Abstract
The thiazolinyl imine reductase from Yersinia enterocolitica (Irp3) catalyzes the NADPH-dependent reduction of a thiazoline ring in an intermediate for the formation of the siderophore yersiniabactin. Two structures of Irp3 were determined in the apo- (1.85 Å) and NADP+-bound (2.31 Å) forms. Irp3 shows structural homology to sugar oxidoreductases such as glucose-fructose oxidoreductase and 1,5-anhydro-D-fructose reductase, as well as to biliverdin reductase. A homology model of the thiazolinyl imine reductase from Pseudomonas aeruginosa (PchG) was generated. Extensive loop insertions are observed in the C-terminal domain that are unique to Irp3 and PchG and not found in the structural homologs that recognize small molecular substrates. These loops are hypothesized to be important for binding of the nonribosomal peptide synthetase modules (found in HMWP1 and PchF, respectively) to which the substrate of the reductase is covalently attached. A catalytic mechanism of proton donation from a general acid (either histidine-101 or tyrosine-128) and hydride donation from C4 of nicotinamide of the NADPH cofactor is proposed for reduction of the carbon-nitrogen double bond of the thiazoline.
Pathogenic bacteria must acquire iron from the environment, since iron is an essential element required as a cofactor by the enzymes of many pathways, including electron transport, DNA synthesis, and the tricarboxylic acid cycle. A typical bacterium requires ~1 μM iron for optimal growth, but the concentration of available iron in the human host is ~10-9 μM (1). In addition, Fe(III) is very insoluble and frequently biologically inaccessible. For pathogenic bacteria, this requirement is a significant obstacle to overcome. Therefore, bacteria have developed elaborate systems to scavenge iron from the environment/host (2). One such system is the use of siderophores: low molecular weight iron chelators, which the bacteria synthesize, secrete, and then selectively take up in the iron-loaded form to survive and colonize human tissues (2-4).
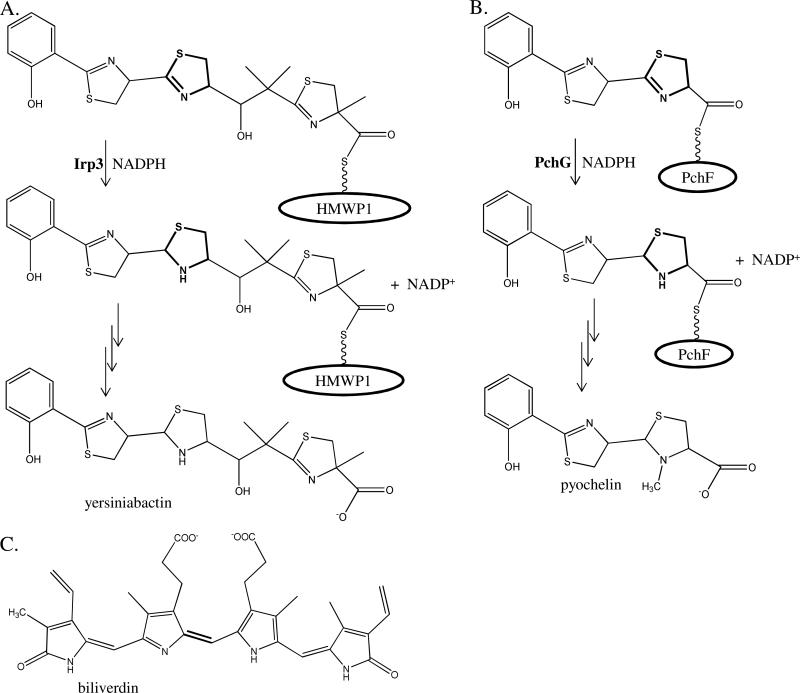
Yersinia and Pseudomonas spp. produce salicyl-capped siderophores that contain thiazoline and thiazolidine rings, both derived from cyclized cysteines (Figure 1). The latter ring is reduced from the thiazoline by an NADPH-dependent reductase, possibly to attain the appropriate geometry to coordinate ferric iron (5). Two enzymes that catalyze this reduction are the focus of this study, in particular Irp3 from Yersinia enterocolitica and PchG from Pseudomonas aeruginosa, which share 26% sequence identity. The homologue from Yersinia pestis, YbtU, is different from Irp3 at 8 of 365 amino acids (98% sequence identity). YbtU was originally identified as required for yersiniabactin formation, but with an unknown role (6). A sequence analysis suggested that there was a possible transmembrane helix from residues 162 to 182 (6).
Figure 1. Yersiniabactin, pyochelin, and biliverdin.
Yersiniabactin, the siderophore produced by Yersinia spp., is the product of the biosynthetic pathway to which Irp3 belongs. Irp3 reduces a double bond in the center thiazoline ring to make the thiazolidine ring of the product. The substrate is reduced while covalently attached to a phosphopantetheinyl post-translational modified serine of the peptidyl carrier domain of the High Molecular Weight Protein 1 (HMWP1). (B) Pyochelin is a siderophore produced by Pseudomonas aeruginosa. PchG performs the analogous reaction as Irp3. The substrate is reduced while covalently attached to a phosphopantetheinyl post-translational modified serine of the peptidyl carrier domain of the nonribosomal peptide synthetase PchF. This ring is subsequently methylated by the methylation domain of PchF. (C) Biliverdin is the substrate of biliverdin reductase, an enzyme which catalyzes the final step in heme degradation, reducing the center methylene bridge to form bilirubin. Biliverdin reductase is a structural homologue of Irp3, as found herein.
