Abstract
Background
Metallothioneins (MT) are low molecular weight, cysteine rich metal binding proteins, found across genera and species, but their function(s) in abiotic stress tolerance are not well documented.
Results
We have characterized a rice MT gene, OsMT1e-P, isolated from a subtractive library generated from a stressed salinity tolerant rice genotype, Pokkali. Bioinformatics analysis of the rice genome sequence revealed that this gene belongs to a multigenic family, which consists of 13 genes with 15 protein products. OsMT1e-P is located on chromosome XI, away from the majority of other type I genes that are clustered on chromosome XII. Various members of this MT gene cluster showed a tight co-regulation pattern under several abiotic stresses. Sequence analysis revealed the presence of conserved cysteine residues in OsMT1e-P protein. Salinity stress was found to regulate the transcript abundance of OsMT1e-P in a developmental and organ specific manner. Using transgenic approach, we found a positive correlation between ectopic expression of OsMT1e-P and stress tolerance. Our experiments further suggest ROS scavenging to be the possible mechanism for multiple stress tolerance conferred by OsMT1e-P.
Conclusion
We present an overview of MTs, describing their gene structure, genome localization and expression patterns under salinity and development in rice. We have found that ectopic expression of OsMT1e-P enhances tolerance towards multiple abiotic stresses in transgenic tobacco and the resultant plants could survive and set viable seeds under saline conditions. Taken together, the experiments presented here have indicated that ectopic expression of OsMT1e-P protects against oxidative stress primarily through efficient scavenging of reactive oxygen species.
Background
Metallothioneins (MTs) are low molecular weight, cysteine-rich metal chelators with an ability to bind heavy metal ions. MTs are able to bind a variety of metal ions by the formation of mercaptide bonds between numerous Cys residues (present in the proteins) and the metal, and thus contribute to metal detoxification by buffering cytosolic metal concentration [1]. MTs typically contain two metal-binding, cysteine-rich domains that give these metalloproteins a dumbbell conformation. They are widely distributed in animals, plants, fungi as well as cyanobacteria. Based on sequence similarities and their phylogenetic relationships, MTs have been broadly classified into three types [2-4]. Class I MTs are widespread in vertebrates and have 20 conserved cysteine residues giving them the dumbbell conformation. Class II MTs do not have this strict arrangement of cysteine residues and are widespread in plants, fungi and invertebrates. On the other hand, phytochelatins, which are enzymatically synthesized metal binding peptides, are described as Class III MTs.
Plant MTs identified so far contain two cysteine-rich domains and a large spacer region (30–50 a.a. residues, devoid of cysteine) [5,6]. These MTs have only a few histidines, while their Cys content varies between 10 and 17 residues. On the other hand, the number of aromatic amino acids in plant MTs varies from none to several. Based on the distribution of cysteine residues, number of aromatic amino acids as well as length of the spacer region, plant MTs are further classified into four types, type 1 through 4 [1,6,7]. Analysis of various EST database shows that MTs are amongst the highly abundant transcripts in plants [8]. Recent studies have established important roles for plant MTs in fruit development, root development and suberization besides heavy metal tolerance [9-12]. Furthermore, the role of plant MTs in abiotic stresses such as oxidative, dehydration, senescence as well as hormonal alterations have also been shown [10,13,14]. The antioxidant function of MTs is attributed to the presence of a large number of cysteine residues, which besides metal binding are also capable of ROS scavenging [6]. Recently, a type 1 MT from mustard i.e. LSC54 has been reported to be induced by ROS production [15]. Further, LSC54 has also been documented to be related to ROS imbalance during leaf senescence. Similarly, in rice, several type 1 and type 2 MTs have been found to play a direct role in antioxidation [16].
Abiotic stresses, a major factor in reducing plant productivity, are proving to be an increasing threat to agriculture. Development of genetically modified abiotic stress tolerant varieties may provide a solution to this problem [17]. Recently, we have characterized the molecular response of rice seedlings towards salinity stress based on their subtractive transcriptome profiling [18]. One of the key members of this response, OsMT1e-P – a type 1 MT isolated from a salinity tolerant genotype i.e. O. sativa cv. Pokkali has been reported to be induced strongly in response to salinity stress. In the present communication, we provide detailed investigations on OsMT1e-P. In essence, we have found that OsMT1e-P is transcriptionally induced in response to salinity stress. Ectopic expression of OsMT1e-P in transgenic tobacco (under the control of 35 S constitutive promoter) provides stress tolerance against salinity, drought, cold, heat and heavy metals (Cu2+ and Zn2+). OsMT1e-P over expressing plants accumulate lower amount of ROS (H2O2) under salinity stress. Further, we also show that at least five members of type 1 MT are tightly clustered on the distal arm of chromosome XII which show co-regulation in response to various abiotic stress conditions. Based on these results, we propose that OsMT1e-P may serve as an important “candidate gene” for raising ‘multi-stress tolerant’ crops.
Results and discussion
MT gene family in rice: highly conserved gene family with 5 members clustered on chromosome XII
The whole genome analysis of rice (O. sativa TIGR database ver. 6.1) revealed the presence of 13 MT genes and 15 protein products in O. sativa sp. Japonica which have been further subdivided into four types (Table 1). In Arabidopsis, nine MT members have been reported previously [4]. The presence of multiple metallothionein genes may indicate their diverse role in plant responses. Their high numbers in plants may also be related to their distinct and overlapping biological roles by the regulation of gene expression and signaling [14]. The classification of these MT proteins was performed based on the conserved cysteine residue positions [6]. The members of the MT protein family were named as OsMT, where the first two initials represent the name of the organism viz. ‘Os’ for Oryza sativa followed by metallothionein abbreviated as ‘MT’. The numeral (1–4) represents the type of MT and alphabet represents the member number identified, numerals following the alphabet represent alternatively spliced products. There are eight members in type 1-OsMT1 (a1, a2, b, c, d, e, f and g); type 2 has five members (OsMT2a, 2b1, 2b2, 2c and 2d); whereas type 3 and type 4 have only one member each (Table 1).
Table 1.
Members of metallothionein family in rice
| Earlier proposed name | Gene | Protein name | Tigr ID | Locus | Coordinate | Amino acids |
|---|---|---|---|---|---|---|
| OsMT-I-4a |
OsMT1a |
OsMT1a1 |
12012.m07613 |
LOC_Os12g38270.1 |
23467876 – 23468536 (+) |
108 |
| |
|
OsMT1a2 |
12012.m56497 |
LOC_Os12g38270.2 |
23467876 – 23468352 (+) |
82 |
| OsMT-I-1b |
OsMT1b |
OsMT1b |
12003.m07212 |
LOC_Os03g17870.1 |
9936480 – 9937064 (+) |
73 |
| OsMT-I-4b |
OsMT1c |
OsMT1c |
12012.m73910 |
LOC_Os12g38051.1 |
23350565 – 23349930 (-) |
80 |
| OsMT-I-4c |
OsMT1d |
OsMT1d |
12012.m07616 |
LOC_Os12g38300.1 |
23481607 – 23482236 (+) |
79 |
| OsMT-I-1a |
OsMT1e |
OsMT1e |
12011.m80109 |
LOC_Os11g47809.1 |
28269831 - 28270196 (+) |
75 |
| |
OsMT1f |
OsMT1f |
12012.m07587 |
LOC_Os12g38010.1 |
23319560 – 23318879 (-) |
79 |
| |
OsMT1g |
OsMT1g |
12012.m07615 |
LOC_Os12g38290.1 |
23479046 – 23479680 (+) |
77 |
| |
OsMT2a |
OsMT2a |
12001.m07196 |
LOC_Os01g05650.1 |
2691774 – 2690331 (-) |
83 |
| OsMT-I-2c |
OsMT2b |
OsMT2b1 |
12005.m28027 |
LOC_Os05g02070.3 |
584010 – 584488 (+) |
85 |
| |
|
OsMT2b2 |
12005.m27672 |
LOC_Os05g02070.4 |
584010 – 584488 (+) |
58 |
| OsMT-I-2b |
OsMT2c |
OsMT2c |
12001.m13456 |
LOC_Os01g74300.1 |
43374103 – 43374519 (+) |
81 |
| |
OsMT2d |
OsMT2d |
13101.m00538 |
LOC_Os01g05585.1 |
2665458-2664412 (+) |
65 |
| OsMT-I-3a |
OsMT3a |
OsMT3a |
12001.m07661 |
LOC_Os01g10400.2 |
5475423 – 5476251 (+) |
61 |
| OsMT-II-1a | OsMT4 | OsMT4 | 12010.m06732 | LOC_Os10g39610.2 | 20838860 – 20839209 (+) | 88 |
The multiple sequence alignment of the various rice MT protein sequences (based on in silico predictions) with the protein sequence of MT gene cloned in our laboratory from Indica rice genotype Pokkali, OsMT1e-P [Genbank: EU684548] has been shown in Additional file 1: Figure S1A. Based on this analysis, it could be concluded that rice MTs share a very high degree of conservation (as high as 98% within their respective types). Two of them i.e. OsMT1a1 and OsMT1a2 have an additional stretch of 17 amino acids which is not present in any other type 1 members. The rooted phylogenetic tree of MTs reported from Arabidopsis and rice (based on the amino acid sequence) showed a very close relationship between them (Additional file 1: Figure S1B). OsMT1e-P of Pokkali was observed to have 100% identity with OsMT1e member of O. sativa sp. Japonica and both of them clustered with other type I members.
