Abstract
Cancer cells must rewire cellular metabolism to satisfy the demands of growth and proliferation. While many of the metabolic alterations are largely similar to those in normal proliferating cells, they are aberrantly driven in cancer by a combination of genetic lesions and non-genetic factors such as the tumor microenvironment. However, a single model of altered tumor metabolism does not describe the sum of metabolic changes that can support cell growth. Instead, the diversity of such changes within the metabolic program of a cancer cell can dictate by what means proliferative rewiring is driven, and can also impart heterogeneity in the metabolic dependencies of the cell. A better understanding of this heterogeneity may enable the development and optimization of therapeutic strategies that target tumor metabolism.
Keywords: Cancer Cell Metabolism, Tumor Heterogeneity
INTRODUCTION
The continuous evolution in our understanding of cancer cell biology has served to demonstrate that cancer is a remarkably complex and heterogeneous collection of diseases (1). Subsequent to the categorization of cancer types by organ or tissue of origin, one can further partition neoplastic diversity into an immense number of unique molecular subtypes, which are marked by varying prognoses, therapeutic regimens, and treatment outcomes (2, 3). Moreover, recent advances in large-scale DNA sequencing and other analytical technologies (4) have enabled recognition of genomic heterogeneities between histologically similar tumors, as well as phenotypic variability among cells within a single tumor population (5–7).
Nevertheless, a series of hallmarks that broadly encompass the distinct biological traits or capabilities that facilitate tumor growth have been proposed and recently revisited (8). One such hallmark stems from the seminal observation, initially described nearly a century ago, that cancer cells exhibit differential aspects of cellular metabolism relative to normal differentiated cells (9). Advancements over the past decade have shown that several features of altered tumor metabolism lie directly downstream of various oncogenes or tumor suppressors (10, 11), and in some cases may even be selected for during transformation (12).
The initial recognition that cancer cells exhibit atypical metabolic characteristics can be traced to the pioneering work of Otto Warburg over the first half of the twentieth century (13–15). In the presence of oxygen, most normal tissues metabolize glucose to pyruvate through glycolysis, and then completely oxidize a large fraction of the generated pyruvate to carbon dioxide in the mitochondria through oxidative phosphorylation. Under anaerobic conditions, normal cells redirect glycolytic pyruvate away from mitochondrial oxidation and instead largely reduce it to lactate (10). The fundamental paradigm stemming from Warburg’s studies was that in contrast to normal cells, rapidly proliferating ascites tumors metabolized glucose to lactate under aerobic conditions despite this process being far less efficient (~18-fold) in terms of net ATP production per molecule of glucose (10). This seemingly paradoxical phenomenon, termed the Warburg effect or aerobic glycolysis, has since been observed across several tumor types and often occurs in parallel with a marked increase in glucose uptake and consumption, as visualized – and clinically exploited – through the use of 18F-deoxyglucose-positron emission tomography (FDG-PET) (16). Though glucose catabolism through aerobic glycolysis has in large part become synonymous with the hallmark designation attributed to altered cancer metabolism, it alone cannot explain all the metabolic changes that may be necessary to support the requirements of cell growth (17). Instead, the importance of glutamine as an additionally critical nutrient in broadly fueling proliferation has also become better understood and appreciated in recent years (18).
The significant progress made towards understanding how glucose and glutamine fuel metabolic rewiring during tumorigenesis has generated optimism that this broad hallmark of cancer may be exploited through therapeutic intervention (19, 20). However, the establishment of therapeutic windows that target tumor metabolism may prove difficult given that normal proliferating cells share similar metabolic requirements and adaptations (21). Moreover, it has become increasingly clear that while a mode of metabolic rewiring necessary to support proliferative requirements is a hallmark of cancer, a single conceptual model of the cancer metabolic program does not exist. Given the incredible biological diversity across cancer types, the variability among tumors of the same subtype, and the heterogeneities present within a single tumor, it is not surprising that several metabolic signatures and distinct dependencies may arise across the spectrum of unique neoplastic cells.
Indeed, both genetic and non-genetic influences can act to not only facilitate metabolic flexibility in terms of tumor cell nutrient utilization, but to also potentiate additional heterogeneous metabolic dependencies or liabilities as well. The continued improvement and integration of multi-omics strategies, as well as the development of novel in vivo systems, are now allowing for the advancement of more sophisticated strategies towards interrogating and better understanding tumor metabolism. Ultimately, such efforts may lead to the identification of more optimal therapeutic intervention points – and thus the maximization of therapeutic windows – dependent upon a given metabolic signature.
In this review, we will first discuss the metabolic adaptations that broadly promote cell growth and how deregulated signaling and transcriptional machinery that may arise during tumorigenesis can aberrantly modulate such alterations. We will then develop the idea that while proliferative metabolism is itself a unifying feature of cancer cells in general, heterogeneities within a given metabolic signature can affect how proliferative reprogramming is achieved, and impart the cell with a specific set of metabolic dependencies or liabilities. Finally, we briefly examine how integrated analytical strategies should allow for an improved understanding of the complexities that do in fact underlie metabolic regulation in a context-dependent manner. Throughout this review, we will also describe the various challenges, efforts, and potential promises in targeting tumor metabolism as a therapeutic strategy.
THE METABOLISM OF CELL PROLIFERATION
The metabolic program of normal resting cells serves to meet the energetic requirements of maintaining homeostatic processes through ATP production (22). In contrast, proliferating cells must not only generate enough energy to support cell replication, but must also satisfy the anabolic demands of macromolecular biosynthesis and maintain cellular redox homeostasis in response to escalated production of toxic reactive oxygen species (ROS) (23) (FIGURE 1). The growth and persistence of tumor cells is also fundamentally dependent upon generating a metabolic solution that satisfies the sum of these requirements. This proliferative solution is primarily fueled by glucose and glutamine, as has been presented in detail by a number of recent excellent reviews (18, 24), and which is described here in brief summary for context in later discussion.
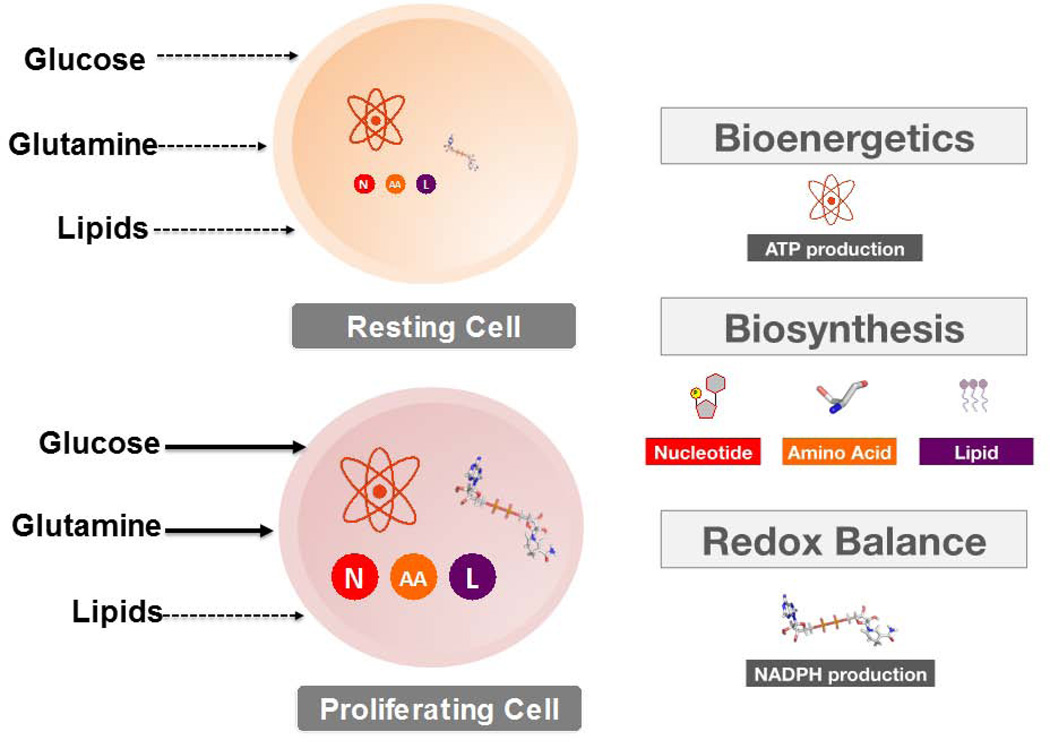
FIGURE 1. Metabolism: Resting versus Proliferating cells.
Normal resting cells employ a catabolic metabolism to satisfy the energetic requirements of homeostasis. This demand is met through fatty acid oxidation and the oxidative metabolism of glucose.
Proliferating cells however, must rewire their metabolic program to not only meet various energetic requirements, but to also satisfy the anabolic demands of macromolecular biosynthesis (nucleotides, lipids, proteins), as well as the maintenance of redox homeostasis. Upon growth factor-mediated stimulation, proliferating cells increase their uptake of glucose and glutamine, which are the two primary substrates that fuel cell growth. Solid arrows are indicative of increased cellular uptake.
Nutrient catabolism for cell growth
Most proliferating human cells metabolize glucose by aerobic glycolysis rather than through the more energetically efficient oxidative phosphorylation used by normal resting cells in the presence of oxygen (10). An initially proposed misconception was that proliferating cells harbored mitochondrial impairments and thus relied on fermentative glucose metabolism to meet their energetic demands. However, it has since been demonstrated that mitochondrial respiration persists in most proliferating cells, and in turn retains its role as the primary source of ATP generation (12, 25). Instead, the increased uptake and subsequent preferential catabolism of glucose to lactate have been proposed to serve more predominantly towards supporting biomass accumulation and redox maintenance in proliferating cells.
Glycolysis does not lie within a metabolic vacuum in which a single input (glucose) is converted through a multi-step process into a single output (pyruvate). Rather, this module of central carbon metabolism is highly interconnected with several other metabolic pathways – particularly those associated with the de novo synthesis of cellular building blocks – within which various glycolytic intermediates serve as substrates (24) (FIGURE 2). It has been noted that under conditions of high glucose uptake, the flux of glycolytic intermediates into these branching biosynthetic pathways could be substantially increased (26), while other mechanisms proposed to support this metabolite diversion will be explored in later sections.
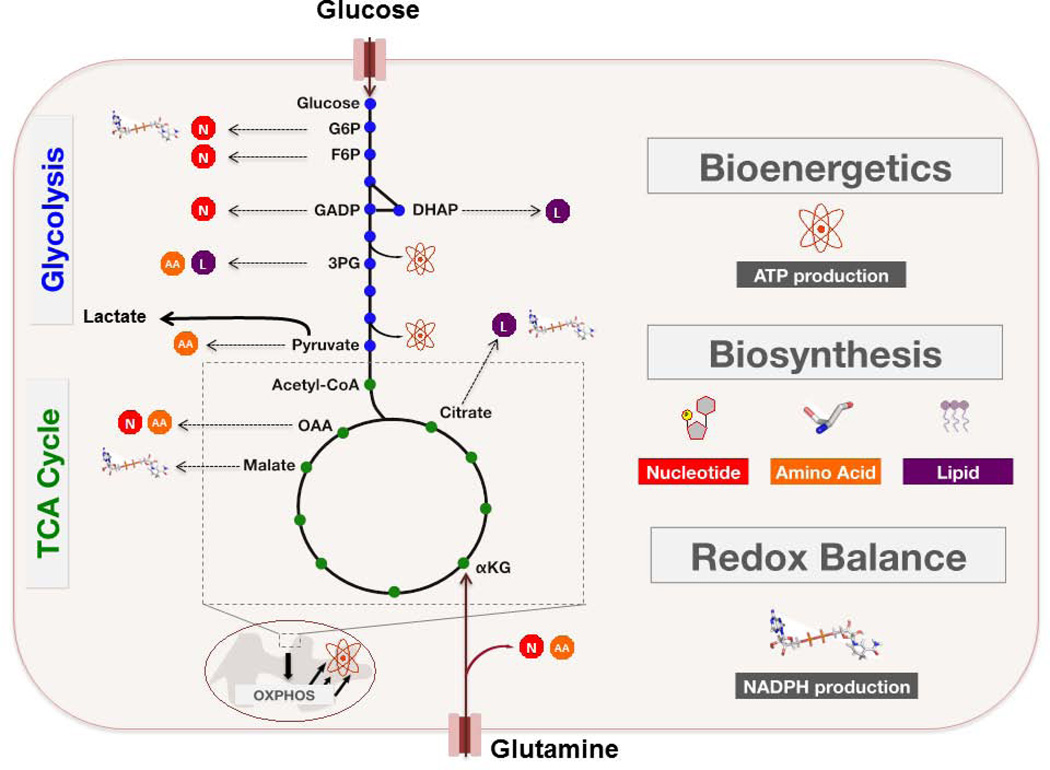
FIGURE 2. Glucose and Glutamine Fuel Proliferation.
