Y, Yang P, Kim H, Du H, Bartholomeusz G, et al. The highly conserved protein methyltransferase, Skb1, is a mediator of hyperosmotic stress response in the fission yeast Schizosaccharomyces pombe. J Biol Chem. 2001;276:14549–52. doi: 10.1074/jbc.C100096200. [DOI] [PubMed] [Google Scholar]
- 79.Yang P, Pimental R, Lai H, Marcus S. Direct activation of the fission yeast PAK Shk1 by the novel SH3 domain protein, Skb5. J Biol Chem. 1999;274:36052–7. doi: 10.1074/jbc.274.51.36052. [DOI] [PubMed] [Google Scholar]
- 80.Yang P, Kansra S, Pimental RA, Gilbreth M, Marcus S. Cloning and characterization of shk2, a gene encoding a novel p21-activated protein kinase from fission yeast. J Biol Chem. 1998;273:18481–9. doi: 10.1074/jbc.273.29.18481. [DOI] [PubMed] [Google Scholar]
- 81.Lee SF, Mahasneh A, de la Roche M, Côteá GP. Regulation of the p21-activated kinase-related Dictyostelium myosin I heavy chain kinase by autophosphorylation, acidic phospholipids, and Ca2+-calmodulin. J Biol Chem. 1998;273:27911–7. doi: 10.1074/jbc.273.43.27911. [DOI] [PubMed] [Google Scholar]
- 82.Brzeska H, Young R, Knaus U, Korn ED. Myosin I heavy chain kinase: cloning of the full-length gene and acidic lipid-dependent activation by Rac and Cdc42. Proc Natl Acad Sci U S A. 1999;96:394–9. doi: 10.1073/pnas.96.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chung CY, Potikyan G, Firtel RA. Control of cell polarity and chemotaxis by Akt/PKB and PI3 kinase through the regulation of PAKa. Mol Cell. 2001;7:937–47. doi: 10.1016/S1097-2765(01)00247-7. [DOI] [PubMed] [Google Scholar]
- 84.Müller-Taubenberger A, Bretschneider T, Faix J, Konzok A, Simmeth E, Weber I. Differential localization of the Dictyostelium kinase DPAKa during cytokinesis and cell migration. J Muscle Res Cell Motil. 2002;23:751–63. doi: 10.1023/A:1024475628061. [DOI] [PubMed] [Google Scholar]
- 85.Brzeska H, Young R, Tan C, Szczepanowska J, Korn ED. Calmodulin-binding and autoinhibitory domains of Acanthamoeba myosin I heavy chain kinase, a p21-activated kinase (PAK) J Biol Chem. 2001;276:47468–73. doi: 10.1074/jbc.M108957200. [DOI] [PubMed] [Google Scholar]
- 86.Lee S, Rivero F, Park KC, Huang E, Funamoto S, Firtel RA. Dictyostelium PAKc is required for proper chemotaxis. Mol Biol Cell. 2004;15:5456–69. doi: 10.1091/mbc.E04-04-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang M, Iijima M, Kamimura Y, Chen L, Long Y, Devreotes P. Disruption of PKB signaling restores polarity to cells lacking tumor suppressor PTEN. Mol Biol Cell. 2011;22:437–47. doi: 10.1091/mbc.E10-06-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hing H, Xiao J, Harden N, Lim L, Zipursky SL. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell. 1999;97:853–63. doi: 10.1016/S0092-8674(00)80798-9. [DOI] [PubMed] [Google Scholar]
- 89.Newsome TP, Schmidt S, Dietzl G, Keleman K, Asling B, Debant A, et al. Trio combines with dock to regulate Pak activity during photoreceptor axon pathfinding in Drosophila. Cell. 2000;101:283–94. doi: 10.1016/S0092-8674(00)80838-7. [DOI] [PubMed] [Google Scholar]
- 90.Pirraglia C, Walters J, Myat MM. Pak1 control of E-cadherin endocytosis regulates salivary gland lumen size and shape. Development. 2010;137:4177–89. doi: 10.1242/dev.048827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vlachos S, Harden N. Genetic evidence for antagonism between Pak protein kinase and Rho1 small GTPase signaling in regulation of the actin cytoskeleton during Drosophila oogenesis. Genetics. 2011;187:501–12. doi: 10.1534/genetics.110.120998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Melzig J, Rein KH, Schäfer U, Pfister H, Jäckle H, Heisenberg M, et al. A protein related to p21-activated kinase (PAK) that is involved in neurogenesis in the Drosophila adult central nervous system. Curr Biol. 1998;8:1223–6. doi: 10.1016/S0960-9822(07)00514-3. [DOI] [PubMed] [Google Scholar]
- 93.Schneeberger D, Raabe T. Mbt, a Drosophila PAK protein, combines with Cdc42 to regulate photoreceptor cell morphogenesis. Development. 2003;130:427–37. doi: 10.1242/dev.00248. [DOI] [PubMed] [Google Scholar]