Chromosomal localization of the various MT members in O. sativa is shown in Figure 1A. These OsMTs were found to be scattered on only six chromosomes i.e. I, III, V, X, XI and XII. Three members of the type 2 MT proteins - OsMT2a (in antisense orientation), OsMT2d and OsMT2c were located on chromosome I, while the fourth, OsMT2b, was located on chromosome V. The type 3 and type 4 classes, having a single member each, were located on chromosome I and chromosome X, respectively. Thus, as far as chromosomal localization is concerned, no specific arrangement of MTs belonging to type 2, 3 and 4 were noticed. In contrast, most of the type 1 MT genes were found to be tightly clustered together (within a segment of 150 kb) on chromosome XII (from 23.3 - 23.4 Mb region; Figure 1A). Two of these genes namely OsMT1f and OsMT1c were found to be on the antisense strand, while other three OsMT1a, OsMT1g and OsMT1d were found to be on the sense strand. In-silico analysis of various OsMT1 genes revealed that they share highly identical gene structure i.e. three exons and two introns per gene, except the alternatively spliced OsMT1a2 which shows the presence of two exons and a single intron (Figure 1A inset). This unique arrangement of MTs in near vicinity on chromosome XII and their high sequence conservation possibly indicate that the present situation is the result of segmental duplication of chromosome. To test this hypothesis, we performed sequence analysis of these regions of chromosomes using Mummer software. This analysis indicated that certain regions of chromosome XII (having MT genes) may have got duplicated and diverged further during the course of evolution. Further, analysis of various orthologous members with respect to the OsMT1e gene showed that OsMT1a, OsMT1b, OsMT1c, OsMT1f and OsMT1g belong to the same orthologous group. Thus, it can be inferred that OsMT1e gene might have got duplicated on chromosome XII at several instances and thus got further diverged from each other. Dot plot analysis (Additional file 2: Figure S2) of the region showed that these genes have diverged enough but during this process, they have preserved their metallothionein signatures as well. Earlier, analysis of genomes of maize, rice and wheat has led to the identification and characterization of shared duplications between rice and wheat [19,20]. Interestingly, MT genes in rice were not observed to be in synteny with maize, sorghum, wheat and barley. However, the extensive duplication and evolution of type 1 MTs in rice reinforces their importance in plants.
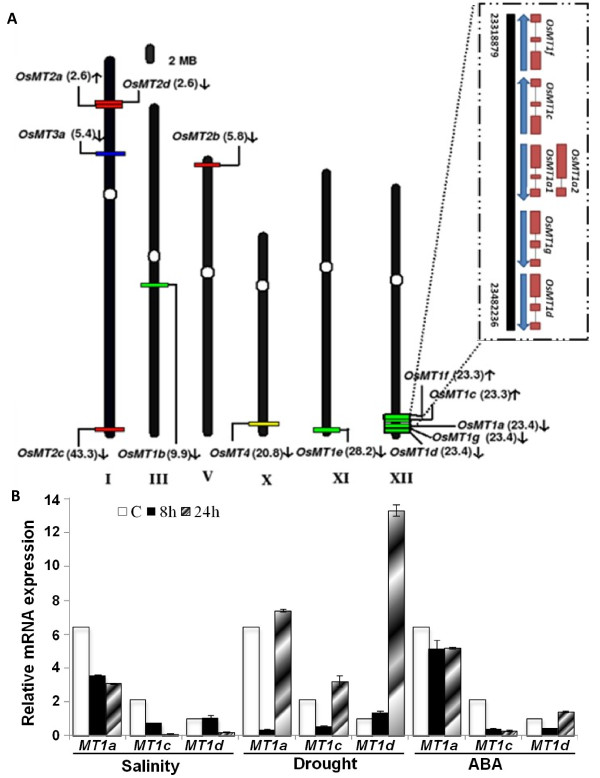
Figure 1 .
Graphical (scaled) representation of location of putative genes for OsMTs on O. sativa chromosomes and qRT PCR analysis of clustered MT genes localized on chromosome XII. (A) The centromeres on chromosomes have been marked in circles. The position of first exon of genes (in Mb) has been marked in the parentheses along with their names at same location on chromosomes. Arrow marks the direction of the ORF specific to the OsMT genes. Enlarged view of a portion of chromosome XII (inset) shows a cluster of MT genes and their distribution revealing the striking similarity in the gene structure of the members. Intron/exon(s) are shown as thin and thick line, respectively. (B) Real-time PCR analysis of OsMT1a, OsMT1c, and OsMT1d expression in shoots of 4-day old rice seedlings under various abiotic stresses such as salinity (200 mM NaCl), drought (air drying) and ABA (100µM) for various durations i.e. 8 h or 24 h. Expression of various MTs have been shown relative to that of the MT1d which is the lowest expressed gene under control conditions. The controls were grown in half strength Yoshida medium. Data are means ± SE. Each data set represents an average of minimum three separate experiments.
Quantitative real-time PCR analysis indicated co-regulation of clustered metallothionein genes
Since OsMT1a, OsMT1c, OsMT1d, OsMT1f and OsMT1g genes were found tightly clustered on chromosome XII, we wanted to check if these genes are ‘transcriptionally co-regulated’ as well. For this purpose, qRT-PCR was carried out with cDNA prepared from shoots of rice seedlings subjected to salinity, drought and ABA stress. Figure 1B shows transcript abundance for three of these genes namely OsMT1a, OsMT1c and OsMT1d. This analysis indicated that these three “tightly placed” genes are down regulated under salinity stress as well as exogenous ABA application while up regulated after 24 h of drought stress (barring OsMT1a and OsMT1c where down regulation in transcript was seen just after 8 h of stress). However, we could not quantify the transcripts for other two genes namely OsMT1g and OsMT1f which may be due to the extremely low abundance of transcripts of these genes under the tested conditions (the low abundance of these transcripts was also confirmed through the analysis of rice MPSS database). This data clearly indicates that these clustered metallothionein genes are co-regulated under various stresses in the shoots of rice seedlings. Similar observation has been made previously where co-expression of nuclear genes, coding for different subunits of photosystem or proteins involved in the transcription/translation of plastome having overlapping functions, has been documented as a novel mechanism for co-ordinated regulation of gene expression [21].
Salinity and development regulated expression of OsMT1e-P
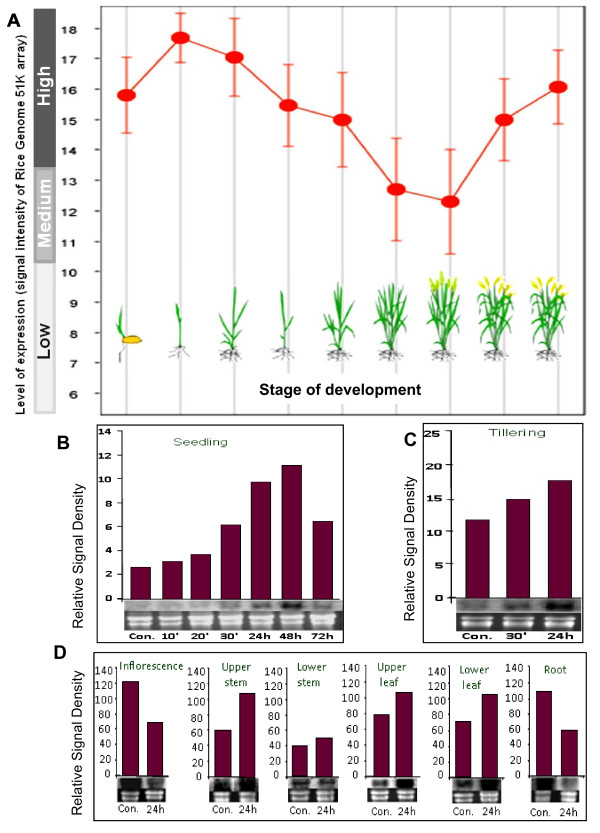
Plant stress responses often mimic certain normal developmental processes [22]. Interaction between development and environmental conditions implies that some genes must be co-regulated by both environmental and developmental cues. Since OsMT1e-P was found to be induced strongly in response to salinity stress [18], detailed investigations on its transcript regulation and functional validation was carried out. To see the effect of development on transcript accumulation of OsMT1eP in rice plants, we looked into publically available microarray data. Very high transcript abundance for OsMT1eP was noted at two most stress sensitive stages of plant development i.e. seedling and reproductive stage (Figure 2A). The vegetative and tillering stages showed relatively low abundance for this transcript. These results are in line with the existing literature where various stress responsive genes have been shown to have developmental regulation and a stage specific pattern of their induction has been reported [22,23].