Proliferating cells must satisfy three metabolic demands: (i) bioenergetics, (ii) macromolecular biosynthesis, and (iii) redox maintenance. The metabolic program of these cells is marked by an increased uptake of glucose and glutamine, and subsequent utilization of these two substrates to support cell growth. Most imported glucose is metabolized to lactate through aerobic glycolysis, although this process is a far less efficient means of ATP production relative to oxidative metabolism. However, the preferential catabolism of glucose to lactate allows proliferating cells to shunt various glycolytic intermediates (blue dots) into branching anabolic pathways that support additional metabolic requirements. Glutamine serves as a nitrogen source for the biosynthesis of nucleotides and various non-essential amino acids. In addition, glutamine is an important carbon source for the replenishment of TCA cycle intermediates (green dots), which are diverted into various anabolic pathways during proliferation. Further detail is found in the main body of the text.
G6P – glucose-6-phosphate. F6P – fructose-6-phosphate. GADP – glyceraldehyde-3-phosphate. DHAP – dihydroxyacetone phosphate. 3PG – 3-phosphoglycerate. αKG – α – ketoglutarate. OAA – oxaloacetate. N – nucleotide biosynthesis. L – lipid biosynthesis. AA – amino acid biosynthesis.
For example, fructose-6-phosphate and glyceraldehyde-3-phosphate may be shunted into the non-oxidative arm of the pentose phosphate pathway (PPP), resulting in the generation of ribose-5-phosphate (R5P), which is a critical intermediate in nucleotide biosynthesis. Alternatively, glucose-6-phosphate can feed into the oxidative arm of the pentose phosphate pathway (PPP) to generate R5P as well as NADPH, which contributes to the cellular defense against oxidative stress. The glycolytic intermediate 3-phosphoglycerate (3PG) provides backbone carbons for multiple nonessential amino acids through its flux into the serine biosynthesis pathway, while one downstream fate of pyruvate is its transamination to alanine. Furthermore, reduction of dihydroxyacetone phosphate to glycerol-3-phosphate effectively provides cells with a critical substrate in the biosynthesis of both phospholipids and triacylglycerols, while 3PG-derived serine can further feed into phospholipid synthesis as well (24). Downstream of glycolysis, glucose-derived pyruvate can enter the TCA cycle and contribute to the production of mitochondrial citrate, which can then feed into de novo fatty acid synthesis upon its export to the cytoplasm (27).
Glutamine is the most abundant free amino acid found in human serum. Proliferating cells use glutamine as the nitrogen donor for biosynthesis of nucleotides, non-essential amino acids, and hexosamines (18). However, many proliferating cells display a glutamine dependence that extends beyond these nitrogen requirements. Similar to the examples of glycolytic branching described above, the TCA cycle also contains intermediates that may act as substrates in various biosynthetic and NADPH-generating pathways (FIGURE 2). Proliferating cells are able to sustain the TCA cycle by replenishing these depleted intermediates through a process called anaplerosis (25). It is well appreciated that glutamine is an important carbon source for anaplerosis in many proliferating cells, through its deamidation to glutamate and subsequent entry into the TCA cycle upon glutamate conversion to α-ketoglutarate (αKG).
Glutamine-derived carbons entering the TCA cycle can, for example, contribute to mitochondrial citrate production. Following its export to the cytoplasm, citrate can be converted to acetyl-CoA and oxaloacetate (OAA) by ATP citrate lyase (ACL). While acetyl-CoA continues the path of exported citrate into fatty acid synthesis as described earlier, OAA can be further metabolized in a multi-step process to yield αKG and NADPH (25). Moreover, TCA cycle-derived OAA may be transaminated to aspartate, which can then be utilized as a carbon source in nucleotide biosynthesis. Finally, glutamine-derived malate can be exported from the mitochondria and then converted to lactate (glutaminolysis), again with concomitant production of NADPH. It is worth noting that beyond its function in redox maintenance, NADPH is also an important reducing agent in multiple biosynthetic pathways – with a particularly critical role in supporting de novo fatty acid synthesis (28, 29).
Derangement of the circuitry linking cellular signaling and metabolism in cancer
In contrast to prokaryotes and unicellular eukaryotes, mammalian cells are not autonomous for initiating the alterations necessary to enable a proliferative metabolic program (10). Instead, normal resting cells typically rely upon growth factor-mediated stimulation of specific signaling cascades, which in turn trigger a transcriptional response driving the expression of genes that promote proliferative adaptations (FIGURE 3A). Therefore, despite exposure to a relatively constant supply of exogenous nutrients, normal resting cells display far less uptake of such nutrients relative to proliferating cells, and generally preserve an oxidative metabolism that facilitates maximal ATP production to maintain homeostasis and survival (10).
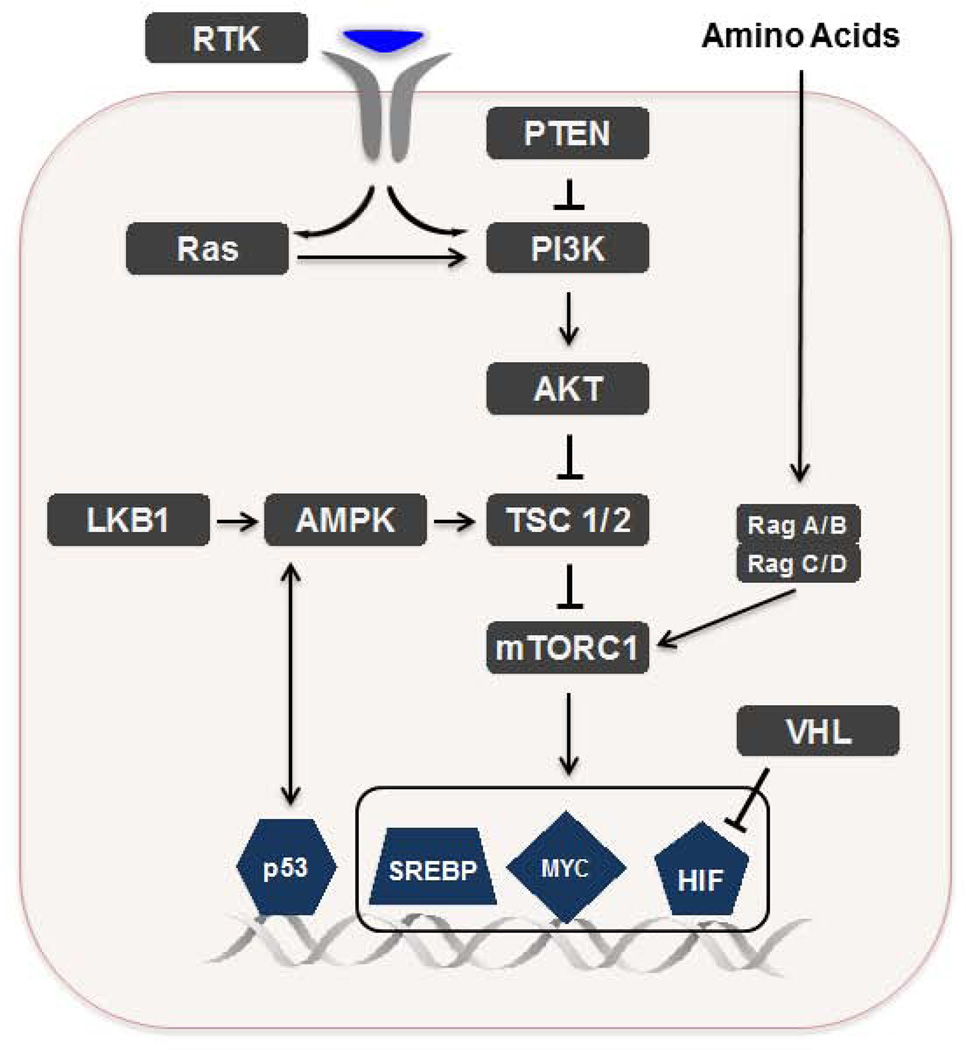
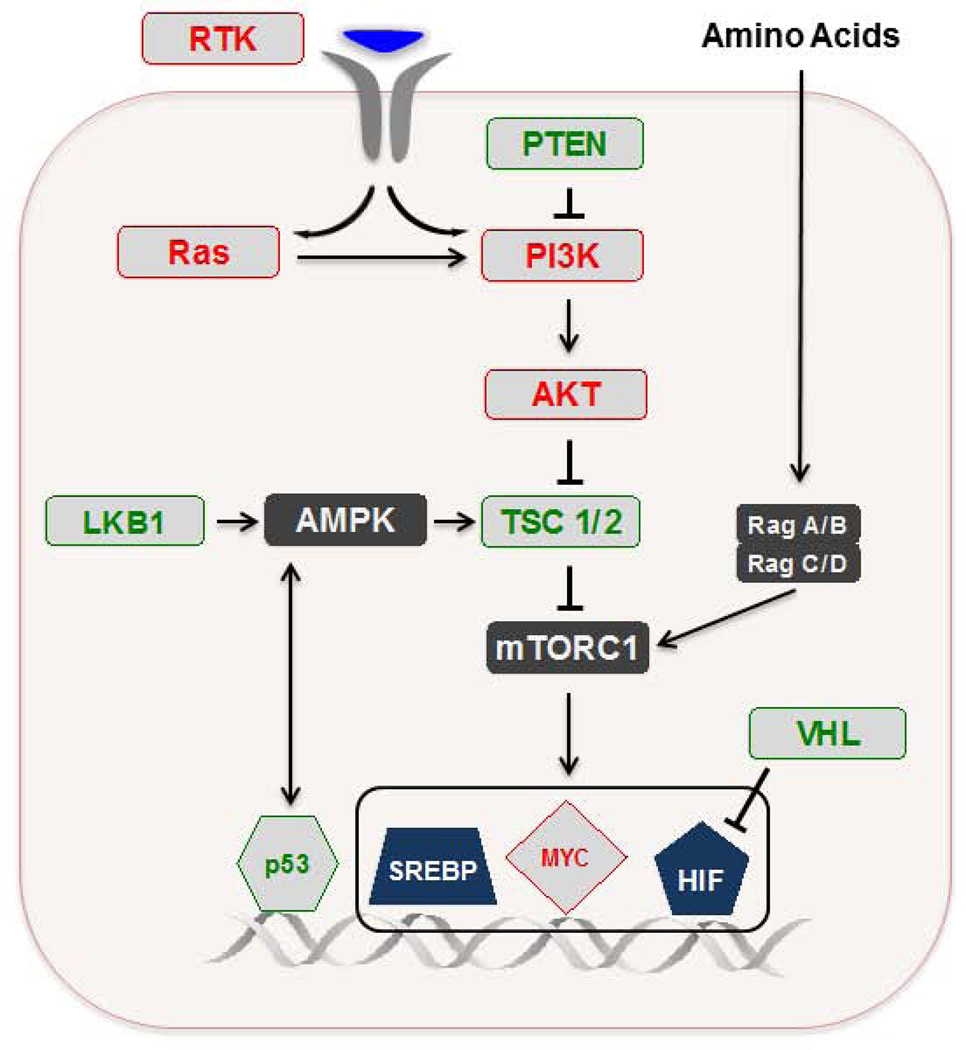
FIGURE 3. Signaling and transcriptional machinery that regulate metabolism.
A. The PI3K/Akt axis can be activated downstream of RTK activation or as a downstream effector of activated Ras. PTEN is a negative regulator of the PI3K/Akt pathway. mTORC1 can become activated upon Akt-mediated phosphorylation of either of two mTORC1 inhibitors: TSC2 (part of the TSC1-TSC2 complex) or PRAS40. Conversely, mTORC1 activity can be suppressed through AMPK-mediated phosphorylation of either TSC2 or RAPTOR. Finally, amino acids can activate mTORC1 by modulating the nucleotide loading states of Rag GTPases, which form obligate heterodimers consisting of RagA or RagB with Rag C or Rag D. Further description of amino acid-dependent activation of mTORC1 and the additional molecular components of this pathway are reviewed elsewhere (41). AMPK itself is activated by the upstream kinase LKB1. Among the downstream targets of mTORC1-dependent translation are the transcription factors HIF-1, Myc, and SREBP-1.
HIF-1 stabilization is repressed by VHL under normoxic conditions. p53 also has a multifaceted role in metabolic control, which includes involvement in a positive feedback loop with AMPK.
RTK – receptor tyrosine kinase. PTEN – phosphatase and tensin homolog. PI3K – phosphatidylinositol-3-phosphate kinase. LKB1 – liver kinase B1. AMPK – AMP-activated protein kinase. TSC – tuberous sclerosis complex. mTOR – mechanistic target of rapamycin. VHL – von Hippel-Lindau. SREBP – sterol regulatory element-binding protein. HIF – hypoxia-inducible factor.