Functional characterization of PchG showed that the enzyme was a required reductase for pyochelin maturation that produced an intermediate, desmethyl-pyochelin, in which the second thiazoline ring is reduced (Figure 1) (7). The substrate for PchG, a hydroxyphenyl-bisthiazolinyl precursor of pyochelin, is covalently attached to the peptidyl carrier domain of PchF, a nonribosomal peptide synthetase (NRPS), through a phosphopantetheinyl (Ppant) post translation modification of a serine (Figure 1b) (7). A complete in vitro reconstitution for the production of pyochelin required PchD (a salicylate adenylase), PchE and PchF (two nonribosomal peptide synthetases modified to have the pPant group), PchG, NADPH, S-adenosylmethionine, ATP, coenzyme A, cysteine and salicylate (5). The authors noted that PchG showed weak sequence similarity to biliverdin reductase (BVR) and proposed a mechanism of hydride transfer from the NADPH cofactor followed by protonation of the nitrogen of the developing thiazolidine (5). The N-terminal portion of PchG was hypothesized to be structurally similar to the C-terminal portion of mouse major urinary protein, which had been determined with 2-(sec-butyl)thiazoline bound (5). A similar role for the reduction of the second thiazoline ring of yersiniabactin was subsequently shown for YbtU when the substrate is Ppant-tethered to the peptidyl carrier domain of the NRPS module of the High Molecular Weight Protein 1 (HMWP1) (Figure 1a) (8). Kinetic measurements for PchG, YbtU, and Irp3 are complicated by the nature of the substrate: an immature siderophore covalently tethered to an NRPS. Therefore, a next viable step for understanding these enzymes is acquiring structural information to provide context for catalysis and protein-protein interactions.
Here we present the first structural characterization of an NADPH-dependent thiazolinyl imine reductase with the structures of apo- and NADP+-bound Irp3. We also present the homology model of PchG. We propose an interaction motif for Irp3 and PchG with the nonribosomal peptide synthetase modules to which their substrates are covalently attached. We further propose a catalytic mechanism based on the structures and by analogy to the closest structural homologues of known function, which are sugar oxidoreductases and biliverdin reductase.
Materials and Methods
Preparation of overexpression plasmids to generate Irp3 protein with N- and C-terminal histidine tags
To generate a plasmid that produces Irp3 protein with a C-terminal histidine tag, the irp3 gene was amplified from Yersinia enterocolitica 33114 genomic DNA (ATCC) by polymerase chain reaction by use of Master Mix (Eppendorf) supplemented with the 25 mM magnesium solution. The forward primer (5’-ATT CTT CAT ATG CCG TCC GCC TCC CCA AAA CA-3’) includes an NdeI site (underlined), whereas the reverse primer (5’- GGA TCC CTC GAG CGC CTC CTT ATC ATC ATC GTT G-3’) contains a XhoI site (underlined). The amplified 1119 basepair fragment was digested with NdeI and XhoI and ligated into the pET29b plasmid (Novagen) digested with the same enzymes. The resulting plasmid encodes the irp3 gene with a C-terminal histidine tag (Irp3-his6-C).
The plasmid to produce Irp3 protein with a N-terminal histidine tag was generated by polymerase chain reaction by use of Herculase II (Agilent) using the Irp3-his6-C plasmid for the template, the Irp3-his6-C forward primer, and a reverse primer (5’- GGA TCC CTC GAG TCA CAG CGC CTC CTT ATC ATC ATC GTT G-3’) containing a XhoI site (underlined) and a stop codon. The amplified 1122 basepair fragment was digested with NdeI and XhoI and ligated into the pET28b plasmid (Novagen) digested with the same enzymes. The resulting plasmid encodes the irp3 gene with an N-terminal histidine tag (N-his6-Irp3).
Irp3-his6-C protein overexpression and purification
BL21 (DE3) E. coli containing the Irp3-his6-C expression plasmid were grown in LB broth containing 50 μg/mL kanamycin at 22 °C with shaking (225 rpm). The cells were harvested by centrifugation (6 000g, 10 min, 4 °C) after 24 h. The cell pellet was resuspended in 15 mL of 25 mM Tris-HCl pH 8, 500 mM NaCl, 5 mM imidazole (buffer A) per liter of culture. Cells were disrupted by use of a French pressure cell (35 000 psi), and cellular debris was removed by centrifugation (12 000g, 30 min, 4 °C). The supernatant was applied to a chelating sepharose fast-flow column (Amersham Biosciences) charged with nickel chloride and pre-equilibrated in buffer A. Irp3-his6-C protein eluted at 300 mM imidazole in a linear gradient of 5-500 mM imidazole in buffer A. The pooled fractions were applied to a Superdex 200 size-exclusion column (Amersham Biosciences) equilibrated with 25 mM Tris-HCl pH 8, 200 mM NaCl, 2 mM dithiothreitol. The fractions containing Irp3-his6-C were pooled and concentrated by use of an Amicon stirred cell with a YM-30 membrane to 19 mg/mL as determined by the Bradford assay and stored at -80 °C.
N-his6-Irp3 protein overexpression and purification
BL21 (DE3) E. coli containing the N-his6-Irp3 expression plasmid were grown in LB broth containing 50 μg/mL kanamycin at 37 °C with shaking (225 rpm). When an optical density of ~0.6 was reached, the temperature was reduced to 25°C. Protein expression was induced with the addition of isopropyl β-D-thiogalactopyranoside to a final concentration of 200 μM. The cells were harvested by centrifugation (6 000g, 10 min, 4 °C) after ~18 h. The cell pellet was resuspended in 10 mL of buffer A per 4 liters of culture and purified similar to Irp3-his6-C with a nickel charged chelating sepharose fast-flow column followed by a Superdex 200 size-exclusion column (Amersham Biosciences) equilibrated with 50 mM potassium phosphate pH 8, 100 mM sodium citrate. The fractions containing N-his6-Irp3 were pooled and concentrated by use of an Amicon stirred cell with a YM-10 membrane to 19 mg/mL as determined by the Bradford assay and stored at -80 °C.