Figure 2 .
Salinity and development regulated expression of OsMT1e-P in rice (Pokkali) grown in green house under standard agronomic practices. (A) Expression pattern of OsMT1e-P at various developmental stages of Oryza sativa based on microarray data (www.genevestigator.com). The expression data shown here is normalized, high quality and manually curated data obtained from several hundreds of independent experiments. The stage of development of plant is also shown diagrammatically in the figure. (B) Time course RNA gel blot analysis of OsMT1e-P transcripts in shoots of Pokkali rice seedlings stressed with 200 mM NaCl for 10′, 20′, 30′, 24 h, 48 h or 72 h. (C) Time course RNA gel blot analysis of OsMT1e-P transcripts in shoots of Pokkali rice tillers stressed with 200 mM NaCl for 30′ or 24 h. (D) RNA gel blot analysis of the OsMT1e-P expression profile in various tissues of mature rice plants (stressed with 200 mM NaCl for 24 h), grown in green house. The 0 h time point served as the control in all the analyses. The RNA gel blot analysis was performed twice with similar results. The EtBr stained RNA gel is shown below each RNA blot as loading control. The scanned intensity of each band is shown above the corresponding blot in the form of a histogram.
It has been established in literature that both ‘very early’ and ‘late’ responses associated with salinity stress have their own significance. The very early response is generally the ‘shock response’ while the late responses may often have genetic basis [18,24]. Since the OsMT1e-P gene was obtained in a salinity stress specific subtractive library [18], RNA gel blot analysis was carried out to study the effect of salt stress on transcript abundance in rice seedlings. We have carried out this transcript abundance analysis twice but data from only one such representative set is presented in Figure 2B. Transcripts for OsMT1eP could be detected in rice seedlings even under non-stress conditions which were further induced during salinity stress (10’, 20’ or 30’). The transcript levels further increased in response to 24 h of stress treatment and continued to increase up to 48 h of stress (as high as 6 folds - as compared to non stress conditions, Figure 2B). Even after 72 h of stress, induced levels of OsMT1e-P could be detected, though lesser as compared to that in response to 48 h of stress.
The salinity stress inducibility of OsMT1e-P was also observed at the tillering stage as its transcript increases within 30’ and 24 h of salinity stress (Figure 2C). In this case, we could again see a clear time-dependent transcript accumulation pattern in response to salinity stress. We have further extended this analysis to various tissues of mature rice plants grown under standard agronomic practices. In Figure 2D, it could be seen that the various tissues of the mature rice plants maintain differential levels for OsMT1e-P transcripts which in turn are differentially regulated by salinity stress. All the tissues analyzed in this study i.e. inflorescence, upper stem, lower stem, upper leaf, lower leaf and roots showed a constitutive level of expression for OsMT1e-P, with roots and inflorescence showing the highest levels. Moreover, the constitutive levels of transcripts were higher at maturity stage than seedling and tillering stage. OsMT1e-P was further found to be induced by salinity in all the tissues except roots and inflorescence, where down regulation of the transcripts could be seen in response to salinity stress. It is interesting to note that both the apical tissue of the mature plant behave differently from all other tissue.
The expression patterns obtained in mature plant tissues are in agreement with the previous studies where type 1 MT-like transcripts have been detected primarily in roots, senescing leaves, stems, leaves and flowers [25-27]. Several rice MT1 and MT2 genes show high transcript accumulation in roots, seedlings as well as stem [26-28]. Probably, the down-regulation of OsMT1e-P expression in the early phase of stress results in an oxidative burst phase, which potentiates ROS to function as a signal in salinity stress tolerance. Roots are the most appropriate site for initiating stresses response as they are the primary site of perception and injury for several types of water-limiting stresses, including salinity and drought. Under many circumstances, it is the stress-sensitivity of the root that limits the productivity of the entire plant. A high level of OsMT1e-P induction was detected in the leaves, while relatively low induction was detected in stem (Figure 2D). Leaves are more sensitive than stem due to the presence of highly developed photosynthetic apparatus which is an active site for ROS production [29] and hence metallothionein induction in leaves is important for protection under abiotic stress.
OsMT1e-P transgenic plants show enhanced tolerance to multiple abiotic stresses including salinity
The response of plants to abiotic stress consists of a coordinated function of several biochemical pathways to regulate cellular ion homeostasis, enabling the plant to show tolerance and yield stability under saline environments. Reports have suggested that although abiotic stress is a multigenic trait, stress tolerant plants could be produced by transgenic approaches by the transfer of a single or multiple genes. Several genes such as barley HVA1[30], rice CDPK[31], alfalfa Alfin1[32], tobacco NPK1[33], and Brassica GlyI and rice GlyII[34-37] have been expressed in transgenic plants to enhance their stress tolerance [38].
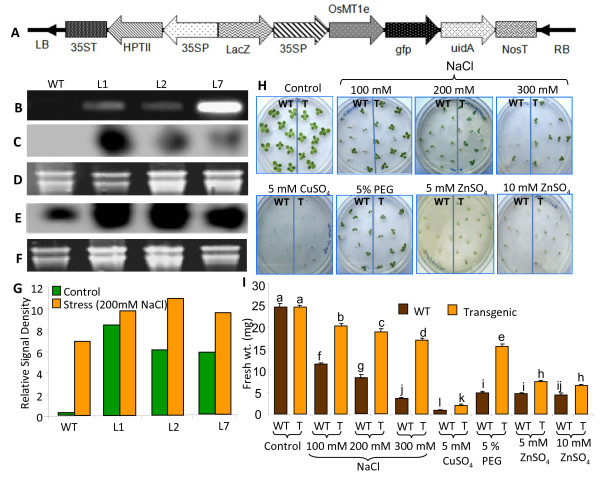
To establish the role of OsMT1e-P gene in abiotic stress tolerance, the full length gene cloned in pCAMBIA1304 vector (513 bp) was introduced into tobacco plants by Agrobacterium mediated transformation (Figure 3A). The preliminary screening of putative tobacco transformants was performed with tissue PCR, employing primers designed based on the T-DNA sequence that flanks the cloned OsMT1e-P gene. Three independently transformed T0 transgenic plants (L1, L2 and L7) were randomly selected for subsequent analysis. The integration of the OsMT1e-P gene into the tobacco genome was further confirmed by PCR analysis. A single band of 513 bp, corresponding to the OsMT1e-P cDNA, was observed in transgenic plants while no band was detected in the WT plants (Figure 3B). Northern blot analysis of wild type and T0 lines showed the presence of higher transcript levels in all the transgenic plants in comparison to WT under normal growth condition (Figure 3C, D). Moreover, increase in the transcript was observed in both WT and transgenic lines when subjected to salinity (200 mM NaCl, Figure 3E, F). These results show that a transcript signal corresponding to the size of OsMT1e-P transcript was detected in WT tobacco plants also, under stress conditions, when rice OsMT1e-P gene was used as probe. This indicated that endogenous gene for MT1e in tobacco gets up regulated under salt stress condition. Since it gets detected when rice OsMT1e-P was used as probe, it indicates that there is a good sequence homology between the two orthologs. Densitometry analysis showed that OsMT1e-P transcript abundance was higher in transgenic lines as compared to wild type maintained under control (no stress) as well as salinity stress conditions (Figure 3G).
Figure 3 .
Confirmation of OsMT1e-P transgenic tobacco plants and assessment of their tolerance towards salinity stress. (A) Schematic representation of construct (pCAMBIA OsMT1e-P ) used to ectopically express OsMT1e-P in tobacco plants. (B) PCR analysis of the wild type (WT) and T0 transgenic lines (L1, L2 and L7) to confirm the integration of the OsMT1e-P gene. (C) Northern blot analysis of OsMT1e-P transcript in shoots of seven days old WT and T0 transgenic lines (L1, L2 and L7) under unstressed condition. (D) Ethidium bromide stained RNA gel used as loading control for northern blot analysis of samples used in C. (E) Northern blot analysis of OsMT1e-P transcript in shoots of seven days old WT and T0 transgenic lines (L1, L2 and L7) under salinity (200 mM NaCl, 48 h) stress condition. (F) Ethidium bromide stained RNA gel used as loading control for northern blot analysis of samples used in E. (G) Histogram depicting signal intensity of OsMT1e-P transcript in WT and T0 transgenic lines (L1, L2 and L7) under unstressed and salinity (200 mM NaCl) stress conditions as obtained from C and E above. (H) Germination assay of OsMT1e-P transgenic seeds (T1) on MS media supplemented with various stressors. For this purpose, medium was supplemented with either NaCl (100 or 200 or 300 mM) or CuSO4 (5 mM) or PEG (5%) or ZnSO4 (5 mM or 10 mM). Seeds sown on plain MS medium served as control. (I) Histograms depicting fresh weight of WT and OsMT1e-P transgenic seedlings grown under various stress conditions. The seedlings were maintained under culture room conditions (28 ± 10 C, 16 h light/8 h dark) and their growth was monitored for 15 days under different stresses and fresh weight of surviving seedlings was measured. The data represent means ± SE of three biological replicates (n = 3). Bars with different letters are statistically significant and those with the same letters are not significantly different (p < 0.05).