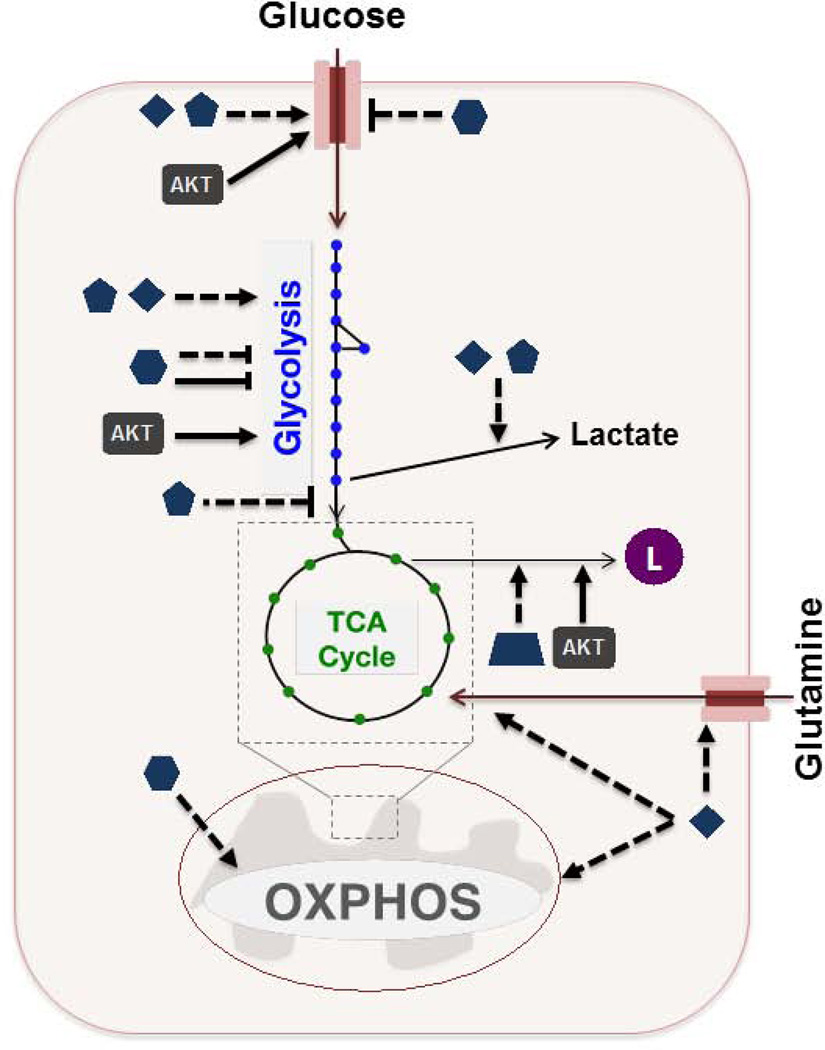
B. HIF induces the expression of various glucose transporters and glycolytic enzymes, and promotes the flux of pyruvate to lactate. Myc affects glucose metabolism in a similar manner to HIF. Additionally, Myc stimulates the expression of glutamine transporters, and promotes mitochondrial biogenesis and entry of glutamine carbon into the TCA cycle. SREBP-1 induces the expression of several genes involved in fatty acid synthesis. p53 affects glucose metabolism through repression of glucose transporters and glycolytic activity, while also transcriptionally promoting oxidative phosphorylation. Akt promotes membrane translocation of glucose transporters and the activation of various glycolytic and fatty acid synthesis enzymes.
Dashed arrow: transcription-mediated effect
C. Various components of the signaling and transcriptional network can genetically behave as oncogenes (red) or tumor suppressors (green) which enable deregulation of the metabolic regulation depicted in (B).
As mentioned above, cancer cells and normal proliferating cells share a similar collection of metabolic demands and adaptations (21, 30). Accordingly, the signaling and transcriptional circuitry that modulates cell growth (FIGURE 3B) is largely conserved across proliferating cells in general. However, whereas normal cells possess a variety of checkpoints that enable correct maintenance of this system, various tumorigenic lesions impart cancer cells with the ability to fracture proper regulation (FIGURE 3C).
PI3K/Akt/mTORC1 and LKB1/AMPK/mTORC1
Opposing regulatory axes of metabolic rewiring
The PI3K/Akt pathway lies downstream of receptor tyrosine kinase (RTK) activation and serves an important role in the tightly controlled regulation of metabolic adaptations that support cell growth (31–33). Inappropriate activation of this pathway is among the most frequent classes of alterations seen across several cancers, owing to the vast number of oncogenes and tumor suppressors identified within this network (34, 35). One downstream effect of PI3K/Akt pathway activation is the promotion of glycolytic metabolism through Akt-mediated membrane translocation of glucose transporters, and Akt-dependent activation of hexokinase and phosphofructokinase (36–39). Additionally, Akt stimulates de novo fatty acid synthesis through direct phosphorylation and corresponding activation of ACL (40), which as noted earlier, catalyzes the conversion of citrate (diverted from the TCA cycle) to acetyl-CoA and OAA.
Perhaps the most dramatic metabolic consequence of PI3K/Akt stimulation however, is downstream activation of the cell growth regulator mechanistic target of rapamycin complex 1 (mTORC1) upon Akt-mediated phosphorylation of either of two mTORC1 inhibitors: tuberous sclerosis 2 (TSC2; part of the TSC1–TSC2 complex) or proline-rich Akt substrate 40 kDa (PRAS40) (41). mTORC1 is the better characterized of two mTOR-containing multiprotein complexes (the other being mTORC2), and its activation status is affected through a variety of environmental cues, including growth factors (as transmitted via the PI3K/Akt pathway for example), energy status (described below), amino acids, and oxygen levels (41, 42). While mTORC1 can regulate many cellular processes, it remains best known for elevating protein synthesis through direct phosphorylation of the translational regulators 4E-binding protein 1 (4E-BP1) and S6 kinase 1 (S6K1) (41). Among the downstream targets of mTORC1-dependent translation are a number of transcription factors (discussed further below) that coordinate metabolic gene expression: (a) hypoxia inducible factor 1α (HIF1α), (b) c-Myc, and (c) sterol regulatory element-binding protein 1 (SREBP-1).
The AMP-activated protein kinase (AMPK) is a protein complex that plays a critical role in regulating the cellular energetic state (43). AMPK senses changes in the cellular ratio of AMP to ATP, and is activated under conditions of metabolic stress that promote ATP consumption and/or inhibit ATP production, such as hypoxia or nutrient deprivation. Upon activation, AMPK stimulates metabolic alterations to limit energy consumption (or enhance energy production) and thus allow for adaptation to a given metabolic stress (44). For example, AMPK can deter ATP-consuming fatty acid synthesis through an inactivating phosphorylation of acetyl-CoA carboxylase (ACC) (45). Moreover, anabolic mTORC1 activity can be suppressed through either of two AMPK-mediated phosphorylation events: (i) activation of the TSC1/2 complex, or (ii) inactivation of the mTORC1 scaffold protein, RAPTOR (46, 47). Under conditions of energetic stress, one of the critical upstream activators of AMPK is liver kinase B1 (LKB1) (48), which is a recognized tumor suppressor, and illustrates another connection between lesion-induced deregulation of a signaling axis and metabolic control.
Key transcriptional effectors of proliferative metabolism
HIF-1
Mammalian cells exposed to hypoxia undergo a metabolic response in which glucose consumption is elevated and glycolytic pyruvate is redirected to lactate, in order to enable net ATP production by an oxygen-independent mechanism (49). This adaptation is coordinated by the HIF-1 transcription factor complex, which induces elevated expression of several genes that support fermentative glucose metabolism, including glucose transporters, glycolytic enzymes, lactate dehydrogenase A (LDHA), and pyruvate dehydrogenase kinase-1 (PDK1) (50). The latter two enzymes divert flux of pyruvate to lactate either (i) directly - LDHA catalyzes conversion of pyruvate to lactate, or (ii) indirectly - PDK1 negatively regulates entry of pyruvate into the mitochondria. HIF-1 activity is dependent upon stabilization of its HIF-1α subunit. Under normoxic conditions, HIF-1α is suppressed through post-translational prolyl hydroxylation, which results in von Hippel-Lindau (VHL) tumor suppressor-mediated proteasomal degradation of HIF-1. However, mTORC1 can increase the transcription and translation of HIF-1α under normoxic conditions, and constitutive activation of HIF-1 can occur in tumor cells through a variety of mechanisms, including (i) loss of VHL, (ii) accumulation of ROS, or (iii) accumulation of the metabolites succinate or fumarate resulting from loss-of-function mutations in the TCA cycle enzymes succinate dehydrogenase (SDH) or fumarate hydratase (FH) (51). This final example will be revisited later.
Myc
In normal cells, the transcription factor c-Myc (Myc) is important in the regulation of cell growth and proliferation, and is activated downstream of growth factor-mediated signaling (52). In several tumors however, Myc is a proto-oncogene that is aberrantly activated by gene amplification, single nucleotide polymorphisms, chromosomal translocations (53), or perhaps as a downstream consequence of mTORC1 hyperactivity. Like HIF-1, Myc stimulates enhanced expression of many genes involved in glucose uptake, glycolysis, and the fate of glycolytic pyruvate (LDHA). Myc also targets genes that support the proliferative utilization of glutamine, including glutamine transporters, and genes involved in both mitochondrial biogenesis and glutaminolysis (23). In fact, Myc-transformed cells undergo apoptosis in the absence of exogenous glutamine, which is a critical carbon source for anaplerosis in these cells (54–56). Moreover, Myc also induces the expression of enzymes in other anabolic pathways, such as serine hydroxymethyltransferase (SHMT) (serine/glycine metabolism) and fatty acid synthase (FAS) (lipid biosynthesis) (57).
SREBP
SREBP-1 is a member of the SREBP family of transcription factors, and induces the expression of several genes involved in fatty acid and sterol biosynthesis in response to growth factors or intracellular sterol levels (58). Recent studies have shown that SREBP-1 is also a downstream effector of mTORC1 (59, 60), thereby affording mTORC1 with an additional mode for regulating cell growth. Consequently, hyperactivation of mTORC1 further potentiates the deregulation of de novo lipid synthesis necessary for sustained membrane production and cell proliferation.
p53
The p53 transcription factor is one of the most vital defenders in the cellular response to a suite of stresses that may otherwise initiate tumorigenic progression. Activation of p53 induces several pathways that impart anticancer mechanisms, including DNA repair, cell cycle arrest, and apoptosis (61). Accordingly, p53 is an especially prominent tumor suppressor, with an estimated 50% of all human cancers harboring either a mutation or deletion in the TP53 encoding gene. More recently, several lines of evidence have uncovered a multifaceted role for p53 in metabolic control as well (62). Given the antitumor regulatory role imparted by p53 in a myriad of other cellular processes, it is unsurprising that p53 also directs metabolic characteristics consistent with those of normal resting cells. Namely, this influence lies in affecting glucose metabolism through repression of glycolysis and concomitant stimulation of oxidative phosphorylation.
For example, p53 transcriptionally induces both synthesis of cytochrome oxidase 2 (SCO2) (63) and TP53-induced glycolysis and apoptosis regulator (TIGAR) (64), while it represses expression of various glucose transporters (65), the glycolytic enzyme phosphoglycerate mutase (66), and PDK-2 (67) – a functionally equivalent isozyme of PDK-1. SCO2 is required for correct assembly of the cytochrome c oxidase complex (COX) in the mitochondrial electron transport chain, while TIGAR is a negative regulator of the glycolytic enzyme phosphofructokinase-1 (PFK1), and thus drives glucose flux through the ox-PPP for production of ROS-titrating NADPH. Further, p53 may remarkably affect glucose metabolism in a transcription-independent manner as well, through its direct binding and inhibition of glucose-6-phosphate dehydrogenase (G6PDH) in the cytoplasm (68). G6PDH catalyzes the first (and rate-limiting) step in the diversion of glycolytic glucose-6-phosphate to the ox-PPP. Therefore, its inactivation could contribute to the dampening of biosynthetic programs, owing to reduction of R5P (nucleotide biosynthesis) and NADPH (lipid biosynthesis) levels. Indeed, p53-deficient cells exhibited an elevated flux of glucose into the ox-PPP, along with increases in both NADPH levels and lipogenic rates, relative to cells expressing wild-type p53 (68). Nonetheless, it is clear that the metabolic control imparted by p53 is likely context-dependent and will require additional investigation given the divergent effects of TIGAR stimulation and G6PDH inhibition in relation to glucose flux. Finally, it is worth noting that p53 also functions in a positive feedback loop with AMPK, whereby p53 can transcriptionally enhance the AMPK-dependent cellular stress response, while AMPK can catalyze the activation-initiating phosphorylation of p53 (69, 70). This p53-AMPK loop can accordingly stimulate catabolic pathways (e.g. fatty acid oxidation), and – through AMPK-mediated effects on mTORC1 – suppress cell growth.