- 94.Menzel N, Schneeberger D, Raabe T. The Drosophila p21 activated kinase Mbt regulates the actin cytoskeleton and adherens junctions to control photoreceptor cell morphogenesis. Mech Dev. 2007;124:78–90. doi: 10.1016/j.mod.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 95.Ozdowski EF, Gayle S, Bao H, Zhang B, Sherwood NT. Loss of Drosophila melanogaster p21-activated kinase 3 suppresses defects in synapse structure and function caused by spastin mutations. Genetics. 2011;189:123–35. doi: 10.1534/genetics.111.130831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen W, Chen S, Yap SF, Lim L. The Caenorhabditis elegans p21-activated kinase (CePAK) colocalizes with CeRac1 and CDC42Ce at hypodermal cell boundaries during embryo elongation. J Biol Chem. 1996;271:26362–8. doi: 10.1074/jbc.271.42.26362. [DOI] [PubMed] [Google Scholar]
- 97.Lucanic M, Kiley M, Ashcroft N, L'etoile N, Cheng HJ. The Caenorhabditis elegans P21-activated kinases are differentially required for UNC-6/netrin-mediated commissural motor axon guidance. Development. 2006;133:4549–59. doi: 10.1242/dev.02648. [DOI] [PubMed] [Google Scholar]
- 98.Lucanic M, Cheng HJA. A RAC/CDC-42-independent GIT/PIX/PAK signaling pathway mediates cell migration in C. elegans. PLoS Genet. 2008;4:e1000269. doi: 10.1371/journal.pgen.1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang H, Landmann F, Zahreddine H, Rodriguez D, Koch M, Labouesse M. A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature. 2011;471:99–103. doi: 10.1038/nature09765. [DOI] [PubMed] [Google Scholar]
- 100.Buchner DA, Su F, Yamaoka JS, Kamei M, Shavit JA, Barthel LK, et al. pak2a mutations cause cerebral hemorrhage in redhead zebrafish. Proc Natl Acad Sci U S A. 2007;104:13996–4001. doi: 10.1073/pnas.0700947104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tay HG, Ng YW, Manser E. A vertebrate-specific Chp-PAK-PIX pathway maintains E-cadherin at adherens junctions during zebrafish epiboly. PLoS One. 2010;5:e10125. doi: 10.1371/journal.pone.0010125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu J, Fraser SD, Faloon PW, Rollins EL, Vom Berg J, Starovic-Subota O, et al. A betaPix Pak2a signaling pathway regulates cerebral vascular stability in zebrafish. Proc Natl Acad Sci U S A. 2007;104:13990–5. doi: 10.1073/pnas.0700825104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu J, Zeng L, Kennedy RM, Gruenig NM, Childs SJ. βPix plays a dual role in cerebral vascular stability and angiogenesis, and interacts with integrin αvβ8. Dev Biol. 2012;363:95–105. doi: 10.1016/j.ydbio.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 104.Lightcap CM, Kari G, Arias-Romero LE, Chernoff J, Rodeck U, Williams JC. Interaction with LC8 is required for Pak1 nuclear import and is indispensable for zebrafish development. PLoS One. 2009;4:e6025. doi: 10.1371/journal.pone.0006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu F, Jia L, Thompson-Baine AM, Puglise JM, Ter Beest MB, Zegers MM. Cadherins and Pak1 control contact inhibition of proliferation by Pak1-betaPIX-GIT complex-dependent regulation of cell-matrix signaling. Mol Cell Biol. 2010;30:1971–83. doi: 10.1128/MCB.01247-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cau J, Faure S, Vigneron S, Labbeá JC, Delsert C, Morin N. Regulation of Xenopus p21-activated kinase (X-PAK2) by Cdc42 and maturation-promoting factor controls Xenopus oocyte maturation. J Biol Chem. 2000;275:2367–75. doi: 10.1074/jbc.275.4.2367. [DOI] [PubMed] [Google Scholar]
- 107.Faure S, Cau J, de Santa Barbara P, Bigou S, Ge Q, Delsert C, et al. Xenopus p21-activated kinase 5 regulates blastomeres adhesive properties during convergent extension movements. Dev Biol. 2005;277:472–92. doi: 10.1016/j.ydbio.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 108.Wu CF, Delsert C, Faure S, Traverso EE, Kloc M, Kuang J, et al. Tumorhead distribution to cytoplasmic membrane of neural plate cells is positively regulated by Xenopus p21-activated kinase 1 (X-PAK1) Dev Biol. 2007;308:169–86. doi: 10.1016/j.ydbio.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 109.Meng J, Meng Y, Hanna A, Janus C, Jia Z. Abnormal long-lasting synaptic plasticity and cognition in mice lacking the mental retardation gene Pak3. J Neurosci. 2005;25:6641–50. doi: 10.1523/JNEUROSCI.0028-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kreis P, Barnier JV. PAK signalling in neuronal physiology. Cell Signal. 2009;21:384–93. doi: 10.1016/j.cellsig.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 111.Peippo M, Koivisto AM, Särkämö T, Sipponen M, von Koskull H, Ylisaukko-oja T, et al. PAK3 related mental disability: further characterization of the phenotype. Am J Med Genet A. 2007;143A:2406–16. doi: 10.1002/ajmg.a.31956. [DOI] [PubMed] [Google Scholar]