Selenomethionine-substituted (SeMet) N-his6-Irp3 protein overexpression and purification
The method for overexpression of SeMet N-his6-Irp3 was adapted from the protocol of Van Duyne et al. (9) with several modifications. BL21 (DE3) E. coli containing the N-his6-Irp3 expression plasmid were grown in M9 minimal medium containing 50 μg/mL kanamycin and inoculated with 20 ml of overnight culture (grown in LB) per liter of medium at 37 °C with shaking (225 rpm). When an optical density of ~0.6 was reached, the temperature was reduced to 25 °C and the methionine biosynthesis pathway was inhibited by the addition of 100 mg each of L-lysine, L-phenylalanine, and L-threonine; 50 mg each of L-isoleucine, L-leucine, and L-valine; and 60 mg of DL-selenomethionine per liter of culture. Fifteen minutes after the addition of the amino acid mixture, protein expression was induced with the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 200 μM. Cultures were grown for ~18 hours. SeMet N-his6-Irp3 was purified using the same protocol as that used for N-his6-Irp3 with the addition of 1 mm β-mercaptoethanol to all buffers.
Crystallization of SeMet N-his6-Irp3
SeMet N-his6-Irp3 protein was crystallized using the hanging drop method at 24 °C with drops containing 1.5 μL of purified protein mixed with equal volumes of a reservoir solution composed of 0.1 M HEPES pH 7.5, 29% PEG 3350 (w/v), 0.225 M ammonium citrate. Large crystals (25 × 75 × 150 m) formed after 1-2 weeks. For data collection, crystals were transferred to the reservoir solution supplemented with 20% ethylene glycol (v/v) as a cryoprotectant and flash cooled by plunging into liquid nitrogen.
Crystallization of Irp3-his6-C
Crystallization was carried out by the hanging drop method at 24 °C. Drops containing 1.5 μL of purified Irp3-his6-C protein supplemented with 1 mM dichloroethylene-diaminoplatinate were mixed with equal volumes of a reservoir solution composed of 0.1 M HEPES pH 7.3, 32% PEG 3350 (w/v), 0.25 M lithium sulfate. Large crystals (50 × 100 × 150 μm) formed after 1-2 weeks. For data collection, crystals were serially washed in reservoir solution supplemented with 20% ethylene glycol (v/v) as a cryoprotectant and flash cooled by plunging into liquid nitrogen.
Crystallization of N-his6-Irp3 with NADP+ bound
N-his6-Irp3 was crystallized using the hanging drop method at 24 °C with drops containing 1.5 μL of purified N-his6-Irp3 protein supplemented with 3 mM NADP+ (Sigma) mixed with equal volumes of a reservoir solution composed of 0.1 M Tris-HCl pH 8.0, 13% PEG 3350 (w/v), 0.11 M magnesium formate. Rod crystals (25 × 25 × 150 μm) formed after 1-2 weeks. For data collection, crystals were transferred to the reservoir solution supplemented with 6 mM NADP+ and 20% ethylene glycol (v/v) as a cryoprotectant and flash cooled by plunging into liquid nitrogen.
Data collection and structure solution of SeMet N-his6-Irp3
Multiwavelength anomalous dispersion data were collected at the Stanford Synchrotron Radiation Laboratory (Stanford, CA) beamline 9-2. Peak (0.97915 Å), inflection (0.97944 Å), and remote (0.91837 Å) wavelengths were determined by a selenium fluorescence scan. Data (180°) were collected as 0.3° oscillation images (2 s exposure per frame) with a crystal to detector distance of 295 mm at 100 K. The crystals were assigned to the space group P212121 with unit cell dimensions a = 83 Å, b = 93 Å, and c = 181 Å. The data were indexed and scaled to 2.60 Å using the XDS program package (10). AutoSolve and AutoBuild from the PHENIX software package (11) were used to determine the location of 23 selenium atoms (figure of merit = 0.41, overall score = 40.5), to calculate initial phase estimates, and to build the initial model with 1275 of the possible 1460 residues. Data processing and phasing statistics are provided in Table 1.
Table 1.
Irp3 SeMet data collection statisticsa
| SeMetb | |||
|---|---|---|---|
| Inflection | Peak | Remote | |
| Wavelength (Å) | 0.97944 | 0.97915 | 0.91837 |
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions; a, b, c (Å) | 82.97, 93.06, 181.37 | 82.93, 92.97, 181.26 | 83.00, 93.09, 181.34 |
| Resolution (Å) | 51.14 - 2.60 (2.74 - 2.60) | 51.11 - 2.60 (2.74 - 2.60) | 51.15 - 2.60 (2.74 - 2.60) |
| R sym c | 0.106 (0.373) | 0.105 (0.366) | 0.107 (0.361) |
| Total observations | 325730 (47180) | 324939 (47099) | 326255 (47276) |
| Total unique observations | 44022 (6307) | 43953 (6305) | 44045 (6306) |
| Mean ((I) / sd(I)) | 12.5 (4.9) | 12.5 (5.1) | 12.6 (5.0) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) |
| Redundancy | 7.4 (7.5) | 7.4 (7.5) | 7.4 (7.5) |
| Anomalous completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) |
| Anomalous redundancy | 3.8 (3.8) | 3.8 (3.7) | 3.8 (3.8) |
| No. of refined selenium sites | 23 | ||
| DelAnom correlation between half-setsd | 0.229 (-0.004) | 0.204 (0.011) | -0.009 (-0.009) |
| Mean figure of merite | 0.41 | ||
| Mid-Slope of Anom Normal Probabilityf | 1.124 | 1.114 | 0.99 |
All data were collected at beamline 9-2 at the Stanford Synchrotron Radiation Lightsource. Values in parenthesis are for the highest resolution shells.
Data indexed and scaled with XDS
Rsym = Σh |Ih - <I>|ΣhIh, where Ih is the intensity of reflection h, and <I> is the mean intensity of all symmetry-related reflections
Anoma lous correlation between half-sets = Σ||FPH ± FP| - FHcalc|/Σ|FPH| reported for all centric reflections.
Mean figure of merit = <aΣPaeia/ΣPa>, where a is the phase and Pa is the phase probability distribution.