General therapeutic targeting of the proliferative metabolic program in cancer
Given the recognition that metabolic reprogramming occurs broadly across cancer, the question of whether facets of tumor metabolism can be therapeutically targeted has garnered significant attention and investigation. As this matter has been addressed and given excellent detailed commentaries in a number of recent reviews (19, 20, 71), we will describe a few key issues here and again briefly revisit the topic in later sections.
One crucial consideration in the development of anticancer therapeutics is to what extent a given drug can achieve its intended mechanism of action without additionally exerting an unacceptable toxicity onto normal cells. The establishment of this “therapeutic window” is especially challenging in the context of targeting various metabolic adaptations that support rapid cell growth because of their relative conservation in properly regulated proliferating cells.
Nonetheless, some of the first chemotherapeutic agents developed – nucleoside analogues referred to as antimetabolites – target nucleotide biosynthesis through the direct inhibition of enzymes used in DNA synthesis (71), and have remained a common and effective component of treatment regimens administered across several types of cancer. However, because their targets are not exclusive to tumor cells, these compounds have somewhat expectedly been linked to toxicities that stem from the unintended effects they exert on non-malignant proliferating cells.
Another therapeutic opportunity that has been explored is the small molecule inhibition of key enzymes involved in metabolic pathways such as glycolysis and fatty acid synthesis. In most cases, the potential for this strategy has stemmed from preclinical studies in which tumor cell proliferation was negatively affected by small molecule or RNAi-mediated knockdown of a particular target enzyme either in vitro or in xenograft models (19). It remains to be determined in most cases whether the potential yielded from these results will remain durable upon downstream evaluation of therapeutic windows that are attainable from the inhibition of enzymes that are likely of similar importance in normal proliferating cells. For example, the glucose analog 2-deoxyglucose (2DG) is a glycolytic inhibitor that has been previously tested as an anticancer agent in phase I clinical trials. Although sufficient amounts of 2DG can potentiate cancer cell arrest and/or death by limiting glucose catabolism, the dosing necessary to achieve such effects in patients resulted in adverse toxicity (19).
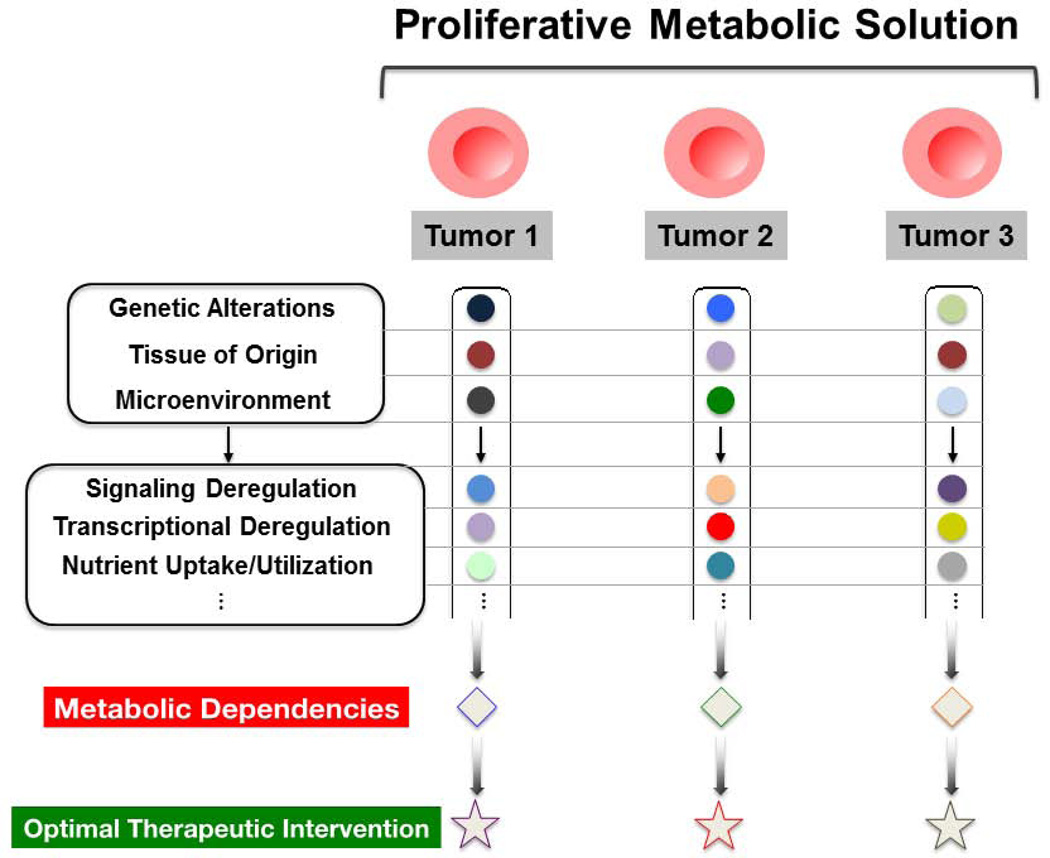
DECIPHERING METABOLIC HETEROGENEITY
In line with the complexity that differentiates cancer into a largely heterogeneous collection of diseases, it has become clear that a single metabolic program cannot be used to globally define altered tumor metabolism. Instead, though rapid cell growth is a general neoplastic feature, variability within the underlying metabolic rewiring of a given cancer cell can dictate not only how proliferative adaptations are driven, but can also confer heterogeneity in the metabolic dependencies of the cell. This diversity may in part clarify why some small molecule compounds, such as antimetabolites and mTOR inhibitors (19, 71), don’t elicit a more general therapeutic efficacy despite the requirement of all malignant cells to promote elevated nucleotide biosynthesis, or the prevalence of mTORC1 hyperactivity in many tumor types.
Ultimately, efforts to unravel metabolic heterogeneity and flexibility may enable the identification of novel therapeutic targets, and as importantly, reveal the metabolic signatures in which intervention through a specific target is most beneficial in terms of potentiating tumor cell death while affording the maximal therapeutic window.
Though cell proliferation is predominantly fueled by glucose and glutamine, the precise extent of their uptake and utilization can vary substantially across different tumors. While ATP generation is broadly attributed to mitochondrial respiration in cancer cells for instance, both cell type and conditional context can affect glycolytic contributions to ATP production by up to two orders of magnitude (72). Tumors also exhibit variable uptake of glutamine and other metabolites such as 18F-labeled amino acid analogues, and display differential levels of lactate secretion (16, 24, 73, 74).
Toward this end, it was recently reported that exogenous glutamine dependence varies across different breast tumor subtypes, as determined by lineage-specific expression of the enzyme glutamine synthetase (encoded by the gene GLUL) (75), which catalyzes the synthesis of intracellular glutamine from glutamate. Another study revealed that glucose-fueled anaplerosis may be the preferred means of replenishing TCA cycle intermediates in some glutamine-independent cancer cell lines, and can also function as a compensatory anaplerotic mechanism in glutaminase-suppressed glutamine-addicted cell lines (76). This glucose-dependent maintenance of the TCA cycle was mediated by the enzyme pyruvate carboxylase (PC), which catalyzes the carboxylation of pyruvate to OAA, and it was those glutamine-addicted cell lines that displayed elevated PC activity that could sustain growth when glutamine-dependent anaplerosis was not an option. Thus, it is becoming clearer that variability exists across different cancers in terms of the glycolytic and glutaminolytic contributions to malignant proliferation, and moreover, that tumors may harbor some extent of metabolic flexibility that allows for the utilization of different anaplerotic precursors or metabolic platforms as a means of dynamic adaptation under stress. Such adaptive strategies could therefore also be considered in the development and optimization of therapies targeting tumor metabolism.
Heterogeneities also exist in the regulation of lipid metabolism in transformed cells. While glucose and glutamine serve as the primary catabolic substrates of proliferative metabolism, fatty acid oxidation (FAO) can also be used as an additional (or alternative) energy source in some leukemia cells and lung tumors (77, 78). In addition, monoacylglycerol lipase (MAGL)-catalyzed hydrolysis of monoacylglycerols may play a role in providing a source of free fatty acids in some higher-grade tumors (79), which would expand the source of free fatty acids in these cells beyond the more typically attributed de novo fatty acid synthesis pathway. Lipidomics analysis further suggested that elevated MAGL activity was particularly responsible for the increased production of specific lipid messengers, such as lysophosphatidic acid (LPA) and prostaglandin E (PGE2) (79), which have been shown to promote tumor cell aggressiveness (80, 81). This result suggests that the deregulation of lipid metabolism extends beyond the commonly noted elevation of lipogenic enzyme levels (ACL, ACC, FAS) (82), and offers the possibility that distinct mechanisms of fatty acid production feed into different downstream pathways.
Metabolic enzymes as oncogenes or tumor suppressors
Glycolytic and oxidative ATP contributions can vary widely between different cancer cells, but mitochondrial activity is generally preserved. Nonetheless, there are examples for which mutations in TCA cycle enzymes have been identified in specific cancers, and these lesions potentiate tumorigenic insults whose mechanisms are now becoming better understood (FIGURE 4A). Loss-of-function germline mutations in SDH and FH have been recognized for over ten years to occur in certain cases of: (i) paraganglioma and pheochromocytoma (SDH), and (ii) leiomyoma and certain cases of renal cell carcinoma (FH) (reviewed in (83)). Ensuing studies revealed that HIF-1 levels were elevated in SDH- and FH-deficient tumors, and could thus elicit transcriptional activation of genes promoting fermentative glucose metabolism even under normoxic conditions. The mechanism underlying this effect was ultimately linked to the accumulation of succinate or fumarate stemming from inactivation of SDH or FH, respectively. Both succinate and fumarate were found to enable aberrant stabilization of HIF-1α through competitive inhibition of prolyl hydroxylase 2 (PHD2) (or PHD3) (84–86), which otherwise suppresses this stabilization under normoxic conditions by marking HIF-1 for VHL-mediated ubiquitination. Therefore, in the cancer types noted above, SDH and FH can genetically act as tumor suppressors.
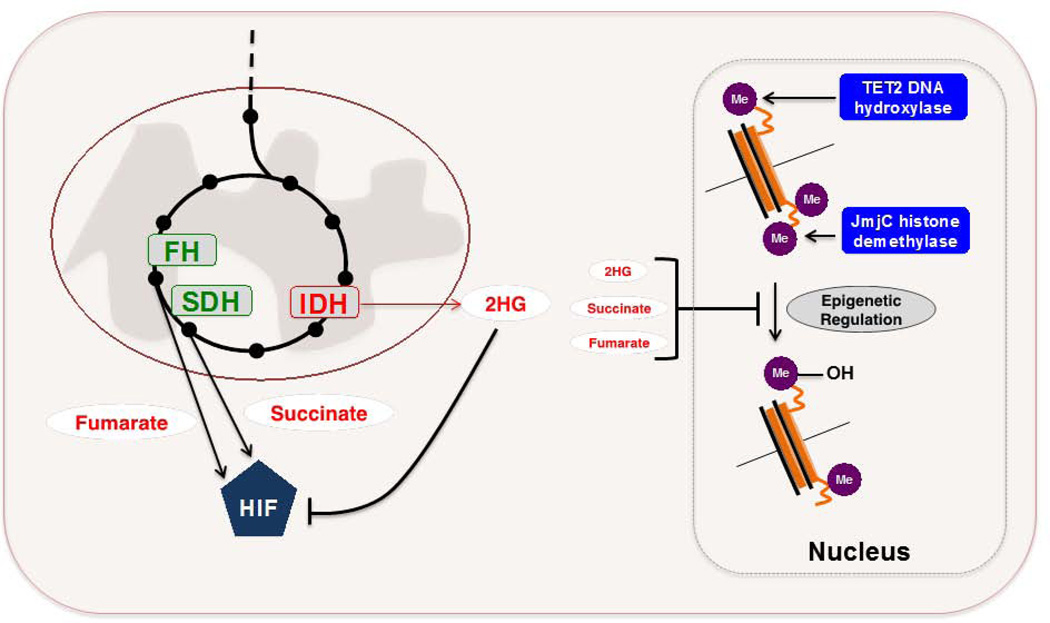
FIGURE 4. Metabolic enzymes as oncogenes or tumor suppressors.
A. SDH and FH can genetically behave as tumor suppressors in specific cancers. The accumulation of succinate or fumarate that arises owing to inactivating mutations in SDH or FH potentiates aberrant stabilization of HIF1 through competitive inhibition of PHDs. IDH mutants arise in a fraction of gliomas, acute myeloid leukemias, and chondrosarcomas. These mutants acquire a neomorphic enzymatic activity that enables the conversion of αKG to 2HG, which can impair normal epigenetic regulation through competitive inhibition of various αKG-dependent dioxygenases, including TET2 DNA hydroxylases and JmjC histone demethylases. Recent evidence suggests that 2HG can also promote HIF1 degradation, and that both succinate and fumarate accumulation may also inhibit various αKG-dependent dioxygenases. Further investigation into the pathophysiological role of 2HG may reveal a context-dependence to its functional role.