- 112.Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–26. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- 113.Asrar S, Meng Y, Zhou Z, Todorovski Z, Huang WW, Jia Z. Regulation of hippocampal long-term potentiation by p21-activated protein kinase 1 (PAK1) Neuropharmacology. 2009;56:73–80. doi: 10.1016/j.neuropharm.2008.06.055. [DOI] [PubMed] [Google Scholar]
- 114.Huang W, Zhou Z, Asrar S, Henkelman M, Xie W, Jia Z. p21-Activated kinases 1 and 3 control brain size through coordinating neuronal complexity and synaptic properties. Mol Cell Biol. 2011;31:388–403. doi: 10.1128/MCB.00969-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao L, Ma QL, Calon F, Harris-White ME, Yang F, Lim GP, et al. Role of p21-activated kinase pathway defects in the cognitive deficits of Alzheimer disease. Nat Neurosci. 2006;9:234–42. doi: 10.1038/nn1630. [DOI] [PubMed] [Google Scholar]
- 116.Hayashi ML, Choi SY, Rao BS, Jung HY, Lee HK, Zhang D, et al. Altered cortical synaptic morphology and impaired memory consolidation in forebrain- specific dominant-negative PAK transgenic mice. Neuron. 2004;42:773–87. doi: 10.1016/j.neuron.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 117.Chen LY, Rex CS, Babayan AH, Kramár EA, Lynch G, Gall CM, et al. Physiological activation of synaptic Rac>PAK (p-21 activated kinase) signaling is defective in a mouse model of fragile X syndrome. J Neurosci. 2010;30:10977–84. doi: 10.1523/JNEUROSCI.1077-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Momboisse F, Ory S, Ceridono M, Calco V, Vitale N, Bader MF, et al. The Rho guanine nucleotide exchange factors Intersectin 1L and β-Pix control calcium-regulated exocytosis in neuroendocrine PC12 cells. Cell Mol Neurobiol. 2010;30:1327–33. doi: 10.1007/s10571-010-9580-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nayal A, Webb DJ, Brown CM, Schaefer EM, Vicente-Manzanares M, Horwitz AR. Paxillin phosphorylation at Ser273 localizes a GIT1-PIX-PAK complex and regulates adhesion and protrusion dynamics. J Cell Biol. 2006;173:587–9. doi: 10.1083/jcb.200509075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nola S, Sebbagh M, Marchetto S, Osmani N, Nourry C, Audebert S, et al. Scrib regulates PAK activity during the cell migration process. Hum Mol Genet. 2008;17:3552–65. doi: 10.1093/hmg/ddn248. [DOI] [PubMed] [Google Scholar]
- 121.Audebert S, Navarro C, Nourry C, Chasserot-Golaz S, Leácine P, Bellaiche Y, et al. Mammalian Scribble forms a tight complex with the betaPIX exchange factor. Curr Biol. 2004;14:987–95. doi: 10.1016/j.cub.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 122.Park E, Na M, Choi J, Kim S, Lee JR, Yoon J, et al. The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the beta PIX guanine nucleotide exchange factor for Rac1 and Cdc42. J Biol Chem. 2003;278:19220–9. doi: 10.1074/jbc.M301052200. [DOI] [PubMed] [Google Scholar]
- 123.Sun Y, Bamji SX. β-Pix modulates actin-mediated recruitment of synaptic vesicles to synapses. J Neurosci. 2011;31:17123–33. doi: 10.1523/JNEUROSCI.2359-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Aghazadeh B, Zhu K, Kubiseski TJ, Liu GA, Pawson T, Zheng Y, et al. Structure and mutagenesis of the Dbl homology domain. Nat Struct Biol. 1998;5:1098–107. doi: 10.1038/4209. [DOI] [PubMed] [Google Scholar]
- 125.Flanders JA, Feng Q, Bagrodia S, Laux MT, Singavarapu A, Cerione RA. The Cbl proteins are binding partners for the Cool/Pix family of p21-activated kinase-binding proteins. FEBS Lett. 2003;550:119–23. doi: 10.1016/S0014-5793(03)00853-6. [DOI] [PubMed] [Google Scholar]
- 126.Minden A. PAK4-6 in cancer and neuronal development. Cellular Log. 2012;2:95–104. doi: 10.4161/cl.21171. [DOI] [PMC free article] [PubMed] [Google Scholar]