Since, OsMT1e-P is also a putative metal binding protein, multiple stress tolerance ability of OsMT1e-P ectopic expressing lines (T1) were assayed at the seedling stage by transferring them to various concentrations of NaCl (salinity), PEG (dehydration), ZnSO4 and CuSO4 (heavy metal stress) and monitoring their growth for 15 days. All OsMT1e-P transgenic seedlings showed comparable growth in the absence of NaCl (Figure 3H). However, in the presence of mild salinity (100 mM NaCl), a strong difference in seedling growth was observed between WT and transgenic lines. When exposed to 200 mM NaCl, strong contrast was observed in growth pattern where the transgenic seedlings grew much better as compared to WT seedlings. WT seedlings just could not grow in the presence of higher levels of salinity (300 mM NaCl), but most of the seedlings of transgenic lines were able to grow well under similar conditions. Similarly, transgenic seedlings outperformed WT plants under 5% PEG or ZnSO4. However, in the presence of 5 mM CuSO4, the WT and transgenic lines behaved almost the same way where growth of the seedlings was almost arrested. Histograms depicting the differences in the fresh weight of transgenic vis-a-vis wild type tobacco lines show a clear advantage of OsMT1e-P ectopic expression, particularly under salinity, PEG and zinc stress (Figure 3I). Stress mitigating capacity of several plant metallothionein genes is mediated through their metal binding capacity [11,13,14]. When released from metallothionein proteins, Zn2+/Cu2+ ions become available for antioxidant metal binding enzymes e.g. Cu, Zn-superoxide dismutase and may also help in spatial regulation of oxidoreductive environment in the cell [39]. MTs have been reported to initiate Zn2+ mediated antioxidant response in mammals and fungi [40]. Improved growth and survival of OsMT1e-P transgenic plants under multiple stresses is proposed to be mediated through maintenance of redox balance and thereby reducing ROS induced injury.
Leaf disc assay has been shown to be one of the quick assays for assessing the tolerance of plants towards salinity stress [35]. We also performed this assay with T1 plants in the presence of high salinity (200 mM and 300 mM NaCl). All the transgenic lines (L1, L2 and L7) showed considerably better chlorophyll retention under high salinity (after five days of stress), while WT lines showed excessive bleaching under similar conditions (Additional file 3: Figure S3A). To explore if the ectopic expression of OsMT1eP is contributing towards better ion homeostasis, ionic measurements were carried out in leaves of the salt-stressed WT and transgenic plants. This study revealed that there was relatively less disturbance in the ion balance in transgenic plants as compared to the WT plants under salinity stress. Under control conditions, a K+/Na+ ratio in the range of 11.5 to 12 was observed for both WT and transgenic plants while a drastic decline was observed in WT when subjected to 200 mM NaCl (4.36) and 300 mM NaCl (0.74) for 5 days (Additional file 3: Figure S3B, statistically significant differences have been marked as different alphabets on the top of the bars). In contrast, the transgenic lines maintained this ratio between 7.6-9.9 and 3.3-4.5 under 200 mM and 300 mM NaCl, respectively. Maintenance of K+/Na+ homeostasis is considered as an important aspect of salinity tolerance in plants and the higher K+/Na+ levels are very well correlated with enhanced salinity tolerance [35,41,42].
Reduced accumulation of H2O2 in the transgenic lines shows their enhanced antioxidant activity
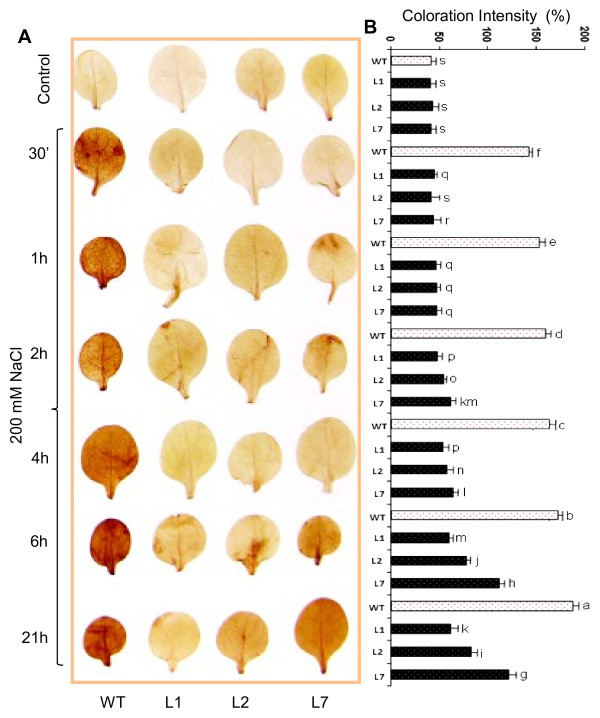
Abiotic stresses like drought, high salinity and low temperature ultimately lead to oxidative stress due to the disturbance in ROS production and scavenging, leading to generation of reactive oxygen species (ROS), such as H2O2[43-45]. In biological systems, ROS are highly reactive and toxic. They partially reduce or activate derivatives of oxygen and thus damage DNA, proteins and carbohydrates, resulting in cell death, ultimately [46,47]. ROS also cause lipid peroxidation and cell membrane damage and their levels can reflect the degree of damage to cellular components [48]. Because ectopic expression of OsMT1e-P improved the salt stress tolerance of transgenic tobacco plants, we examined whether OsMT1e-P functions in stress tolerance through detoxification of ROS. For this purpose, leaves of the WT and transgenic lines (T2), subjected to salt stress for 30 min, 1 h, 2 h, 4 h, 6 h and 21 h were stained with DAB to reveal in planta accumulation of H2O2. Histochemical staining showed that under control conditions, transgenic lines had ROS levels similar to WT. Salinity stress resulted in notable increase in ROS levels in WT within 30 min as these leaves showed intense brown colored polymer accumulation in midribs and veins (Figure 4A). However under similar condition, the leaves of transgenic lines showed remarkably less accumulation of brown colored polymer implying less ROS production. This contrast in H2O2 levels between WT and transgenic lines was noticed even after longer durations of salinity stress (4-6 h). The visual observation of the quantitative difference in the H2O2 accumulation was reconfirmed by densitometric scanning of the DAB stained leaves (Figure 4B). After 21 h of stress, transgenic lines also show a slight increase in H2O2 level but still much lower than that of WT plants (the differences were statistically significant as shown by different alphabets on the bars in the Figure 4B). Out of the three transgenic lines studied here, line L1 was found to be relatively more tolerant to salinity stress than the other two. As ROS levels during stress greatly relies on the homeostasis between ROS generation and removal [49], accumulation of less ROS in the transgenic lines seems to indicate that ROS scavenging systems in these plants might work more effectively as compared to WT. MTs can protect plants by participating in signaling (early response) or adaptation (late response). Up regulation of the antioxidant thiol protein, MT during late phases of oxidative stress, helps in mitigating stress by scavenging the ROS probably through the oxidation of cysteine thiols [50]. Recently, a number of reports have demonstrated MTs as efficient ROS scavengers in animals [51-53]. Based on this analysis, we propose that OsMT1e-P may enhance plant stress tolerance through the up regulation of antioxidative enzymes and the reduction of ROS.
Figure 4 .
Assessment of relative H2O2 levels in tobacco plants under salinity stress. (A) Constitutive and salinity stress (200 mM NaCl) induced levels of H2O2 in leaves of WT as well as OsMT1e-P ectopically expressing lines (T1 - 15-d-old plants) were visualized by DAB staining. (B) Histograms depicting relative quantity of H2O2 in these leaves in terms of their colouration intensity. The data represents the means ± SE of three biological replicates (n = 3). Bars with different letters are statistically significant and those with same letter are not significantly different (p < 0.05).