SDH – succinate dehydrogenase. FH – fumarate hydratase. HIF – hypoxia-inducible factor. PHD – prolyl hydroxylase. αKG – α – ketoglutarate. 2HG – 2-hydroxyglutarate.
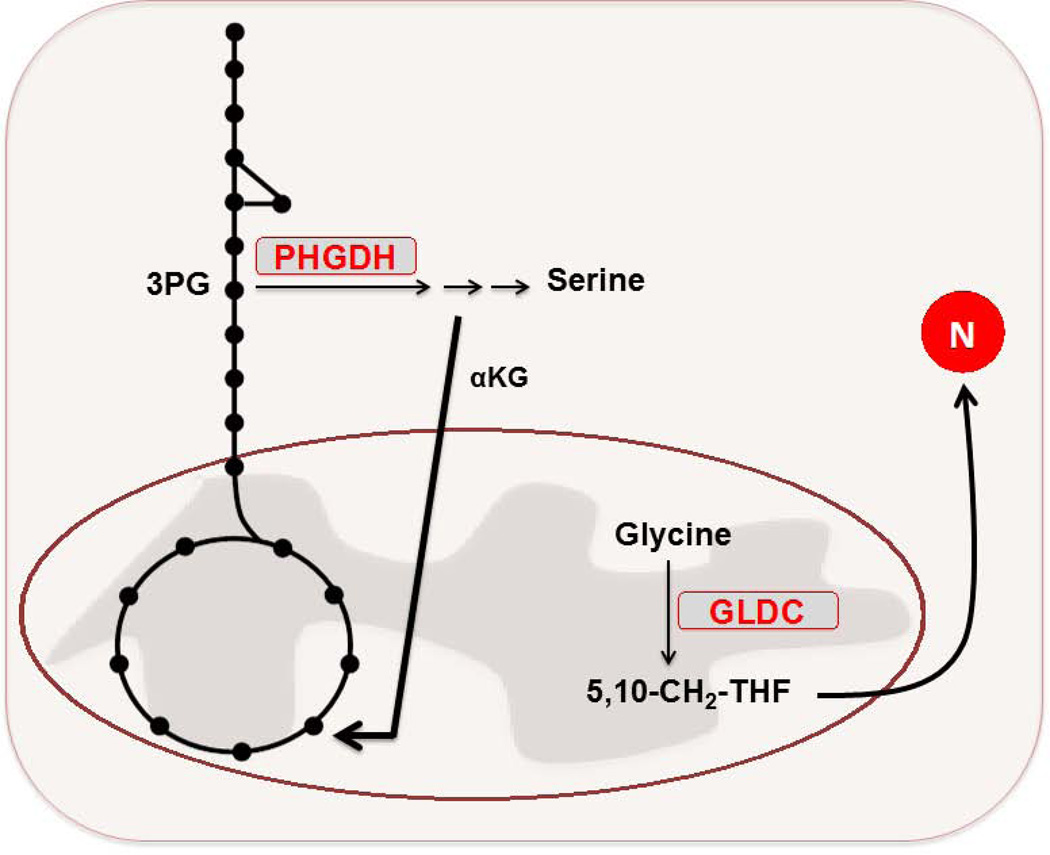
B. PHGDH is elevated in a fraction of malignant breast and melanoma cells. This elevation promotes flux of glucose into the serine biosynthesis pathway. Suppression of PHGDH in those cell lines that had elevated expression of the enzyme caused a strong decrease in cell proliferation and serine synthesis. Moreover, it was revealed that the serine pathway was responsible for nearly 50% of the net conversion of glutamate to αKG for glutamine-driven anaplerosis in these PHGDH-overexpressing cells. GLDC is overexpressed in the TIC population of NSCLC cells. Suppression of GLDC effectively reduced proliferation in the TICs. Among the alterations driven by enhanced GLDC expression in these cells was an increase in pyrimidine biosynthesis, which made these cells particularly sensitive to treatment with low doses of the antimetabolite methotrexate.
PHGDH – 3-phosphoglycerate dehydrogenase. GLDC – glycine decarboxylase. NSCLC – non-small cell lung cancer. TIC – tumor-initiating cell.
More recently, whole-genome sequencing efforts led to the identification of recurrent mutations in isocitrate dehydrogenase 1 (IDH1) and IDH2 in a fraction of gliomas, acute myeloid leukemias, and chondrosarcomas (87–89). However, unlike the SDH and FH examples above, IDH mutations are somatically acquired and don’t strictly abolish the wild-type function of IDHs, which normally catalyze the decarboxylation of isocitrate to αKG. Instead, IDH mutants acquire the neomorphic ability to reduce αKG to 2-hydroxyglutarate (2HG) – a metabolite that is present only at trace levels under normal conditions (90). Collective evidence stemming from several reports have highlighted that the pathophysiological role of 2HG is, at least in part, the competitive inhibition of various αKG-dependent dioxygenases, including TET2 DNA hydroxylases and JmjC histone demethylases (91, 92). Therefore, the tumorigenic effect linked to IDH mutations appears in part to be at the level of epigenetic deregulation. For example, tumor-associated IDH mutations are sufficient to block the differentiation of non-transformed cells through 2HG-mediated inhibition of histone demethylation (93). Further, the introduction of mutant IDH1 into primary human astrocytes can trigger remodeling of the DNA methylome in a fashion that recapitulates patterns similarly observed in mutant IDH-harboring glioma cells (94). Similarly, conditional knock-in mice in which the most common IDH1 mutant (R132H) was expressed in myeloid-lineage hematopoietic cells, showed among a number of alterations, changes in DNA methylation similar to those seen in IDH-mutant AML (95). Interestingly, recent evidence has further suggested that the accumulation of both succinate and fumarate that arise in SDH- and FH-deficient tumors respectively (as described above), may also inhibit various αKG-dependent dioxygenases, resulting in analogous deregulation of epigenetic modifications (96). An additional report described an enantiomer-specific mechanism by which (R)-2HG, but not (S)-2HG, stimulated EGLN prolyl hydroxylase activity and consequent HIF-1α degradation (97). (R)-2HG-mediated elevation of EGLN activity promoted the growth and proliferation of human astrocytes, and this result may explain why IDH-mutated adult tumors seem to select for the (R)-2HG enantiomer. At this point, it appears that 2HG-mediated effector functions are complex and seemingly extend beyond induced deregulation of epigenetic signatures. Further investigation into the mechanisms potentiated by SDH, FH, and IDH1/2 mutations may uncover a context-dependence for 2HG function, inhibition of various αKG-dependent dioxygenases, and the role of HIF-1α stabilization in these particular tumors. Nonetheless, 2HG may serve as a useful biomarker in disease monitoring, and the development of small molecule inhibitors specific to mutant IDH1/2 could be an attractive treatment strategy given the potential therapeutic window afforded by tumor-specific expression of the neomorphic enzymes.
The recognition that metabolic enzymes can genetically behave as tumor suppressors (SDH, FH) or oncogenes (IDH1/2) in certain cancer subsets has been extended over the past year to include two additional examples in which tumorigenic effects are linked to genome-level alteration of a metabolic gene (FIGURE 4B).
A fraction of malignant breast and melanoma cells are dependent on elevated expression of phosphoglycerate dehydrogenase (PHGDH), which catalyzes the conversion of 3-phosphoglycerate to 3 phosphohydroxypyruvate in the first step of the serine biosynthesis pathway (98, 99). The PHGDH gene is commonly amplified within a number of these tumors, leading to elevated PHGDH protein expression. Increases in PHGDH expression at both the mRNA and protein levels were observed in nearly 70% of estrogen receptor (ER)-negative breast cancers, despite the lack of PHGDH amplification among a fraction of cell lines within this subtype (99). Cell lines that exhibited PHGDH overexpression displayed increased flux of glucose carbon through the serine biosynthesis pathway branching from glycolysis (98, 99). Further, RNAi-mediated suppression of PHGDH in cell lines designated as having increased levels of the enzyme, but not in those lacking such status, resulted in a marked decrease in cell growth and reduced serine synthesis. Strikingly, suppression of PHGDH in the knockdown-sensitive breast cell lines did not affect intracellular serine levels, but rather, caused a reduction in αKG levels (99). In fact, the serine pathway was responsible for nearly 50% of the net conversion of glutamate to αKG for glutamine-driven anaplerosis in PHGDH-overexpressing cells. The results illustrated by these studies suggest a potential utility in the therapeutic inhibition of PHGDH in those cancers that express elevated levels of the enzyme.
Another recent report described the identification of a molecular signature for the tumor initiating cells (TICs) of non-small lung cancer (NSCLC) (100). Among the signature features described was overexpression of glycine decarboxylase (GLDC) – a component of the glycine cleavage system – relative to NSCLC cells lacking the TIC-status marker specified in the study, and thus demonstrates an example of intratumor metabolic heterogeneity. The RNAi-mediated knockdown of GLDC effectively diminished proliferation and tumorigenicity in the lung cancer TICs, whereas analogous effects were not observed upon similar treatment in normal human lung fibroblasts. Moreover, GLDC overexpression was identified across a variety of other tumor types, and remarkably, overexpression of active GLDC alone could induce transformation of 3T3 cells in vitro and promote tumor formation from these same cells in vivo. Metabolomics profiling revealed that among the alterations driven by enhanced GLDC expression was an increase in pyrimidine biosynthesis – an effect that imparted elevated sensitivity of GLDC-overexpressing cells to treatment with low doses of methotrexate, with minimal effects seen in control cells. Perhaps additional investigation will determine whether GLDC overexpression can be more broadly used as a predictor of antifolate sensitivity, thus informing for contexts in which these chemotherapeutics may enjoy a greater therapeutic window. The methotrexate result further suggests a possible route of increased therapeutic efficacy through the synergistic use of antifolates with a GLDC inhibitor for treatment of relevant GLDC-overexpressing tumors.
Systemic depletion of tumor-essential amino acids
Altered tumor metabolism need not consist entirely of changes or adaptations that are driven to satisfy the demands of cell growth and proliferation. Instead, metabolic rewiring during tumorigenesis may also result in the development of specific metabolic liabilities that, while likely not selected for nor acting to promote proliferation, still arise as additional dependencies that must be met to maintain cell survival. In particular, several tumor types are auxotrophic for one or more amino acids owing to deficiencies in a corresponding endogenous biosynthesis or salvage pathway (101). Consequently, these cells have a particular reliance on the import of a given amino acid(s) from the extracellular serum pool. Extensive in vitro, in vivo, and clinical evaluations conducted over the past sixty years have established that this type of metabolic liability can be exploited through enzyme-mediated systemic depletion of a circulating “tumor-essential” amino acid, resulting in the selective starvation and death of targeted auxotrophic malignant cells with minimal effect on normal cells (101).
The most notable example of this strategy in a therapeutic setting is certainly the success of L-Asparaginase in the treatment of acute lymphoblastic leukemia (ALL) (102). Whereas normal cells can endogenously synthesize the non-essential amino acid L-asparagine (L-Asn) through the catalytic action of asparagine synthetase (ASNS), certain ALL lymphoblasts lack or express very low levels of ASNS and therefore require uptake of serum L-Asn (103–105). The enzyme L-Asparaginase catalyzes the hydrolysis of L-Asn to L-aspartate (L-Asp) and ammonia, resulting in depletion of ALL-essential L-Asn from the extracellular pool, which subsequently induces selective apoptosis of the auxotrophic lymphoblasts (106, 107). Though initially evaluated for clinical efficacy as a single agent, Escherichia coli L-Asparaginase II (EcAII) has since become a standard component in a combination chemotherapy regimen that now provides a remarkable survival rate in pediatric ALL, but still garners further optimization for the treatment of adult ALL (108, 109). While EcAII is currently approved for use in the front-line treatment of ALL, other cancer types (e.g. certain adult non-Hodgkin’s lymphomas (110) and ovarian carcinomas (111)) have since been shown to have a similar dependence on the uptake of serum L-Asn as well. EcAII can also hydrolyze glutamine, though at efficiencies considerably lower than those observed for L-Asn deamidation (112). However, the clinical implications and utility of EcAII-mediated glutamine degradation remain unclear. While glutamine depletion could enhance the efficacy of EcAII given the critical role of glutamine as a proliferative fuel, others have suggested that glutamine hydrolysis may instead contribute to clinical toxicities associated with EcAII administration (113).