Mid-slope of anomalous probability = <|FH|/||FP + FH| - |FPH||> reported for all reflections.
Data collection and structure solution for Irp3-his6-C, considered the “native” structure
Irp3-his6-C diffraction data (0.5° oscillation images for a total of 90.5°) were collected at the Stanford Synchrotron Radiation Laboratory (Stanford, CA) beamline 9-2 with a wavelength of 1.000034 Å at 100 K. The exposure time per frame was 5 s with a crystal to detector distance of 250 mm. The data were indexed with MOSFLM (12) and scaled with SCALA in the CCP4 program package (13) to 1.85 Å. The crystals were assigned to the space group P212121 with unit cell dimensions a = 83.87 Å, b = 93.90 Å, and c = 181.12 Å. Because the SeMet and native data were isomorphous, the initial SeMet model and phase estimates were used to build the higher resolution native Irp3 model. Model building and refinement of the structure were accomplished using Coot (14) and Refmac5 (15) from the CCP4 software suite. Waters were added manually using the water addition function in Coot, and suggested positions were manually verified in Coot following a refinement cycle in Refmac. Molecules from the crystallization solution were also modeled (sulfate, ethylene glycol and HEPES). Alternate conformations were tested for several residues, but in the end are not found in the final model since the density did not justify their inclusion. Data collection and refinement statistics are found in Table 2.
Table 2.
Irp3 native and NADP+ data collection and refinement statisticsa
| Native | NADP+ bound | |
|---|---|---|
| Data collectionb | ||
| Wavelength (Å) | 1.000034 | 0.97944 |
| Space group | P212121 | P212121 |
| Cell dimensions; a, b, c (Å) | 83.87, 93.90, 181.12 | 83.42, 93.82, 181.99 |
| Resolution (Å) | 76.11 - 1.85 (1.95 - 1.85) | 39.89 - 2.31 (2.43 - 2.31) |
| R sym c | 0.054 (0.395) | 0.087 (0.389) |
| Total observations | 465318 (67058) | 234222 (33388) |
| Total unique observations | 122197 (17701) | 63447 (9044) |
| Mean ((I) / sd(I)) | 12.4 (3.1) | 11.2 (3.2) |
| Completeness (%) | 99.8 (100.0) | 99.6 (98.5) |
| Redundancy | 3.8 (3.8) | 3.7 (3.7) |
| Refinement | ||
| Resolution (Å) | 40.97 - 1.85 (1.90 - 1.85) | 39.89 - 2.31 (2.37 - 2.31) |
| R cryst d | 19.6 (26.8) | 19.7 (25.8) |
| R free | 24.2 (32.8) | 26.0 (33.1) |
| Total unique observations | 115,840 | 60,174 |
| No. of non-hydrogen atoms | 10,938 | 11,156 |
| Protein | 10,397 | 10,854 |
| Ligand | 20 | 192 |
| Solvent | 23 | 0 |
| Water | 498 | 110 |
| rms deviation bonds (Å) | 0.02 | 0.015 |
| rms deviation angles (°) | 2.01 | 1.93 |
| Overall mean B-factor (Å2) | 28.3 | 28.7 |
| Ra machandran plot analysise | ||
| Favored region | 98.4 | 96.9 |
| Allowed region | 1.4 | 2.9 |
| Outlier region | 0.2 | 0.3 |
All data were collected at beamline 9-2 at the Stanford Synchrotron Radiation Lightsource. Values in parenthesis are for the highest resolution shells.
native data indexed with MOSFLM and scaled with Scala, NADP+ data indexed and scaled with XDS
Rsym = Σh |Ih - <I>|/Σh Ih, where Ih is the intensity of reflection h, and <I> is the mean intensity of all symmetry-related reflections
Rcryst = Σ ||Fo| - |Fc||/Σ |Fo|, Fo and Fc are observed and calculated structure factor amplitudes. Five percent of the relections were reserved for the calculation of Rfree.
Calculated with RAMPAGE
Data collection and structure solution for NADP+-bound N-his6-Irp3
Oscillation images of N-his6-Irp3 crystals grown in the presence of NADP+ (90° with an oscillation angle of 0.6°) were collected at the Stanford Synchrotron Radiation Laboratory (Stanford, CA) beamline 9-2 with a wavelength of 0.97949 Å at 100 K. The exposure time per frame was 25 s with a crystal to detector distance of 330 mm. The data were indexed and scaled to 2.3 Å with XDS (10). The crystals were assigned to the space group P212121 with unit cell dimensions a = 83.42 Å, b = 93.82 Å, and c = 181.99 Å. Molecular replacement calculations were performed using the program PHASER (16) and the data from 38.0 to 2.3 Å. Molecule A of the Irp3-his6-C native structure was used as a search model yielding a clear solution with a log likelihood gain of 12,806.61. The map generated with this solution showed unmodeled electron density in the active site corresponding to the NADP+ and an additional 15 residues at the N-terminus of molecule A compared with the native Irp3-his6-C model. These differences in electron density relative to the molecular replacement model indicate a correct solution. Iterative rounds of model building and refinement were performed with Coot (14) and Refmac5 (15). Waters were added manually using the water addition function in Coot, and suggested positions were manually verified in Coot following a refinement cycle in Refmac. Data collection and refinement statistics are found in Table 2.
Native Irp3 crystallographic model
The final native Irp3 model contains four monomers per asymmetric unit. The ordered residues in the structure can be found in Table 3. The structure also includes four sulfate ions, one HEPES molecule, and two ethylene glycol molecules from the crystallization conditions and 498 water molecules. Ramachandran analysis as calculated by RAMPAGE (17) showed good geometry (Table 2). The amino acids in the outlier region were glycine-276 in monomers A, B and C.
Table 3.