Ectopic expression of OsMT1e-P in tobacco provides multiple stress tolerance in subsequent generations
Since we found that transgenic tobacco seedlings (T1) outperform the WT plants in the leaf disc assays under salinity stress (Additional file 3: Figure S3), we decided to extend the analysis to T2 generation and also various abiotic stresses such as dehydrations (PEG), cold, heavy metals (ZnSO4 and CuSO4) and high temperature. Leaf disc assays showed a clear advantage of transgenic lines over WT plants in overcoming the deleterious effects of NaCl toxicity. The WT plants showed rapid senescence and extensive bleaching reflecting symptoms of injury due to stress, while the transgenic lines performed better under similar conditions (Figure 5A). Chlorosis was also less in transgenic lines under PEG stress (dehydration stress), cold stress, ZnSO4, and copper (CuSO4) while the WT was completely bleached under similar conditions. However, under high temperature stress, even the transgenic lines showed injury symptoms (chlorophyll depletion) but were still better than WT plants. The results of leaf disc assay were confirmed biochemically by estimating the chlorophyll content. There was nearly complete loss of chlorophyll in the WT plants, whereas the OsMT1e-P transformants showed relatively less reduction in total chlorophyll content at 200 mM NaCl (Figure 5B). Similar observations were made in response to other stresses and these results were also validated statistically as shown in different alphabets on top of the bars in Figure 5B. These observations establish a positive relationship (though not absolute) between the ectopic expression of OsMT1e-P and salinity tolerance, dehydration stress as well as heavy metal tolerance (Zn2+ and Cu2+) in leaf tissues. Another rice type I metallothionein, OsMT1a has been reported to provide tolerance against drought and oxidative stress besides being involved in Zn2+ homeostasis and its over expression leads to higher Zn2+ accumulation in rice seeds [13]. In Arabidopsis, all 4 types of MT proteins provide Cu2+ tolerance in yeast cup1 mutant [12]. Similar metal toxicity alleviation has been reported in Arabidopsis where the MT1 knockdown lines were hypersensitive to Cd2+ and accumulated several fold lower levels of Cd2+, and Zn2+ than WT [4].
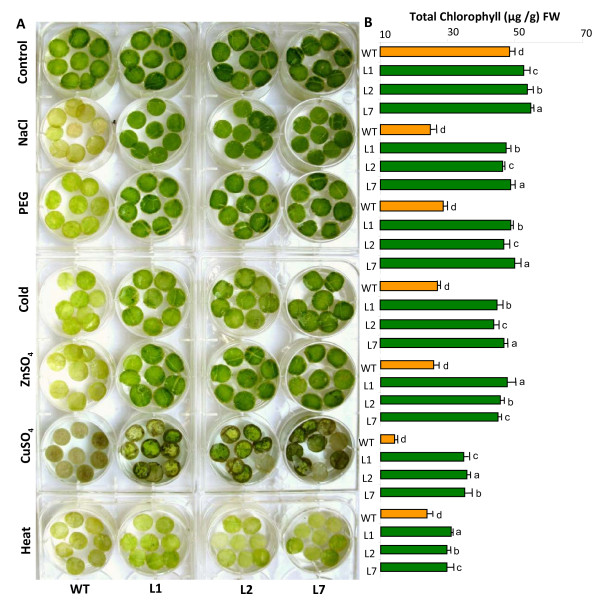
Figure 5 .
Assessment of OsMT1e-P transgenic tobacco plants for their tolerance towards various abiotic stresses and heavy metal toxicity. (A) Floating leaf disc senescence assay for abiotic stress tolerance in transgenic tobacco plants (T2). Experiments were performed on three transgenic lines (L1, L2 and L7) subjected to either 420 C for heat stress, 10 mM CuSO4, 10 mM ZnSO4, 40 C for cold stress, 5% PEG or 200 mM NaCl stress in Hoagland medium. Heat stress was given for 8 h only while other stresses were given for 5 days. Leaf discs floated in Hoagland medium served as the experimental controls. (B) Histogram depicting chlorophyll content retained in corresponding leaf discs shown in A. The data represent mean ± SE of three biological replicates (n = 3). Bars with different letters are statistically significant and those with same letter are not significantly different (p < 0.05).
OsMT1e-P transgenic plants flower normally and set viable seeds under high salinity
Since OsMT1eP seedlings were found to perform better than WT seedlings in response to salinity stress, we decided to test the performance of these plants under extended durations of stress. For this purpose, we used saline water for watering of plants till maturity. We found that while transgenic plants grew normally, flowered and produced seeds even in the presence of 200 mM NaCl, WT plants exhibited stunted growth, failed to flower and completely senesced before reaching maturity (Figure 6A). Several growth parameters viz. plant height, fresh weight, days to flower and pod weight of transgenic plants grown under salinity stress were compared with the similar parameters obtained from WT plants irrigated with water instead of salinity (WT plants could not sustain growth under saline conditions). Plant height and fresh weight measurements clearly suggests the capacity of OsMT1e-P transgenic plants in retaining higher biomass than WT under saline conditions or even WT plants under non saline conditions (Figure 6B). Flowering time was slightly delayed (2–7 days) in OsMT1e-P transgenic plants grown under stress vis-à-vis WT plants maintained under control conditions. However, it did not affect the seed setting and the overall yield of transgenic plants growing under salinity stress and the yield was comparable (or even higher) to that obtained from WT plants growing under non stress conditions (Figure 6B). Accumulation of various elements was analyzed in OsMT1eP transgenic plants using energy dispersive X-ray fluorescence (EDXRF) spectrometry which is a very useful technique for the identification of trace elements from the plant tissues [54]. OsMT1e-P ectopically expressing plants accumulate negligible Na+ as well as heavy metals Cu2+ and Zn2+ in upper leaf and pods as compared to older leaves and roots (Figure 6C, D). High accumulation of Na+ and Cu2+ in root indicates the ability of OsMT1e-P plants to use roots and lower leaves as sink thereby restricting the mobilization of deleterious amounts of these elements in the upper vegetative tissue and pods. These parameters thus clearly establish a positive correlation of OsMT1eP ectopic expression with salinity and heavy metal tolerance in tobacco plants.
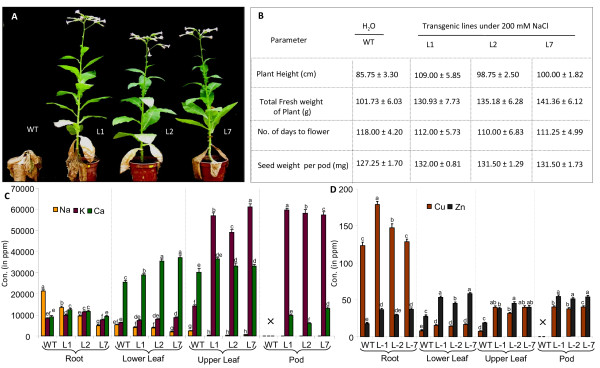
Figure 6 .
Comparison of various yield parameters of WT and OsMT1e-P ectopically expressing tobacco plants, continuously grown under saline conditions. (A) Growth of WT and OsMT1e-P tobacco plants grown in the presence 200 mM NaCl as assessed visually (B) Various growth and yield parameters of the WT and OsMT1e-P tobacco plants grown in the presence of water or 200 mM NaCl, respectively. (C) Na+, K+ and Ca2+ (D) Cu2+ and Zn2+ content [conc. as ppm], in various tissues of the OsMT1e-P ectopically expressing transgenic plants grown under continued presence of 200 mM NaCl, measured using EDXRF analysis. For each analysis, roots, old leaf, young leaf, or pods were collected from three different plants of each line. Similar data for WT plants could not be obtained as these plants did not grow further in the presence of 200 mM NaCl. The data represents mean ± SE of three biological replicates (n = 3). Bars with different letters are statistically significant and those with same letter are not significantly different (p < 0.05).
Conclusions
Expression of OsMT1e-P gene is regulated by multiple abiotic stresses in a development and organ specific manner and is therefore an important candidate gene for abiotic stress tolerance. It is proposed that OsMT1e-P helps in detoxification and cellular repair while maintaining the cellular homeostasis via ROS scavenging directly or indirectly via other antioxidants. However, further studies are warranted to work out the precise mechanism to link their metal binding affinity with abiotic stress tolerance. OsMT1e-P probably interacts with other physiological processes in the cell for selective uptake and sequestration of ions during salinity stress.
Methods
Isolation of cDNA clone and sequencing
The OsMT1e-P clone (acc. no. EU684548) was isolated during subtractive hybridization of differentially expressed cDNAs from 4-day old seedlings of O. sativa L. cv Pokkali treated with 200 mM NaCl for 30 min [18].
Sequence analysis of rice MT genes
The MT sequence protein profile was built using MT sequences obtained from NCBI database using psi-BLAST [55]. The sequences thus obtained were used to make profile using hmmbuild program of HMMER2 package (version 2.3.2; http://hmmer.janelia.org). This profile was further used for searching the presence of MT proteins in rice [TIGR version 6.1]. The sequences were aligned using MUSCLE [56] and the parsimonious tree was plotted using the alignment with Phylip package (version 3.6) with default parameters [57]. Pairwise alignment of the OsMT genomic sequences were carried out using Needle program of EMBOSS package (5.0.0) [58]. Mummer software package (v 3.20) [59] was used for analyzing the duplication of genes present on chromosome XII. The final figures for alignment were prepared using Jalview [60].