It is worth noting that while ASNS transcript level has acted as the historical predictor of ALL sensitivity to EcAII treatment, a number of studies over the past decade have alternatively demonstrated that additional ASNS-independent genetic and metabolic determinants may also contribute to EcAII efficacy (114–116). One example is described through the correlation between ASNS expression and asparaginase sensitivity in patients either harboring or lacking the t(12;21)(p13;q22) chromosomal translocation, which results in expression of the TEL-AML1 fusion protein. This chromosomal alteration occurs in approximately 25% of pediatric B-cell lineage ALL cases (117). Although ASNS expression was expectedly correlated with asparaginase sensitivity in TEL-AML1-negative patients, a similar pattern was absent in the converse TEL-AML1-positive cohort (118). Strikingly, while TEL-AML1-positive patients were generally more sensitive to asparginase treatment, one report showed that samples isolated from these patients exhibited nearly 5-fold greater levels of ASNS mRNA relative to TEL-AML1-negative patients and healthy controls (119). Therefore, while native EcAII received FDA approval nearly forty years ago, the mechanisms dictating its efficacy are still not entirely understood.
Another emergent example that highlights the therapeutic exploitation of a tumor-specific amino acid dependency stems from the recognition that a large fraction of hepatocellular carcinomas, metastatic melanomas, and renal cell carcinomas express low levels of the enzyme argininosuccinate synthetase (ASS) or harbor other urea cycle defects that render the host malignant cells auxotrophic for L-arginine (L-Arg) (120). Two arginine-degrading enzymes (bacterial arginine deimidase (ADI) and human arginase I) have since been evaluated in various xenograft models and/or early phase clinical trials for their ability to inflict selective toxicity upon urea cycle-defective tumors, with promising results to date (101, 121). The systemic depletion of L-Arg as a therapeutic strategy is of particular interest because the prognoses for malignant melanomas and hepatocellular carcinomas are generally poor, given the difficulty in treating these aggressive tumors with conventional chemotherapeutics (120).
Unraveling selective expression of metabolic enzymes
It is well recognized that many metabolic reactions can be effectively catalyzed by multiple, distinct members of a given enzyme family, albeit with some level of variability in kinetic efficiency. Moreover, members of the same enzyme family can possess other distinguishing molecular or biochemical characteristics that extend beyond catalysis of the otherwise unifying reaction. This diversity arises through either: (i) expression of functionally similar enzymes encoded by different genes, or (ii) post-transcriptional alternative splicing from a single encoding gene. It has become increasingly appreciated that cancer cells preferentially express specific members of a metabolic enzyme family in either a broad or context-dependent manner.
For instance, some cancers such as glioblastoma multiforme (GBM) exhibit increased expression and/or reliance on hexokinase 2 (HK2) (122, 123). Hexokinase catalyzes the phosphorylation of glucose to glucose-6-phosphate in the initiating step of glycolysis. In general, HK2 expression is restricted to skeletal and muscle tissues, while normal brain and lower-grade gliomas for example, predominantly express HK1. Perhaps the elevated expression of HK2 in some tumors is indicative of this particular HK imparting some additional functional role necessary in the context of these malignancies.
Downstream of HK in the glycolytic pathway is the enzyme PFK1, which catalyzes the rate-limiting conversion of fructose-1-phosphate to fructose-1,6-bisphosphate. PFK1 is a target of positive allosteric regulation by the metabolite 2,6-fructose bisphosphate (F2,6P), whose intracellular levels are modulated in part by PFK2. Most members of the PFK2 family have dual kinase and phosphatase activity, and can therefore catalyze the generation or depletion F2,6P, depending on the ratio of the two activities (124). Accordingly, the four identified PFK2 enzymes are more appropriately designated as PFKFB1–4 to reflect the additional F2,6Pase activity. The PFKFB3 isozyme has almost negligible phosphatase activity and can therefore activate PFK1 to promote flux through glycolysis. PFKFB3 is highly expressed in many cancers (125), and its small molecule-mediated inhibition was shown to induce a cytostatic effect in RAS-transformed cells (126).
In contrast to PFKFB3, the PFKFB4 isozyme has a greater phosphatase to kinase activity ratio and can therefore affect a shunt of glucose-6-phosphate flux into the PPP by decreasing PFK1 activity. It was recently reported that PFKFB4 is essential to the survival of certain prostate cancer cell lines because its modulation of F2,6P levels was critical for the maintenance of redox homeostasis in these cells (127). In fact, while PFKFB4 depletion in prostate cancer cell lines induced a large increase in ROS and subsequent cell death, similar PFKFB4 silencing in normal human prostate epithelial cells had little effect on ROS levels and cell growth.
Glutaminase-catalyzed deamidation of glutamine to glutamate is a key step in both glutaminolysis and synthesis of ROS-scavenging glutathione (GSH). There are two predominant human isozymes of glutaminase: kidney-type (GLS1) and liver-type (GLS2) that, despite sharing some functional similarities, are regulated quite differently (128, 129). Whereas Myc specifically potentiates elevated expression of GLS1 and promotes the downstream utilization of glutamine carbon in anaplerosis (55, 56), p53 specifically activates GLS2 to support cellular defense against oxidative stress (130). It is thus likely that GLS1 and GLS2 have different roles in tumorigenesis as well. For example, GLS2 levels are markedly decreased in hepatocellular carcinoma relative to normal liver tissues, while GLS2 overexpression in tumor cells reduced colony formation (130). Moreover, an in vivo model of Myc-transformed hepatocellular carcinoma demonstrated that progression from the pretumor to tumor state was marked with changes in the expression of several genes, including a considerable downregulation of GLS2 and substantial upregulation of GLS1 (131). Additional lines of evidence have further described a link between GLS1 inhibition and the growth suppression of various cancer cells (e.g. lymphoma, breast) as well (132–134). The sum of these results suggest that selective inhibition of GLS1 may provide a tractable treatment approach (135), though the corresponding attainable therapeutic window will likely be context-dependent.
The most extensively studied example of selective isoform expression described to date focuses on the enzyme pyruvate kinase (PK), which catalyzes the final irreversible step of glycolysis – conversion of phophoenolpyruvate (PEP) to pyruvate with concomitant generation of ATP. There are four members of the PK family in mammals: PKL and PKR are splice variant isoforms encoded by the PK-LR gene, and PKM1 and PKM2 are splice variant isoforms encoded by the PK-M gene. Products of the PK-LR gene are normally restricted to expression in the liver (PKL) and red blood cells (PKR). The M1 and M2 isoforms differ by a single exon and share ~96% sequence identify at the amino acid level. Nonetheless, PKM2 is the major PK isoform expressed in an essentially universal fashion across all proliferating cells evaluated to date (30, 136). Moreover, PKM2-expressing cells show a selective growth advantage relative to PKM1-expressing cells as evaluated in xenograft models (137), despite the seeming paradox that PKM2 possesses lower specific PK activity relative to PKM1 (138). However, whereas PKM1 is constitutively active, PKM2 can be negatively regulated through its binding to tyrosine-phosphorylated proteins downstream of cellular kinase signaling (139). One prominent hypothesis for rationalizing the selective expression of PKM2 in proliferating cells is that its propensity for modulation enables cellular tuning of glycolytic intermediate flux into branching anabolic pathways rather than to pyruvate (24). It was further reported recently that elevated levels of ROS could inactivate PKM2 in a human lung cancer cell line (140). This additional regulatory mechanism could promote diversion of glycolytic flux into the PPP as a means to generate NADPH for combatting oxidative stress. Remarkably, PKM2-expressing cells also paradoxically generate more glucose-derived lactate relative to cells expressing PKM1 (137). A potential resolution to this observation may lie in a recently described alternative glycolytic pathway in which PEP conversion to pyruvate could be catalyzed without the corresponding generation of ATP that marks the PK-mediated catalysis of this conversion otherwise (138).
One complicating factor in further understanding the selection for PKM2 in proliferating cells stems from a recent study, which while confirming PKM2 as the major PK expressed in primary tumor tissues and cell lines, further offered evidence suggesting that PKM2 was also the predominant isoform expressed in matched control tissues and normal cell lines (141). This result presents disparity to a commonly cited model that proposes a PK isoform “switch”, whereby normal differentiated cells specifically select for PKM1 expression, but shift to expression of PKM2 upon rewiring to a proliferative metabolic program (136). It is worth noting however, that while PKM2 is not necessarily selected for specifically in proliferating cells, the expression of PKM1 in such cells is essentially absent or detected at only very low levels (141).
PKM2 may have roles beyond that of its regulated PK activity. In particular, a number of non-metabolic activities mediated by PKM2 upon translocation to the nucleus have been described, including the transactivation of β-catenin (142) and HIF1 (143). Moreover, modulation of active tetrameric PKM2 to its inactive dimeric form may impart proliferative advantages beyond those described earlier. The dimeric form of PKM2, which is predominantly nuclear-localized in highly proliferative tumor cells, can remarkably act as a protein kinase that catalyzes phosphorylation of the STAT3 transcription factor, subsequently leading to the elevated expression of cancer-relevant genes such as MEK5 (144). Certainly, much remains to be unraveled in terms of how the glycolytic and non-glycolytic functions PKM2 may be balanced, regulated, and manipulated in both differentiated and proliferating cells. Interestingly, inhibitors and activators of PKM2 are each being explored as therapeutic points of intervention (19).
Non-genetic contributions to tumor metabolism
Altered tumor metabolism is not simply the final outcome of some combination of cell-autonomous genetic alterations. Instead, a non-genetic component in the form of the tumor microenvironment must additionally be considered as a component in the equation that influences metabolic changes in cancer cells (7). Solid tumors are poorly vascularized, and therefore their surrounding environment can subject distinct regions of the tumor to spatial and temporal gradients of oxygenation, pH, and nutrient availability (145). Consequently, metabolic alterations are likely also stimulated to some extent as a means for a given tumor cell to adapt to these dynamic and energetically stressful conditions.
The relationship between genetic and non-genetic determinants in the regulation of tumor cell metabolism appears to be complex and not simply one of cause and effect. For example, fluctuating oxygen gradients across the microenvironment can drive sporadic hypoxia, the stabilization of HIF-1α, and a corresponding induction of the HIF1-induced transcriptional program (50). Regardless of whether HIF-1-induced transcriptional effects are promoted through inappropriate genetic regulation or in response to hypoxic stress, one of its downstream consequences remains the conversion of a large percentage of glycolytic pyruvate to secreted lactate. The secreted lactate in turn triggers additional metabolic responses as a result of local acidification within the tumor microenvironment. It has also been suggested that this lactate-driven acidification can promote both tumor invasion and immune evasion (146, 147), which are among the other denoted hallmarks of cancer (8). Moreover, lactate secretion may have a functional role within a larger system of metabolic cooperation and symbiosis between cells in the microenvironment. Described as a “two-compartment” model of tumor metabolism, the symbiosis is characterized as the potential for anabolic malignant cells to extract high-energy metabolites (e.g. lactate, glutamine, fatty acids) from adjacent catabolic cells (within the tumor or neighboring stromal cells) through a network of nutrient sharing that can stimulate tumor proliferation and metastasis (148, 149). Studies reporting two-compartment tumor metabolism have recently emerged in the context of breast cancer cells and their neighboring fibroblasts (150, 151), as well as for ovarian cancer cells and their neighboring adipocytes (152). A further understanding of this concept may lead to the development of therapies that inhibit this process of energy transfer in the tumor microenvironment, and could demonstrate one example of a means to overcome potential barriers posed by intratumor heterogeneity in targeting cancer cell metabolism.
Non-genetic factors may also have additional roles in affecting energy metabolism. Under hypoxic conditions, several cell lines utilize glutamine carbon as the major source of de novo lipogenesis through the reductive carboxylation of glutamine-derived αKG (153, 154). This IDH-mediated process generates isocitrate, which is then isomerized to citrate, thus effectively redirecting the traditional direction of the TCA cycle. Glutamine-fueled reductive carboxylation of αKG serves a similar role in cells harboring defective mitochondria (155), which would possess a comparable impairment in glucose-dependent lipogenesis. In another study, the interplay between oncogenic alteration and tumor tissue of origin was shown to influence a differential utilization of glucose and glutamine as proliferative fuels (156). While MET-transformed murine liver tumors were marked by increased glucose metabolism, little glutaminolysis and net glutamine synthesis, Myc-induced liver tumors exhibited significant increases in both glucose and glutamine catabolism. Further, Myc-induced lung tumors displayed increases in both glucose and glutamine catabolism, however; similar to MET-driven liver tumors, had a net accumulation of glutamine.
TECHNOLOGICAL ADVANCEMENTS TO STUDY TUMOR METABOLISM
Tumor metabolism is proving to be a general term used to describe a complex collection of diverse metabolic signatures, comprised of various metabolic changes that arise as an overall response to some combination of genetic and non-genetic determinants. This metabolic program must, by definition, satisfy the multiple demands of cell proliferation; however, there is some degree of heterogeneity in the precise collection of metabolic dependencies and liabilities within a given tumor cell.