Ordered amino acids in structures by monomeric chain
| Residues in model | Native | NADP+ bound |
|---|---|---|
| Monomer A | 7 - 75, 78 - 255, 270 - 357 | -7 - 254,a 262 - 357 |
| Monomer B | 8 - 26, 31 - 74, 78 - 255, 270 - 357 | 7 - 255, 262 - 357 |
| Monomer C | 7 - 27, 31 - 74, 78 - 254, 270 - 357 | 7 - 254, 270 - 357 |
| Monomer D | 7 - 74, 78 - 254, 271 - 357 | 7 - 255, 264 - 357 |
includes 7 N-terminal amino acids of the purification tag
NADP+-bound Irp3 crystallographic model
The final native Irp3 model contains four monomers per asymmetric unit. The ordered residues in the structure can be found in Table 3. The structure also includes one NADP+ in each active site and 110 water molecules. Ramachandran analysis as calculated by RAMPAGE (17) showed good geometry (Table 2). The amino acids in the outlier region were glycine-276 in all monomers, and proline-2 in monomer A.
Structural analysis
Root mean square deviations (r.m.s.d.) were calculated using LSQMAN program suite (18). Structural comparisons (including r.m.s.d. calculations therein) were carried out using the PDBeFold (19) and protein interaction interfaces were analyzed using PDBePISA (20), both of the European Bioinformatics Institute server (http://www.ebi.ac.uk/pdbe/). Protein structure figures were generated using PyMOL (21). The atomic coordinates and structure factors (codes 4GMF and 4GMG) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (www.rcsb.org).
Homology model of PchG
Using monomer A of the Irp3 native structure, a model of the Pseudomonas aeruginosa thiazolinyl imine reductase, PchG, was built with the SWISS-MODEL fully automated protein structure homology modeling server (http://swissmodel.expasy.org) (22, 23).
Results
Preparation of Irp3
Irp3 protein was cloned into both the pET28b and pET29b vectors and overproduced in BL21(DE3) E. coli to produce protein with either an N- or C-terminal histidine tag (N-his6-Irp3 and Irp3-his6-C, respectively). Protein purification was completed in two steps using nickel chelating chromatography followed by gel filtration chromatography. The ~40 kDa protein eluted from the gel filtration column at a molecular weight consistent with an 80 kDa protein, indicating dimer formation in solution. This purification protocol yielded 100 mg N-his6-Irp3 per liter of culture and 24 mg Irp3-his6-C per liter of culture at a purity estimated to be >90% by polyacrylamide gel electrophoresis. The N-his6-Irp3 was also produced in a selenomethionine-substituted form, with a yield of 24 mg per liter of culture. The SeMet protein was purified using a similar protocol, with the addition of a reducing agent to all buffers. All three forms of the protein were crystallized using PEG 3350 as the precipitant, but with differing buffers and salt additives. The Irp3-his6-C and SeMet N-his6-Irp3 were crystallized without ligands. N-his6-Irp3 without selenomethionine substitution was crystallized in the presence of NADP+.
Overall architecture of Irp3
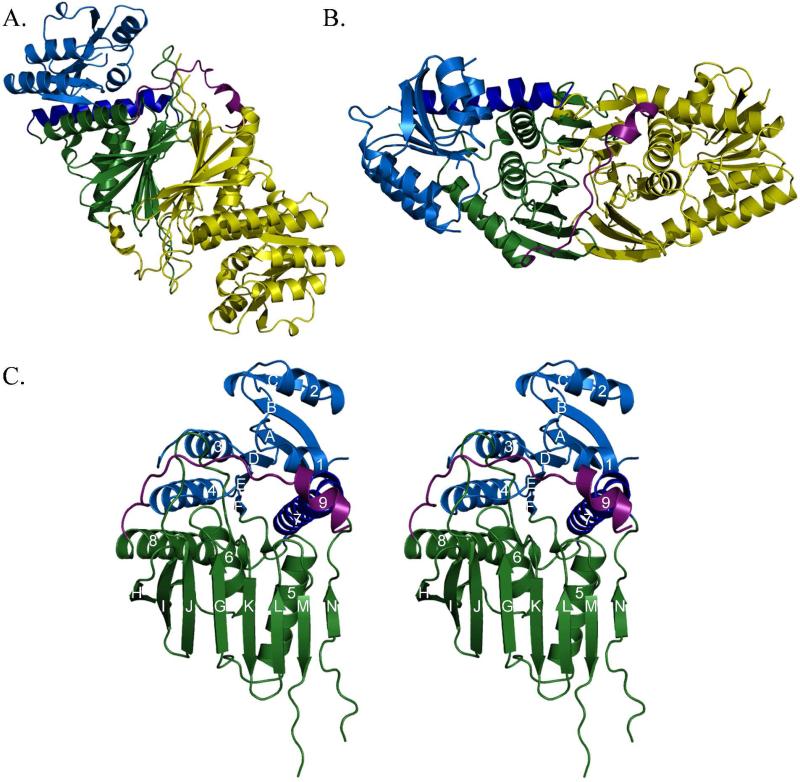
Initial phase estimates for N-his6-Irp3 were determined by multiwavelength anomalous dispersion phasing using a selenomethionine-substituted form of the protein (Table 1). The Se-Met model was used as a starting point for model building and structure refinement with the 1.85 Å isomorphous native (Irp3-his6-C) dataset. The high resolution native model was used for molecular replacement to solve the NADP+-bound (N-his6-Irp3) structure to 2.31 Å (Table 2). In both structures, the asymmetric unit contains four monomers of Irp3, arranged as two dimers (Figures 2A and 2B). For each dimer, the two monomers are related by a non-crystallographic two-fold axis, and the two dimers are related by a translation roughly along this axis.
Figure 2. Overall architecture of Irp3.