Expression analysis based on microarray data
Genevestigator 4 (http://www.genevestigator.com) was used to fetch the expression data for OsMT1eP using default parameters [23]. The gene query was the LOC (Os.3445.1.S1_at). The output of the analysis was exported in the pdf format for presentation.
Plant material and stress treatments for transcript analysis of OsMT1e-P under various conditions
For the analysis of transcript abundance under various stress conditions at the seedling stage, rice seeds were rinsed with distilled water and germinated in a hydroponic system for 7-days in half strength Yoshida medium. Salinity stress treatment was given by transferring the seedlings to 200 mM NaCl solution for 10 min, 20 min, 30 min, 8 h, 24 h, 48 h, and 72 h. Further, seedlings were exposed to drought/dehydration (air drying), and ABA (100 μM) for 8 h and 24 h. The untreated samples were taken as control.
Leaves of field grown plants at the tillering stage, while in the mature plant, tissues from various plant parts viz. stem and leaves (upper and lower), inflorescence and roots subjected to 200 mM salinity stress for 30 min or 24 h in ½ Yoshida medium and untreated samples were taken as control [18].
Total RNA isolation, mRNA purification and cDNA synthesis
Total RNA was isolated using Raflex solution (Bangalore Genei) according to the manufacturer’s instructions. The concentration of RNA was determined using spectrophotometer and its integrity was checked on agarose gel. Enrichment of polyA+ RNA and cDNA synthesis was carried out as described earlier [61].
Real-time PCR
Primers for real time PCR analysis of the OsMT gene members were designed using Primer Express 3.0 software (PE Applied Biosystems, USA). The sequences for these primers are given in additional file 4: Table S2 and real time PCR was performed as described earlier [61]. The specificity of amplification was tested by dissociation curve analysis and agarose gel electrophoresis. The expression of each gene in different RNA samples was normalized with the expression of internal control gene, actin. The mRNA levels for each candidate gene in different tissue samples were calculated relative to its expression in control seedlings using ΔΔCT method of SDS 1.4 software (Applied Biosystems). Three technical as well as biological replicates were analyzed for each sample (n = 3).
Northern hybridization
Northern blot was prepared using 20 μg total RNA and probes were prepared by labeling the PCR amplicons with α32P-dATP using HexaLabel DNA labeling kit (Fermentas Life Sciences, USA). Hybridization, washing and scanning of RNA blots were carried out as described [18].
Construction of plant transformation vector and generation of transgenic plants
For ectopic expression of OsMT1e-P, the complete ORF (513 bp) was PCR amplified by using primers OsMT1eP-F1 (5′-GAAGATCTTCATGTCTTGCAGCTGTGGATC-3′) and OsMT1eP-R1 (5′-GACTAGTCTTAACAGTTGCAAGGGTTGC-3′), and cloned at the BglII and SpeI sites of plant expression vector pCAMBIA1304. For tobacco transformation, the pCAMBIAOsMT1e-P construct was mobilized into Agrobacterium tumefaciens (GV3101) by liquid nitrogen freeze-thaw method. Tobacco leaf discs (Nicotiana tabaccum L. cv Xanthi) were transformed using the standard protocol [62] and the transformants were selected on hygromycin (25 mg/L).
Genomic DNA PCR of transgenic plants
Putative transformants were screened by PCR analysis using tobacco genomic DNA (from WT and various transgenic lines) as template and vector specific primers (forward primer 5'-CAAGACCCTTCCTCTATATAAG-3' and reverse primer 5'-CAAGAATTGGGACAACTCCAG-3'). The pCAMBIAOsMT1e-P vector was used as template for positive control.
Testing transgenic tobacco seedlings for their stress tolerance
To assess the relative stress tolerance of various plants, WT and transgenic seeds ectopically expressing OsMT1e-P were germinated on MS medium supplemented with NaCl (100, 200 or 300 mM) or 5 mM CuSO4 or ZnSO4 (5 or 10 mM) or 5% PEG for imposing different abiotic stresses or onto plain MS medium that served as the experimental control. The seedlings were maintained under culture room conditions (28 ± 1°C, 16 h light/8 h dark) and their growth was monitored for 15 days under different stresses and fresh weight of surviving seedlings was measured.
Leaf disc assay and measurement of chlorophyll contents
Leaf discs of 1 cm diameter were excised from healthy and fully expanded tobacco leaves of similar age from transgenic and WT plants were kept in half strength Hoagland media containing 200 mM NaCl or 5 mM CuSO4 or 10 mM ZnSO4 or 5% PEG. For cold stress, leaf discs were exposed to 4°C for 5 days while heat stress was given for 8 h at 42°C. Leaf discs kept in half strength Hoagland were taken as control. The chlorophyll content was measured spectrophotometrically after extraction in 80% acetone [63]. The experiment was repeated thrice with three different transgenic lines (n = 3).
In planta histochemical estimation of H2O2
Accumulation of H2O2 was examined based on histochemical staining by 3, 3'-diaminobenzidine (DAB) as described earlier [48]. WT and transgenic leaves of 15 days old seedlings were kept in 200 mM NaCl stress for different time intervals (30 min, 1 h, 2 h, 4 h, 6 h and 21 h). These leaves were vacuum infiltrated into 1 mg/ml fresh DAB solution (pH 3.8) prepared in 10 mM phosphate buffer (pH 7.8) and placed in a plastic box under high humidity and light until brown spots were observed (5 to 6 h). The stained leaves were then fixed with a solution of 3:1:1 ethanol: lactic acid: glycerol and photographed. The experiment was repeated thrice with three different transgenic lines (n = 3).
Testing transgenic tobacco plants for their tolerance towards salinity stress
In addition to the experiments at seedling stage, we carried out the assessment for the tolerance of transgenic plants when they are grown in the presence of salinity throughout their life cycle. For this purpose, the seedlings were transferred to earthern pots and grown in a greenhouse until maturity (16 h light/8 h dark and 25°C ± 2°C) either in absence or presence of 200 mM NaCl. Various parameters such as plant height, total fresh weight of plant, number of days to flower and pod weight were taken into consideration for assessment of salinity stress tolerance. Three plants from each transgenic line were randomly picked up for this analysis.
EDXRF analysis
Three plants each of transgenic lines and WT grown under control or salinity stress conditions (as described above) were harvested and washed with distilled water three times. Plants were separated into root, lower leaf, upper leaf and pod. Equal amount of these plant tissue samples were dried at 105 °C in an oven, crushed to fine powder using a mortar-pestle grinder. Dry plant powder thus obtained was pressed by using 15-ton pressure and tablets of 100.0 mg were made. EDXRF measurements were performed on the EDXRF spectrometer (PANalytical, Netherland) with a Ge solid state detector. The source of X-ray was 100 keV Gadolinium tube which allows fluorescence efficiency for K-lines higher than for L-lines of elements. All samples were measured for a period of 2000 seconds on the sample holders made of Titanium rings. For relative quantitative analysis of element, Epsilon software was used. The experiment was repeated thrice with three different transgenic lines (n = 3).
Statistical analysis
Data analysis was performed in Microsoft excel and analysis of variance (ANOVA) and presented as mean ± standard error (SE) of three biological replicates. Statistical analysis was performed using One-way-Analysis-of Variance (ANOVA). This was followed by Tukey’s post-hoc multiple comparison test using SPSS (version 19.0) for data statistics at different time points. Different letters were used (p < 0.05) to present statistically significant differences and similar letters were considered as statistically non significant.
Abbreviations
MT: Metallothionein; ROS: Reactive oxygen species; Os: Oryza sativa; At: Arabidopsis thaliana; PCR: Polymerase chain reaction; DAB: Diaminobenzidine; WT: Wild type.
Competing interests
The author(s) declare that they have no competing interests.
Authors’ contributions
GK performed all physiological and histochemical analysis of transgenic plants. The bioinformatics related computing work was done by HRK. VP-S and SK involved in amplification, cloning and transformation. RJ and RK participated in all RNA extractions and transcriptome analyses, qRT-PCR experiments. SM helped in statistical analysis of the data. SLSP and AP made contributions to the conception of the study and AP in the preparation of the final draft of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Sequence and phylogenetic analysis of MT members in O. sativa. (A) Multiple alignment of the amino acid sequences of OsMT proteins of O. sativa. The OsMT1e-P sequence was from the O. sativa sp. Pokkali, while rest of the sequences was from O. sativa sp. japonica. The (*) above the sequences shows the conserved amino acid residues present in OsMT proteins. The figures were prepared using Jalview multiple alignment editor. (B) Rooted tree of OsMTs from O. sativa sp. japonica and OsMT1e-P from O. sativa sp. Pokkali. MUSCLE program was used for alignment, and PHYLIP was used for performing bootstrapping analysis and graphical presentation of the relationship among the OsMTs. (TIFF 3114 kb)
Small gene segment repeat on chromosome XII showing that certain regions may have got duplicated and diverged further during the course of evolution. The analysis was performed using Mummer software (Kurtz et al., 2004). (A) Mummer plot of 23.3 Mbp to 23.6 Mbp region of chromosome XII. (B) Dotplot analysis of 23.3 Mbp to 23.6 Mbp region of chromosome 12. Dotplot was plotted using dottup program using EMBOSS package. (TIFF 1444 kb)
Relative stress tolerance of WT and OsMT1e-P over expressing transgenic T1 generation tobacco plants at seedling level. (A) Leaf disc senescence assay for salinity stress tolerance in transgenic tobacco plants. Experiments were performed on three transgenic lines (L1, L2 and L7). Leaf discs of uniform size were floated on either NaCl solution (200 or 300 mM) for salinity stress or on water which served as control. Photographs were taken after 5 days of treatments. (B) K+/Na+ ratio of the leaves of the OsMT1e-P transgenic plants incubated in 200 mM and 300 mM NaCl. Data are means ± SE. Each data set represents an average of minimum three separate experiments. Bars with different letters are statistically significant and those with the same letters are not significantly different (p < 0.05). (TIFF 4089 kb)
List of primers used for qRT-PCR analysis in the present study. (PDF 9 kb)
Contributor Information
Gautam Kumar, Email: kumargautamroy@gmail.com.