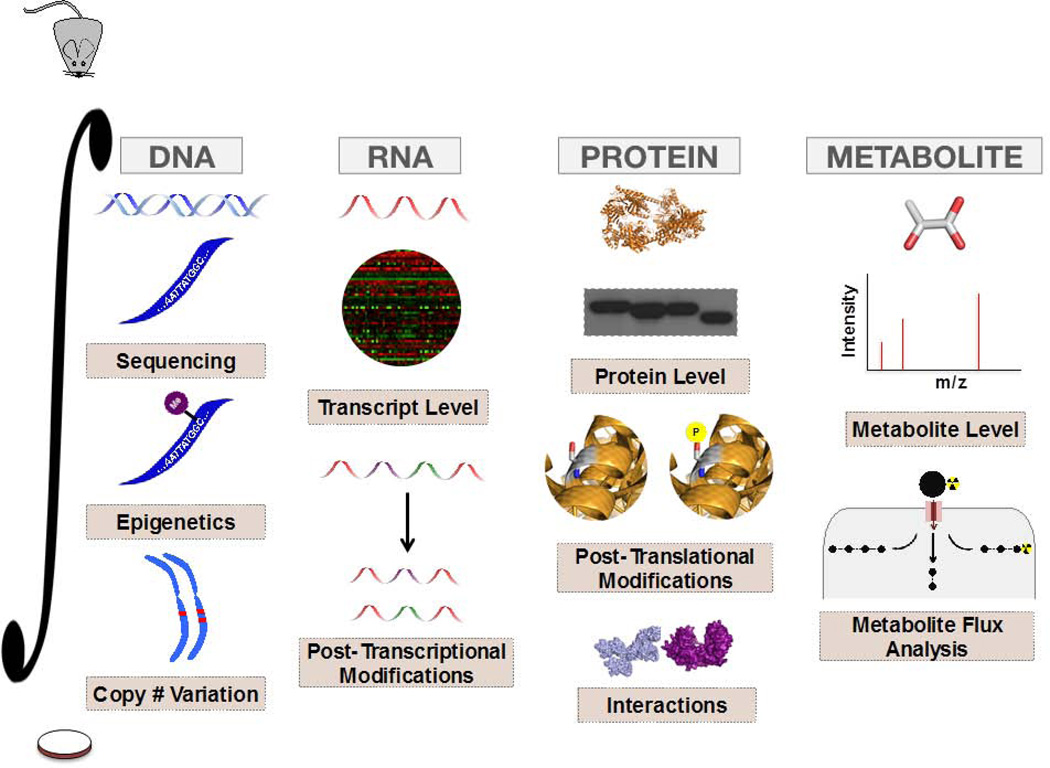
Recognition of this complexity has in large part been aided by the progress of various omics-based strategies over the past decade. Ideally, the continued exploitation and integration (FIGURE 5) of these approaches will result in the identification of novel or context-dependent therapeutic targets that optimally balance drug efficacy and therapeutic window. Moreover, efforts to model tumor metabolism in vivo should better mimic physiological conditions that are met in clinical settings, and may afford the development of new approaches to in vivo metabolic analysis. In addition to several examples cited throughout this review, two recent studies have further demonstrated the power of integrative approaches and in vivo modeling in the study of tumor metabolism going forward.
Figure 5. Application and Integration of Tools to study Tumor Metabolism.
The exploitation and integration of various components of the omics cascade can provide a new depth of insight into the study of tumor metabolism. Moreover, these approaches can be employed not only for the interrogation of cell lines in culture, but can also be incorporated with in vivo systems used to better model human metabolism.
In one example, a pancreatic ductal adenocarcinoma (PDAC) mouse model was utilized and integrated with downstream biochemical, transcriptomic, and metabolomics analyses to determine the mechanisms by which the KRasG12D lesion promoted tumor maintenance within this particular carcinoma (157). It was ultimately determined that KRasG12D had an important role in altering glucose metabolism in PDAC, namely by stimulating glucose uptake and promoting diversion of its flux from glycolysis to branching anabolic pathways.
In the other example, mouse models of genetically diverse primary human glioblastoma (GBM) were each infused with 13C-labeled nutrients to ultimately allow for evaluation of the mechanisms by which these tumors were fueled (158). One finding was that flux through the pyruvate dehydrogenase complex (PDH) was the major source of carbon flux into the TCA cycle, despite a prior proposal that PDH was suppressed in high-grade tumors (159). This observation served to perhaps indicate that differences in metabolism could stem from whether evaluation was conducted using an in vivo system versus in the context of cultured cell lines. Additionally, glucose uptake and oxidation did not appear to vary among GBM tumors with distinct oncogenic driver mutations, further implicating complexities in tumor metabolism that in part lie beyond genomic footprints.
CONLCUSIONS
The collection of advances made in our understanding of tumor metabolism in recent years has not only afforded a better understanding of the metabolic changes that help satisfy proliferative demands, but as critically, has revealed the diversity of mechanistic inputs and context-dependent determinants that can drive metabolic rewiring. Moreover, numerous studies have illustrated that metabolic adaptations can in fact be selected for during transformation as well.
While the general uniformity of altered tumor metabolism lies in the shared ability of neoplasms to induce adaptations that stimulate rapid cell growth, we are now recognizing that the metabolic signature of cancer cells is one marked by the same complexities and diversity that characterize the disease as a whole. Namely, we are now beginning to unravel the heterogeneities that exist within the metabolic program of tumors that arise from different tissues, among different tissue subtypes, and even between cells populating a single tumor.
Our understanding of tumor metabolism continues to evolve as advances in several analytical technologies and modeling strategies are affording the implementation of systems-level and integrated strategies for use in metabolic studies. Ultimately, these efforts will ideally facilitate further progress in capitalizing upon the exploitation of atypical metabolic features in cancer as a means of therapeutic intervention. Deciphering the interplay between genetic and non-genetic components that together contribute to metabolic reprogramming in a given setting, may serve as the critical factor in determining therapeutic targets that enable maximal drug efficacy with minimal deleterious effect on normal cells (FIGURE 6).
Figure 6. Unraveling Metabolic Diversity.
The commonality of metabolic rewiring in tumor cells is that the sum of alterations and adaptations must ultimately provide a means to support the various demands of cell proliferation. However, the proliferative solution arises in an integrated response to some combination of genetic and non-genetic determinants, which in turn, dictate the precise metabolic signature and dependencies of a given tumor cell. A better understanding of this heterogeneity should promote the continued development of therapeutic strategies that best exploit metabolic liabilities while achieving maximal therapeutic windows.
Indeed, altered metabolism appears to constitute a unique hallmark of cancer. However, now we must embrace, dissect, and improve upon our understanding of the variability that exists beneath this encompassing designation.
STATEMENT OF SIGNIFICANCE.
Altered tumor metabolism is now a generally regarded hallmark of cancer. Nevertheless, the recognition of metabolic heterogeneity in cancer is becoming clearer as a result of advancements in several tools used to interrogate metabolic rewiring and dependencies. Deciphering this context-dependent heterogeneity will supplement our current understanding of tumor metabolism and may yield promising therapeutic and diagnostic utilities.
ACKNOWLEDGEMENTS
The authors thank members of the Sabatini lab for helpful discussion, especially William Comb for review of the manuscript. J.R.C. is supported by the American Cancer Society (PF-12-099-01-TBG). D.M.S. is in part supported by the NIH grant CA129105, and is an investigator at the Howard Hughes Medical Institute.
Footnotes
The authors disclose no potential conflicts of interest.
REFERENCES
- 1.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Kelloff GJ, Sigman CC. Cancer biomarkers: selecting the right drug for the right patient. Nat Rev Drug Discov. 2012;11:1–14. doi: 10.1038/nrd3651. [DOI] [PubMed] [Google Scholar]
- 4.Wong KM, Hudson TJ, McPherson JD. Unraveling the Genetics of Cancer: Genome Sequencing and Beyond. Annu Rev Genomics Hum Genet. 2011;12:407–430. doi: 10.1146/annurev-genom-082509-141532. [DOI] [PubMed] [Google Scholar]
- 5.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yap T, Gerlinger M, Futreal P, Pusztai L. Intratumor Heterogeneity: Seeing the Wood for the Trees. Sci Transl Med. 2012;4:1–4. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 7.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 12.Ward PS, Thompson CB. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 14.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 15.Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 16.Groves AM, Win T, Haim SB, Ell PJ. Non-[18F] FDG PET in clinical oncology. Lancet Oncol. 2007;8:822–830. doi: 10.1016/S1470-2045(07)70274-7. [DOI] [PubMed] [Google Scholar]
- 17.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deberardinis RJ, Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:1–14. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 20.Jones NP, Schulze A. Targeting cancer metabolism – aiming at a tumour's sweet-spot. Drug Discov Today. 2012;17:1–10. doi: 10.1016/j.drudis.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 21.Wang R, Green DR. The immune diet: meeting the metabolic demands of lymphocyte activation. F1000 Biol Rep. 2012;4:9. doi: 10.3410/B4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vander Heiden MG, Lunt SY, Dayton TL, Fiske BP, Israelsen WJ, Mattaini KR, et al. Metabolic pathway alterations that support cell proliferation. Cold Spring Harb Symp Quant Biol. 2012;76:325–334. doi: 10.1101/sqb.2012.76.010900. [DOI] [PubMed] [Google Scholar]
- 23.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 24.Lunt SY, Vander Heiden MG. Aerobic Glycolysis: meeting the metabolic eequirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 25.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Locasale JW, Cantley LC. Metabolic Flux and the Regulation of Mammalian Cell Growth. Cell Metab. 2011;14:443–451. doi: 10.1016/j.cmet.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–193450. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cairns RA, Harris I, McCracken S, Mak TW. Cancer cell metabolism. Cold Spring Harb Symp Quant Biol. 2012;76:299–311. doi: 10.1101/sqb.2011.76.012856. [DOI] [PubMed] [Google Scholar]
- 30.Michalek RD, Rathmell JC. The metabolic life and times of a T-cell. Immunol Rev. 2010;236:190–202. doi: 10.1111/j.1600-065X.2010.00911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franke TF, Hornik CP, Segev L, Shostak GA, Sugimoto C. PI3K/Akt and apoptosis: size matters. Oncogene. 2003;22:8983–8998. doi: 10.1038/sj.onc.1207115. [DOI] [PubMed] [Google Scholar]
- 32.Plas DR, Thompson CB. Akt-dependent transformation: there is more to growth than just surviving. Oncogene. 2005;24:7435–7442. doi: 10.1038/sj.onc.1209097. [DOI] [PubMed] [Google Scholar]
- 33.Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell. 2003;4:257–262. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 34.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 35.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohn AD, Summers SA, Birnbaum MJ, Roth RA. Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J Biol Chem. 1996;271:31372–31378. doi: 10.1074/jbc.271.49.31372. [DOI] [PubMed] [Google Scholar]
- 37.Deprez J, Vertommen D, Alessi DR, Hue L, Rider MH. Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J Biol Chem. 1997;272:17269–17275. doi: 10.1074/jbc.272.28.17269. [DOI] [PubMed] [Google Scholar]
- 38.Gottlob K, Majewski N, Kennedy S, Kandel E, Robey RB, Hay N. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev. 2001;15:1406–1418. doi: 10.1101/gad.889901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rathmell JC, Fox CJ, Plas DR, Hammerman PS, Cinalli RM, Thompson CB. Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Mol Cell Biol. 2003;23:7315–7328. doi: 10.1128/MCB.23.20.7315-7328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J Biol Chem. 2002;277:33895–33900. doi: 10.1074/jbc.M204681200. [DOI] [PubMed] [Google Scholar]
- 41.Laplante M, Sabatini DM. mTOR Signaling in Growth Control and Disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 44.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carling D, Zammit VA, Hardie DG. A common bicyclic protein kinase cascade inactivates the regulatory enzymes of fatty acid and cholesterol biosynthesis. FEBS Lett. 1987;223:217–222. doi: 10.1016/0014-5793(87)80292-2. [DOI] [PubMed] [Google Scholar]
- 46.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 47.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vazquez DS, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greer SN, Metcalf JL, Wang Y, Ohh M. The updated biology of hypoxia-inducible factor. EMBO J. 2012;31:2448–2460. doi: 10.1038/emboj.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Semenza GL. Regulation of Metabolism by Hypoxia-Inducible Factor 1. Cold Spring Harb Symp Quant Biol. 2012;76:347–353. doi: 10.1101/sqb.2011.76.010678. [DOI] [PubMed] [Google Scholar]
- 51.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dang CV. Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. 2010;70:859–862. doi: 10.1158/0008-5472.CAN-09-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dang CV, Le A, Gao P. MYC-Induced Cancer Cell Energy Metabolism and Therapeutic Opportunities. Clin Cancer Res. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wise DR, DeBeradinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: Sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eberlé D, Hegarty B, Bossard P, Ferré P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 59.Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 62.Gottlieb E, Vousden KH. p53 Regulation of Metabolic Pathways. Cold Spring Harb Perspect Biol. 2010;2:a001040. doi: 10.1101/cshperspect.a001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 64.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 65.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 66.Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–185. [PubMed] [Google Scholar]
- 67.Contractor T, Harris CR. p53 negatively regulates transcription of the pyruvate dehydrogenase kinase Pdk2. Cancer Res. 2012;72:560–567. doi: 10.1158/0008-5472.CAN-11-1215. [DOI] [PubMed] [Google Scholar]