(A and B) Dimeric structure of Irp3. The A monomer is shown in yellow. The B monomer is shown in blue (N-terminal domain) and green (C-terminal domain), with the C-terminal tail shown in magenta. Parts (A) and (B) are related by a 90° rotation about the x-axis. The dimerization interface is evident where the open sides of the β-sheet in the C-terminal domains pack together. The magenta helix of the C-terminal tail packs against the opposing monomer, also contributing to the dimerization interface. Monomers A and B of the native structure are depicted. (C) Stereoview of monomer. The N-terminal, NADP(H) binding domain (residues 1-125) is blue, with the helix donated from the C-terminus shown in dark blue (residues 285-310). The C-terminal substrate binding domain is shown in green (residues 126-284 and 311 – 338). The C-terminal tail is shown in magenta. Strands are denoted by letters A – N, starting from the N-terminus. Helices are counted from the N-terminus, 1-8. Monomer A of the native structure is depicted.
Monomer architecture
Each monomer is composed of two domains. The N-terminal domain is a Rossmann fold (24) comprising residues 7-125 with a helix donated from the C-terminal domain (residues 285-310) packing against the central 6-stranded parallel β-sheet (Figure 2B). The faces of the β-sheet are packed with two or three α-helices. The C-terminal domain (residues 126 – 284 and 311 – 337) is an eight stranded mixed (nearly antiparallel) β-sheet with three α-helices packed on one side. The N-his6-Irp3 NADP+-bound structure includes electron density for residues 1-6 and eight residues of the N-terminal purification tag (residues -7 to 0) of monomer A. Residues -7 to 1 form an additional crystal contact with monomer C. Residues 339 to 357 are evident in all four monomers of both the Irp3-his6-C native and N-his6-Irp3 NADP+-bound structures. Residues 338 to 350 are a random coil with the last seven residues (351 to 357) forming helix 9, which contributes to the dimerization interface. In Irp3, a further eight amino acids are disordered at the C-terminus, as well as one (NADP+-bound structure) or two or three (native structure) internal loops (Table 3). Finally, we note that the previously hypothesized transmembrane region (residues 162-182, (6)) comprises helix 6 and the 6-H loop, and does not play the role suggested.
Comparison of native and NADP+-bound structures
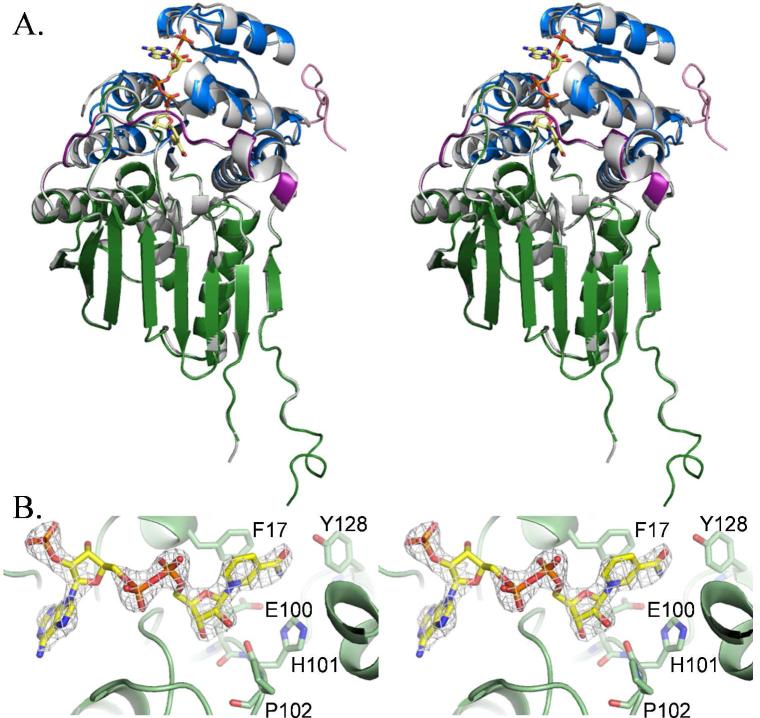
The overall architecture of the native and NADP+-bound structures is quite similar (Figure 3A). The root mean square deviations for comparison between monomers of the same structure (native monomer A to native monomer B, for example) range from 0.22 to 0.55 Å for 328 to 343 Cα residues. Comparison of monomers between structures (native monomer A to NADP+-bound monomer A, etc.) yielded values ranging from 0.31 to 0.60 Å for 326 to 331 Cα residues. Binding of NADP+ causes the ordering of the loop connecting strand D and helix 3, which is disordered in all four monomers of the native structure (Table 3).
Figure 3. NADP+ binding.
(A) Superposition of the native Irp3 structure (grey) with the NADP+-bound structure. The domains of the NADP+-bound structure are colored as in Figure 2, with the addition of the N-terminal tail, show in pink. The NADP+ is depicted as yellow sticks. The sulfate ion bound in the NADP + binding site of the native structure is also shown in sticks. The A monomers of both structures are overlayed. (B) Fo – Fc difference density contoured at 3σ (grey cages) for the NADP+. Phenylalanine-17 is part of the glycine rich 14GAKFGE19 sequence for nucleotide binding. The functionally important 100EHP102 sequence is also shown, as is tyrosin-128. Oxygen atoms are red, nitrogen atoms are dark blue, and phosphorous atoms are orange. Carbon atoms are colored as per the molecule: Irp3, light green; NADP+, yellow.