Hemant Ritturaj Kushwaha, Email: ritturajhemant@gmail.com.
Vaishali Panjabi-Sabharwal, Email: vaishali18@gmail.com.
Sumita Kumari, Email: sumitaslsjnu@gmail.com.
Rohit Joshi, Email: joshirohit6@gmail.com.
Ratna Karan, Email: karan.ratna@gmail.com.
Shweta Mittal, Email: shwetamittal22@gmail.com.
Sneh L Singla Pareek, Email: sneh@icgeb.res.in.
Ashwani Pareek, Email: ashwanip@mail.jnu.ac.in.
Acknowledgements
Work carried out in this paper has been supported by funds available from UGC Resource Networking, Capacity building funds from JNU, Department of Science and Technology, Department of Biotechnology, Ministry of Science and Technology, Government of India. Authors acknowledge AIRF, JNU for EDXRF measurements. Award of research fellowship from UGC (GK and RK) and CSIR (HRK and SK) is also gratefully acknowledged.
References
- Cobbett CS, Goldsbrough P. Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Ann Rev Plant Physiol Mol Biol. 2002;53:159–182. doi: 10.1146/annurev.arplant.53.100301.135154. [DOI] [PubMed] [Google Scholar]
- Cherian GM, Chan HM. In: Metallothionein III: Biological Roles and Medical Implications. Suzuki KT, Imura N, Kimura M, editor. Birkhauser Verlag, Boston; 1993. Biological functions of metallothioneins-a review; pp. 87–109. [Google Scholar]
- Binz PA, Kagi JHR. In: Metallothionein IV. Klaassen C, editor. Basel, Switzerland: Birkhauser Verlag; 1999. Metallothionein: molecular evolution and classification; pp. 7–13. [Google Scholar]
- Zimeri AM, Dhankher OP, McCaig B, Meagher RB. The plant MT1 metallothioneins are stabilized by binding cadmium and are required for cadmium tolerance and accumulation. Plant Mol Biol. 2005;58:839–855. doi: 10.1007/s11103-005-8268-3. [DOI] [PubMed] [Google Scholar]
- Freisinger E. Metallothioneins in plants. Met Ions Life Sci. 2009;5:107–153. doi: 10.1007/978-94-007-5179-8_11. [DOI] [PubMed] [Google Scholar]
- Hassinen VH, Tervahauta AI, Schat H, Karenlampi SO. Plant metallothioneins-metal chelators with ROS scavenging activity? Plant Biol. 2011;13:225–232. doi: 10.1111/j.1438-8677.2010.00398.x. [DOI] [PubMed] [Google Scholar]
- Palmiter RD. The elusive function of metallothioneins. Proc Natl Acad Sci USA. 1998;95:8428–8430. doi: 10.1073/pnas.95.15.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura H, Nirasawa S, Terauchi R. Transcript profiling of rice (Oryza sativa L.) seedlings using serial analysis of gene expression (SAGE) Plant J. 1999;20:719–726. doi: 10.1046/j.1365-313X.1999.00640.x. [DOI] [PubMed] [Google Scholar]
- Chiang HC, Lo JC, Yeh KC. Genes associated with heavy metal tolerance and accumulation in Zn Cd hyperaccumulator Arabidopsis halleri: a genomic survey with cDNA microarray. Environ Sci Technol. 2006;40:6792–6798. doi: 10.1021/es061432y. [DOI] [PubMed] [Google Scholar]
- Zhigang A, Cuijie L, Yuangang Z, Yejie D, Wachter A, Gromes R, Rausch T. Expression of BjMT2, a metallothionein 2 from Brassica juncea, increases copper and cadmium tolerance in Escherichia coli and Arabidopsis thaliana, but inhibits root elongation in Arabidopsis thaliana seedlings. J Exp Bot. 2006;57:3575–3582. doi: 10.1093/jxb/erl102. [DOI] [PubMed] [Google Scholar]
- Xu YF, Zhou GK, Zhou L, Li YQ, Liu JY. Expression patterns of the rice class I metallothionein gene family in response to lead stress in rice seedlings and functional complementation of its members in lead-sensitive yeast cells. Chinese Sci Bull. 2007;52:2203–2209. doi: 10.1007/s11434-007-0335-5. [DOI] [Google Scholar]
- Guo W, Meetam M, Goldsbrough PB. Examining the specific contributions of individual Arabidopsis metallothioneins to copper distribution and metal tolerance. Plant Physiol. 2008;146:1697–1706. doi: 10.1104/pp.108.115782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wu Y, Li Y, Ling HQ, Chu C. OsMT1a, a type 1 metallothionein, plays the pivotal role in zinc homeostasis and drought tolerance in rice. Plant Mol Biol. 2009;70:219–229. doi: 10.1007/s11103-009-9466-1. [DOI] [PubMed] [Google Scholar]
- Xue T, Li X, Zhu W, Wu C, Yang G, Zheng C. Cotton metallothionein GhMT3a, a reactive oxygen species scavenger, increased tolerance against abiotic stress in transgenic tobacco and yeast. J Exp Bot. 2009;60:339–349. doi: 10.1093/jxb/ern291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navabpour S, Morris K, Allen R, Harrison E, A-H-Mackerness S, Buchanan-Wollaston V. Expression of senescence-enhanced genes in response to oxidative stress. J Exp Bot. 2003;54:2285–2292. doi: 10.1093/jxb/erg267. [DOI] [PubMed] [Google Scholar]
- Zhou GK, Xu YF, Liu JY. Characterization of a rice class II metallothionein gene: tissue expression patterns and induction in response to abiotic factors. J Plant Physiol. 2005;162(6):686–696. doi: 10.1016/j.jplph.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Ranold P. Plant Genetics, Sustainable Agriculture and Global Food Security. Genetics. 2011;188:11–20. doi: 10.1534/genetics.111.128553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S, Panjabi Nee Sabharwal V, Kushwaha HR, Sopory SK, Singla-Pareek SL, Pareek A. Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct Integr Genomics. 2009;9:109–123. doi: 10.1007/s10142-008-0088-5. [DOI] [PubMed] [Google Scholar]
- Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, Calcagno T, Cooke R, Delseny M, Feuillet C. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell. 2008;20:11–24. doi: 10.1105/tpc.107.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse J, Abrouk M, Bolot S, Guilhot N, Courcelle E, Faraut T, Waugh R, Close TJ, Messing J, Feuillet C. Reconstruction of monocotelydoneous proto-chromosomes reveals faster evolution in plants than in animals. Proc Natl Acad Sci USA. 2009;106:14908–14913. doi: 10.1073/pnas.0902350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biehl A, Richly E, Noutsos C, Salamini F, Leister D. Analysis of 101 nuclear transcriptomes reveals 23 distinct regulons and their relationship to metabolism, chromosomal gene distribution and co-ordination of nuclear and plastid gene expression. Gene. 2005;344:33–41. doi: 10.1016/j.gene.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Cooper B, Clarke JD, Budworth P, Kreps J, Hutchison D, Park S, Guimil S, Dunn M, Luginbuhl P, Ellero C, Goff SA, Glazebrook J. A network of rice genes associated with stress response and seed development. Proc Natl Acad Sci USA. 2003;100:4945–4950. doi: 10.1073/pnas.0737574100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas H, Oliver L, Gabor S. 420747 et al. “Genevestigator V3: A Reference Expression Database for the Meta-Analysis of Transcriptomes,”. Advances in Bioinformatics. 2008;2008:5. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G, Purty RS, Sharma MP, Singla-Pareek SL, Pareek A. Physiological responses among Brassica species under salinity stress show strong correlation with transcript abundance for SOS pathway-related genes. J Plant Physiol. 2009;166:507–520. doi: 10.1016/j.jplph.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Evans IM, Gatehouse LN, Gatehouse JA, Robinson NJ, Croy RRD. A gene from pea (Pisum sutivum L.) with homology to metallothionein genes. FEBS Lett. 1990;262:29–32. doi: 10.1016/0014-5793(90)80145-9. [DOI] [PubMed] [Google Scholar]
- Hsieh HM, Liu WK, Huang PC. A novel stress-inducible metallothionein-like gene from rice. Plant Mol Biol. 1995;28:381–389. doi: 10.1007/BF00020388. [DOI] [PubMed] [Google Scholar]
- Zhou G, Xu Y, Li J, Yang L, Liu JY. Molecular analyses of the metallothionein gene family in rice (Oryza sativa L.) J Biochem Mol Biol. 2006;39(5):595–606. doi: 10.5483/BMBRep.2006.39.5.595. [DOI] [PubMed] [Google Scholar]
- Yu LH, Umeda M, Liu JY, Zhao NM, Uchimiya H. A novel MT gene of rice plants is strongly expressed in the node portion of the stem. Gene. 1998;206:29–35. doi: 10.1016/S0378-1119(97)00577-5. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol Plant. 2003;119:355–364. doi: 10.1034/j.1399-3054.2003.00223.x. [DOI] [Google Scholar]