- 68.Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 70.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 71.Tennant DA, Durán RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 72.Zu XL, Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochem Biophys Res Commun. 2004;313:459–465. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 73.Ben-Haim S, Ell P. 18F-FDG PET and PET/CT in the evaluation of cancer treatment response. J Nucl Med. 2009;50:88–99. doi: 10.2967/jnumed.108.054205. [DOI] [PubMed] [Google Scholar]
- 74.Tessem MB, Swanson MG, Keshari KR, Albers MJ, Joun D, Tabatabai ZL, et al. Evaluation of lactate and alanine as metabolic biomarkers of prostate cancer using 1H HR-MAS spectroscopy of biopsy tissues. Magn Reson Med. 2008;60:510–516. doi: 10.1002/mrm.21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kung H-N, Marks JR, Chi J-T. Glutamine synthetase is a genetic determinant of cell type–specific glutamine independence in breast epithelia. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002229. e1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng T, Sudderth J, Yang C, Mullen AR, Jin ES, Mates JM, et al. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc Natl Acad Sci U S A. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest. 2010;120:142–156. doi: 10.1172/JCI38942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zaugg K, Yao Y, Reilly PT, Kannan K, Kiarash R, Mason J, et al. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011;25:1041–1051. doi: 10.1101/gad.1987211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nomura DK, Lombardi DP, Chang JW, Niessen S, Ward AM, Long JZ, et al. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chem Biol. 2011;18:846–856. doi: 10.1016/j.chembiol.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nature Rev Cancer. 2003;3:582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 81.Gupta GP, Nguyen DX, Chiang AC, Bos PD, Kim JY, Nadal C, et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 82.DeBerardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. 2005;5:857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 84.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 85.Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–153. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 86.Lee S, Nakamura E, Yang H, Wei W, Linggi MS, Sajan MP, et al. Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromocytoma genes: developmental culling and cancer. Cancer Cell. 2005;8:155–167. doi: 10.1016/j.ccr.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 87.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 90.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brüstle A, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012 doi: 10.1038/nature11323. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsory K. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 101.Cantor JR, Panayiotou V, Agnello G, Georgiou G, Stone EM. Engineering reduced-immunogenicity enzymes for amino acid depletion therapy in cancer. Methods Enzymol. 2012;502:291–319. doi: 10.1016/B978-0-12-416039-2.00015-X. (2012) [DOI] [PubMed] [Google Scholar]
- 102.Rytting M. Peg-asparaginase for acute lymphoblastic leukemia. Expert Opin Biol Ther. 2010;10:833–839. doi: 10.1517/14712591003769808. [DOI] [PubMed] [Google Scholar]
- 103.Horowitz B, Madras BK, Meister A, Old LJ, Boyes EA, Stockert E. Asparagine synthetase activity of mouse leukemias. Science. 1968;160:533–535. doi: 10.1126/science.160.3827.533. [DOI] [PubMed] [Google Scholar]
- 104.Haskell CM, Canellos GP. l-asparaginase resistance in human leukemia--asparagine synthetase. Biochem Pharmacol. 1969;18:2578–2580. doi: 10.1016/0006-2952(69)90375-x. [DOI] [PubMed] [Google Scholar]
- 105.Cooney DA, Handschumacher RE. L-asparaginase and L-asparagine metabolism. Annu Rev Pharmacol. 1970;10:421–440. doi: 10.1146/annurev.pa.10.040170.002225. [DOI] [PubMed] [Google Scholar]
- 106.Story MD, Voehringer DW, Stephens LC, Meyn RE. L-asparaginase kills lymphoma cells by apoptosis. Cancer Chemother Pharmacol. 1993;32:129–133. doi: 10.1007/BF00685615. [DOI] [PubMed] [Google Scholar]
- 107.Ueno T, Ohtawa K, Mitsui K, Kodera Y, Hiroto M, Matsushima A. Cell cycle arrest and apoptosis of leukemia cells induced by L-asparaginase. Leukemia. 1997;11:1858–1861. doi: 10.1038/sj.leu.2400834. [DOI] [PubMed] [Google Scholar]
- 108.Avramis VI, Tiwari PN. Asparaginase (native ASNase or pegylated ASNase) in the treatment of acute lymphoblastic leukemia. Int J Nanomedicine. 2006;1:241–254. [PMC free article] [PubMed] [Google Scholar]
- 109.Pui C-H, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 110.Winter SS, Holdsworth MT, Devidas M, Raisch DW, Chauvenet A, Ravindranath Y, et al. Antimetabolite-based therapy in childhood T-cell acute lymphoblastic leukemia: a report of POG study 9296. Pediatr Blood Cancer. 2006;46:179–186. doi: 10.1002/pbc.20429. [DOI] [PubMed] [Google Scholar]
- 111.Lorenzi PL, Llamas J, Gunsior M, Ozbun L, Reinhold WC, Varma S, et al. Asparagine synthetase is a predictive biomarker of L-asparaginase activity in ovarian cancer cell lines. Mol Cancer Ther. 2008;7:3123–3128. doi: 10.1158/1535-7163.MCT-08-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kurtzberg J. Asparaginase. In: Bast R, Kufe D, Pollock R, Weichselbaum R, Holland J, Frei E, editors. Cancer Medicine. 5th Edition. Hamilton (ON): BC Decker; 2000. pp. 699–705. [Google Scholar]
- 113.Derst C, Henseling J, Röhm KH. Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase activity by amino acid replacements at position 248. Protein Sci. 2000;9:2009–2017. doi: 10.1110/ps.9.10.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Iwamoto S, Mihara K, Downing JR, Pui C-H, Campana D. Mesenchymal cells regulate the response of acute lymphoblastic leukemia cells to asparaginase. J Clin Invest. 2007;117:1049–1057. doi: 10.1172/JCI30235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Su N, Pan Y, Zhou M, Harvey R, Hunger S, Kilberg M. Correlation between asparaginase sensitivity and asparagine synthetase protein content, but not mRNA, in acute lymphoblastic leukemia cell lines. Pediatr. Blood Cancer. 2008;50:274–279. doi: 10.1002/pbc.21213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen SH, Yang W, Fan Y, Stocco G, Crews KR, Yang JJ, et al. A genome-wide approach identifies that the aspartate metabolism pathway contributes to asparaginase sensitivity. Leukemia. 2010;25:66–74. doi: 10.1038/leu.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zelent A, Greaves M, Enver T. Role of the TEL-AML1 fusion gene in the molecular pathogenesis of childhood acute lymphoblastic leukaemia. Oncogene. 2004;23:4275–4283. doi: 10.1038/sj.onc.1207672. [DOI] [PubMed] [Google Scholar]
- 118.Stams WA, den Boer ML, Holleman A, Appel IM, Beverloo HB, van Wering ER, et al. Asparagine synthetase expression is linked with L-asparaginase resistance in TEL-AML1-negative but not TEL-AML1-positive pediatric acute lymphoblastic leukemia. Blood. 2005;105:4223–4225. doi: 10.1182/blood-2004-10-3892. [DOI] [PubMed] [Google Scholar]
- 119.Stams WA, den Boer ML, Beverloo HB, Meijernik JP, Stigter RL, van Wering ER, et al. Sensitivity to L-asparaginase is not associated with expression levels of asparagine synthetase in t(12;21)+ pediatric ALL. Blood. 2003;101:2743–2747. doi: 10.1182/blood-2002-08-2446. [DOI] [PubMed] [Google Scholar]
- 120.Feun L, Savaraj N. Pegylated arginine deiminase: a novel anticancer enzyme agent. Expert Opin Investig Drugs. 2006;15:815–822. doi: 10.1517/13543784.15.7.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kuo MT, Savaraj N, Feun LG. Targeted cellular metabolism for cancer chemotherapy with recombinant arginine-degrading enzymes. Oncotarget. 2010;1:246–251. doi: 10.18632/oncotarget.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wolf A, Agnihotri S, Micallef J, Mukherjee J, Sabha N, Cairns R, et al. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011;208:313–326. doi: 10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mathupala SP, Ko YH, Pedersen PL. Hexokinase-2 bound to mitochondria: cancer's stygian link to the "Warburg Effect" and a pivotal target for effective therapy. Semin Cancer Biol. 2009;19:17–24. doi: 10.1016/j.semcancer.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yalcin A, Telang S, Clem B, Chesney J. Regulation of glucose metabolism by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatases in cancer. Exp Mol Pathol. 2009;86:174–179. doi: 10.1016/j.yexmp.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 125.Atsumi T, Chesney J, Metz C, Leng L, Donnelly S, Makita Z, et al. High expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (iPFK-2; PFKFB3) in human cancers. Cancer Res. 2002;62:5881–5887. [PubMed] [Google Scholar]
- 126.Clem B, Telang S, Clem A, Yalcin A, Meier J, Simmons A, et al. Small-molecule inhibition of 6-phosphofructo-2-kinase activity suppresses glycolytic flux and tumor growth. Mol Cancer Ther. 2008;7:110–120. doi: 10.1158/1535-7163.MCT-07-0482. [DOI] [PubMed] [Google Scholar]
- 127.Ros S, Santos CR, Moco S, Baenke F, Kelly G, Howell M, et al. Functional metabolic screen identifies 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 as an important regulator of prostate cancer cell survival. Cancer Discov. 2012;2:328–343. doi: 10.1158/2159-8290.CD-11-0234. [DOI] [PubMed] [Google Scholar]
- 128.Vousden KH. Alternative fuel--another role for p53 in the regulation of metabolism. Proc Natl Acad Sci U S A. 2010;107:7117–7118. doi: 10.1073/pnas.1002656107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Márquez J, López de la Oliva AR, Matés JM, Segura JA, Alonso FJ. Glutaminase: A multifaceted protein not only involved in generating glutamate. Neurochem Int. 2006;48:465–471. doi: 10.1016/j.neuint.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 130.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hu S, Balakrishnan A, Bok RA, Anderton B, Larson PE, Nelson SJ, et al. 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab. 2011;14:131–142. doi: 10.1016/j.cmet.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, et al. Inhibition of Glutaminase Preferentially Slows Growth of Glioma Cells with Mutant IDH1. Cancer Res. 2010;70:8981–8987. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B Cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dang CV. Therapeutic targeting of Myc-reprogrammed cancer cell metabolism. Cold Spring Harb Symp on Quant Biol. 2012;76:369–374. doi: 10.1101/sqb.2011.76.011296. [DOI] [PubMed] [Google Scholar]
- 136.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 137.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 138.Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 140.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bluemlein K, Grüning NM, Feichtinger RG, Lehrach H, Kofler B, Ralser M. No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis. Oncotarget. 2011;2:393–400. doi: 10.18632/oncotarget.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, et al. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, et al. Pyruvate Kinase M2 Is a PHD3-Stimulated Coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst. 2007;99:1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 146.Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nat Rev Cancer. 2008;8:56–61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- 147.Parks SK, Chiche J, Pouyssegur J. pH control mechanisms of tumor survival and growth. J Cell Physiol. 2011;226:299–308. doi: 10.1002/jcp.22400. [DOI] [PubMed] [Google Scholar]
- 148.Sotgia F, Martinez-Outschoorn UE, Howell A, Pestell RG, Pavlides S, Lisanti MP. Caveolin-1 and cancer metabolism in the yumor microenvironment: markers, models, and mechanisms. Annu Rev Pathol. 2012;7:423–467. doi: 10.1146/annurev-pathol-011811-120856. [DOI] [PubMed] [Google Scholar]
- 149.Nakajima EC, Van Houten B. Metabolic symbiosis in cancer: Refocusing the Warburg lens. Mol Carcinog. 2012 doi: 10.1002/mc.21863. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Martinez-Outschoorn UE, Goldberg A, Lin Z, Ko YH, Flomenberg N, Wang C, et al. Anti-estrogen resistance in breast cancer is induced by the tumor microenvironment and can be overcome by inhibiting mitochondrial function in epithelial cancer cells. Cancer Biol Ther. 2011;12:924–938. doi: 10.4161/cbt.12.10.17780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sotgia F, Martinez-Outschoorn UE, Pavlides S, Howell A, Pestell RG, Lisanti MP. Understanding the Warburg effect and the prognostic value of stromal caveolin-1 as a marker of a lethal tumor microenvironment. Breast Cancer Res. 2011;13:213. doi: 10.1186/bcr2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Metallo CM, Gamerio PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15:827–837. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Michelakis ED, Sutendra G, Dromparis P, Webster L, Haromy A, Niven E, et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med. 2010;12:31ra34. doi: 10.1126/scitranslmed.3000677. [DOI] [PubMed] [Google Scholar]