Structural homologues
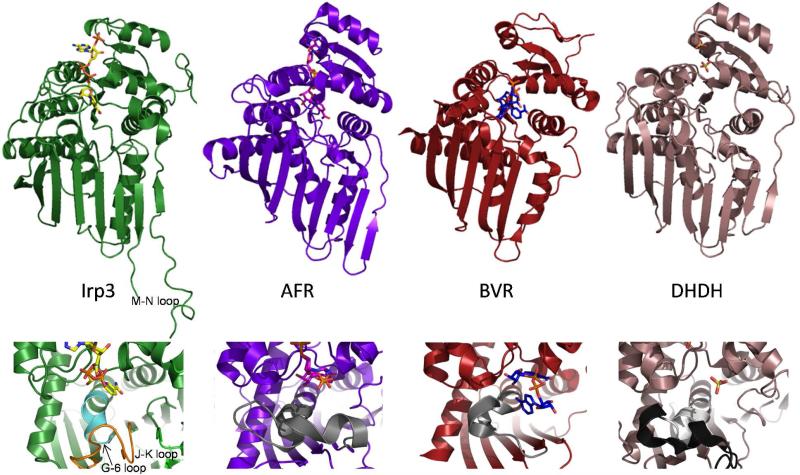
A search with the PDBeFold server (19) revealed that the closest structurally related enzymes for which a function has been determined belong predominantly to the GFO_IDH_MocA oxidoreductase family and also the dimeric dihydrodiol dehydrogenase. There are many structures with similarity to Irp3 that have been deposited by structural genomics consortia with little annotation, but labeled “oxidoreductase” or “dehydrogenase.” The fold similarity is evident, even by eye (Figure 4). For the top 100 hits, the Z-scores range from 12.2 to 9.7, with a range in r.m.s.d. for these same comparisons of 2.2 to 2.9 Å aligning 235-265 α-carbons with 8-17% sequence identity. More simply, this means that the structure of Irp3 falls into the general structural class of oxidoreductase. This comparison led us to compare the structure Irp3 to the sugar oxidoreductases of known function, with hopes of deducing functional significance. These enzymes include the glucose-fructose oxidoreductase from Zymomonas mobilis (GFOR, (25, 26)), the 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense (AFR, (27)), monkey dimeric dihydrodiol dehydrogenase (DHDH, (28-30)), WlbA from Bordetella pertussis and Chromobacterium violaceum, which are required to oxidize a C-3’ hydroxyl group of a UDP-linked sugar for formation of 2,3-diacetamido-2,3-dideoxy-D-manuronic acid (31, 32), and KijD10, a ketoreductase in the biosynthetic pathway of L-digitoxose derived from the bacterium Actinomadura kijaniata (33). Irp3 was also compared to biliverdin reductase (34, 35) and mouse major urinary protein (36), since Irp3 had previously been hypothesized to be structurally similar to these proteins (5). Biliverdin reductase is roughly as structurally similar to Irp3 as the oxidoreductases identified by PDBeFold, with an r.m.s.d. of 1.88 Å for 199 Cα's. Mouse major urinary protein, which is a β-barrel, shares no structural similarity with Irp3.
Figure 4. Cartoon representations of the NADP+-bound Irp3, 1,5-anhydro-D-fructoase reductase (AFR, Protein Data Bank code 2GLX), biliverdin reductase (BVR, code 1LC3), dimeric dihyrodiol dehydrogenase (DHDH, code 2O48).
The upper panels show the overall fold in cartoon and the NADP+ (or sulfates in DHDH) in sticks. It should be noted that the NADP+ binds in a similarly extended conformation in Irp3 and AFR. In BVR, the NADP+ is folded in a U-shaped conformation that is hypothesized to be the result of crystal packing (35). The DHDH structure does not contain NADP+, but has sulfates bound in locations similar to where the phosphates of NADP+ are anticipated to bind (30). In Irp3, the M-N loop, which has 6 to 15 residues disordered depending on monomer, is labeled. The lower panels are a close-up of the putative substrate binding region. In DHDH, the region of the GGX3DX3(Y/H) consensus sequence is shown in light grey and the remainder of the loop containing this sequence is black. The corresponding loops in AFR and BVR are also grey. In Irp3, the much shorter loop connecting strand G and helix 6 is shown in cyan. The extended loop connecting the J and K strands that occupies the corresponding space in three-dimensions is shown in orange.
Dimerization interface
The open side of the β-sheet of the C-terminal domain comprises the most extensive part of the dimerization interface, a common oligomerization motif in this class of enzymes (25, 29, 31, 33, 37), with an additional interaction from the C-terminal tail (Figures 2A and 2B). Interestingly, GFOR has an N-terminal “arm” that seems to perform a similar role, controlling oligomerization (26). Analysis in PDBePISA (20) shows that the interface between the A and B monomers (dimer 1) and C and D monomers (dimer 2) range from 3260 – 3300 Å2, with an average of 3280 Å2. No higher order oligomers are evident in the structure, consistent with the dimer observed in solution.
NADP+ binding
The N-terminal, NADP+-binding domain of Irp3 is a Rossmann fold with a topology like that found in lactate and malate dehydrogenase (38). None of the canonical consensus sequences for cofactor binding (for example, GXGXXG) are found (38-40). Nevertheless, there is a glycine rich sequence 14GAKFGE19 in the first loop of the first βαβ motif to which the NADP+ binds. As is seen in GFOR, an aromatic amino acid stacks with the nicotinamide ring on the side away from the substrate binding site. A tyrosine is observed in GFOR (25, 37) and a phenylalanine (residue 17) is found at this site in Irp3 (Figure 3B). Unlike the GFOR and AFR, Irp3 does not have a kink in the first helix from the inclusion of a proline or glycine in the third turn, which is proposed to influence the helix dipole moment (27).
The NADP+-bound structure has clear density for the cofactor (Figure 3B). We assume that the cofactor remained oxidized, but the electron density of the nicotinamide ring is insufficient to determine whether the cofactor is oxidized (planar) or became reduced (not planar). The density is sufficient for a supposition of stereochemistry. The nicotinamide ring is bound in the anti conformation, which is consistent with the A-side specific oxidoreductases (27, 33) and in contrast to the B-side specific biliverdin reductase (35), suggesting that Irp3 may also be A-side specific. In Irp3, the D-3 loop becomes ordered upon binding of NADP+ (yellow loop in Figure 5A). KijD10 with NADP+ and a substrate analogue bound crystallized in “open” and “closed” conformations in which the nicotinamide ring was syn and anti, respectively. An extended loop in KijD10 (grey in Figure 5) undergoes a disorder to order transition from an open to a closed conformation, which is hypothesized to be responsible for the change in conformation of the nicotinamide ring (from syn to anti). Note that the loops that become ordered upon binding of NADP+ in Irp3 and KijD10 are not comparable. The loop that became ordered in KijD10 is comparable to the G-6 loop of Irp3. The adenine ring is bound in the anti conformation in Irp3. In the determined structures of the oxidoreductases (27, 31-33), the conformation of the adenine is varied, with both anti and syn observed.