- Xu D, Duan X, Wang B, Hong B, Ho T-HD WR. Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 1996;110:249–257. doi: 10.1104/pp.110.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000;23:319–327. doi: 10.1046/j.1365-313x.2000.00787.x. [DOI] [PubMed] [Google Scholar]
- Winicov I. Alfin1 transcription factor overexpression enhances plant root growth under normal and saline conditions and improves salt tolerance in alfalfa. Planta. 2000;210:416–422. doi: 10.1007/PL00008150. [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA. 2000;97:2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veena K, Reddy VS, Sopory SK. Glyoxalase I from Brassica juncea: molecular cloning, regulation, and its over expression confer tolerance in transgenic tobacco under stress. Plant J. 1999;17:385–395. doi: 10.1046/j.1365-313X.1999.00390.x. [DOI] [PubMed] [Google Scholar]
- Singla-Pareek SL, Reddy MK, Sopory SK. Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc Natl Acad Sci USA. 2003;100:14672–14677. doi: 10.1073/pnas.2034667100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla-Pareek SL, Yadav SK, Pareek A, Reddy MK, Sopory SK. Transgenic tobacco overexpressing glyoxalase pathway enzymes grow and set viable seeds in zinc-spiked soils. Plant Physiol. 2006;140:613–623. doi: 10.1104/pp.105.073734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla-Pareek SL, Yadav SK, Pareek A, Reddy MK, Sopory SK. Enhancing salt tolerance in a crop plant by overexpression of glyoxalase II. Transgenic Res. 2008;17:171–180. doi: 10.1007/s11248-007-9082-2. [DOI] [PubMed] [Google Scholar]
- Singh AK, Ansari MW, Pareek A, Singla-Pareek SL. Raising salinity tolerant rice: recent progress and future perspectives. Physiol Mol Biol Plants. 2008;14:137–154. doi: 10.1007/s12298-008-0013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liochev SI, Fridovich I. CO2, not HCO3-, facilitates oxidations by Cu, Zn superoxide dismutase plus H2O2. Proc Natl Acad Sci USA. 2004;101:743–744. doi: 10.1073/pnas.0307635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker SL, Thornton CR, Tasker K, Jacob C, Giles G, Egan M, Talbot NJ. A fungal metallothionein is required for pathogenicity of Magnaporthe grisea. Plant Cell. 2004;16:1575–1588. doi: 10.1105/tpc.021279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus AM, Estañ MT, Gisbert C, Garcia-Sogo B, Serrano R, Caro M, Moreno V, Bolarín MC. Expressing the yeast HAL1 gene in tomato increases fruit yield and enhances K+/Na+ selectivity under salt stress. Plant Cell Environ. 2001;24:875–880. doi: 10.1046/j.1365-3040.2001.00719.x. [DOI] [Google Scholar]
- Garg AK, Kim JK, Owens TG, Ranwala AP, Choi YD, Kochian LV, Wu RJ. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA. 2002;99:15898–15903. doi: 10.1073/pnas.252637799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Plant salt tolerance. Trends Plant Sci. 2001;6:66–72. doi: 10.1016/S1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- Mittler R. Oxidative stress, antioxidants and stress tolerance. Trend Plant Sci. 2002;7:405–410. doi: 10.1016/S1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14(S):165–183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Ouyang SQ, Liu YF, Liu P, Lei G, He SJ, Ma B, Zhang WK, Zhang JS, Chen SY. Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 2010;62:316–329. doi: 10.1111/j.1365-313X.2010.04146.x. [DOI] [PubMed] [Google Scholar]
- Huang XS, Liu JH, Chen XJ. Overexpression of PtrABF gene, a bZIP transcription factor isolated from Poncirus trifoliata, enhances dehydration and drought tolerance in tobacco via scavenging ROS and modulating expression of stress-responsive genes. BMC Plant Biol. 2010;10:230. doi: 10.1186/1471-2229-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ. 2010;33:453–457. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- Chubatsu LS, Meneghin R. Metallothionein protects DNA from oxidative damage. Biochem J. 1993;291:193–198. doi: 10.1042/bj2910193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chen H, Epstein PN. Metallothionein and catalase sensitize to diabetes in nonobese diabetic mice: reactive oxygen species may have a protective role in pancreatic beta-cells. Diabetes. 2006;55:1592–1604. doi: 10.2337/db05-1357. [DOI] [PubMed] [Google Scholar]
- Dong F, Li Q, Sreejayan N, Nunn JM, Ren J. Metallothionein prevents high-fat diet-induced cardiac contractile dysfunction: role of peroxisome proliferator activated receptor gamma coactivator 1alpha and mitochondrial biogenesis. Diabetes. 2007;56:2201–2212. doi: 10.2337/db06-1596. [DOI] [PubMed] [Google Scholar]
- Peng Z, Peng L, Fan Y, Zandi E, Shertzer HG, Xia Y. A critical role for IkappaB kinase beta in metallothionein-1 expression and protection against arsenic toxicity. J Biol Chem. 2007;282:21487–21496. doi: 10.1074/jbc.M702510200. [DOI] [PubMed] [Google Scholar]
- Dogan O, Tirasoglub E. Determination of potassium, calcium and chlorine in some vegetables by EDXRF. J Quant Spectrosc Radiat Transfer. 2006;101:141–145. doi: 10.1016/j.jqsrt.2005.11.012. [DOI] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) Version 36. Department of Genetics, University of Washington, Seattle; 2004. Distributed by the author. [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The european molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu S, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Karan R, Singla-Pareek SL, Pareek A. Histidine kinase and response regulator genes as they relate to salinity tolerance in rice. Funct Integr Genomics. 2009;9:411–417. doi: 10.1007/s10142-009-0119-x. [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Arnon DI. Copper enzymes in isolated chloroplasts: polyphenol oxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence and phylogenetic analysis of MT members in O. sativa. (A) Multiple alignment of the amino acid sequences of OsMT proteins of O. sativa. The OsMT1e-P sequence was from the O. sativa sp. Pokkali, while rest of the sequences was from O. sativa sp. japonica. The (*) above the sequences shows the conserved amino acid residues present in OsMT proteins. The figures were prepared using Jalview multiple alignment editor. (B) Rooted tree of OsMTs from O. sativa sp. japonica and OsMT1e-P from O. sativa sp. Pokkali. MUSCLE program was used for alignment, and PHYLIP was used for performing bootstrapping analysis and graphical presentation of the relationship among the OsMTs. (TIFF 3114 kb)
Small gene segment repeat on chromosome XII showing that certain regions may have got duplicated and diverged further during the course of evolution. The analysis was performed using Mummer software (Kurtz et al., 2004). (A) Mummer plot of 23.3 Mbp to 23.6 Mbp region of chromosome XII. (B) Dotplot analysis of 23.3 Mbp to 23.6 Mbp region of chromosome 12. Dotplot was plotted using dottup program using EMBOSS package. (TIFF 1444 kb)
Relative stress tolerance of WT and OsMT1e-P over expressing transgenic T1 generation tobacco plants at seedling level. (A) Leaf disc senescence assay for salinity stress tolerance in transgenic tobacco plants. Experiments were performed on three transgenic lines (L1, L2 and L7). Leaf discs of uniform size were floated on either NaCl solution (200 or 300 mM) for salinity stress or on water which served as control. Photographs were taken after 5 days of treatments. (B) K+/Na+ ratio of the leaves of the OsMT1e-P transgenic plants incubated in 200 mM and 300 mM NaCl. Data are means ± SE. Each data set represents an average of minimum three separate experiments. Bars with different letters are statistically significant and those with the same letters are not significantly different (p < 0.05). (TIFF 4089 kb)
List of primers used for qRT-PCR analysis in the present study. (PDF 9 kb)