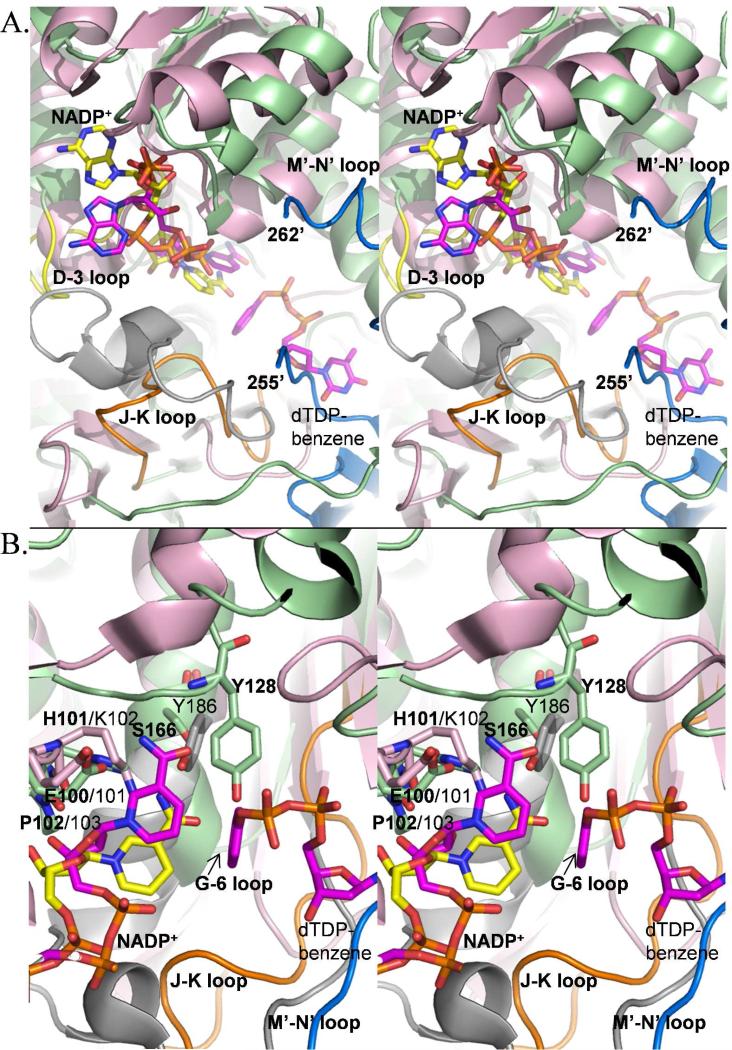
Figure 5. Overlay of the Irp3 and KijD10 active sites, highlighting differences in loops (A) and residues (B).
The colors are maintained in the (A) and (B) panels: NADP+-bound Irp3 is shown in green cartoon. KijD10 (PDB code 3RC1) is shown in pink cartoon. The NADP+ bound in the active sites are shown as yellow (Irp3) and magenta (KijD10) sticks. The substrate analogue, dTDP-benzene, for KijD10 is also shown in magenta sticks. KijD10 is a member of the glucose-fructose oxidoreductase family, and contains the GGX3DX3(Y/H) consensus and following loop shown as the grey cartoon, similar to Figure 4. The D-3 loop of Irp3 is shown in yellow. The J-K loop of Irp3 is shown in orange. The blue cartoon is for the M’-N’ loop of the opposing dimer, which would be connected by six disordered residues (256-261). (A) The grey loop in KijD10 undergoes a disorder to order transition when the protein goes from the open to the closed conformation, and with this transition, the nicotinamide of the NADP+ flips from syn to anti (33). The nicotinamide ring in Irp3 is anti as seen in the closed conformation of KijD10 shown here. The loop in Irp3 that undergoes a disorder-to-order transition upon NADP+-binding is the D-3 loop (yellow). The orange (J-K) loop of Irp3 is in a comparable position to the grey loop of KijD10, but does not contain the consensus sequence nor does it connect comparable secondary structure (this is the G-6 loop in Irp3) as discussed in Figure 4. We hypothesize that the very long M’-N’ loop (blue) resides in a different location in the presence of substrate and aids in binding the protein HMWP1 to which the substrate is covalently attached. (B) The EKP sequence of KijD10 and the corresponding EHP of Irp3 are shown in sticks and labeled. The K/H at the center of this loop is hypothesized to be the general acid, which is supported by mutagenic analyses in KijD10 (33). The Y186 of the GGX3DX3(Y/H) consensus sequence in KijD10 is displayed in sticks to show the relative location to S166, the structurally equivalent amino acid, and Y128, found in the linker between the N-terminal and C-terminal domains (the F-5 loop), both in Irp3. Labels in bold are for Irp3 and those in plain text are for KijD10.
In GFOR, AFR and DHDH, there is a second consensus sequence for cofactor binding. In GFOR and AFR, this sequence is GX2VXCEKP (25, 27) and in DHDH it is HX2HX16EKP (28-30), with the last residues being most important (EKP). The glutamic acid hydrogen bonds to the carboxyamide of the nicotinamide ring. The proline residue is in the cis configuration which allows for positioning of the lysine residue into the active site to form a cation-π interaction with the nicotinamide ring (27). In WlbA and KijD10, the EKP sequence is conserved although the remainder of the longer consensus sequence is not found (31, 33), whereas biliverdin reductase has an EYP sequence at this site (35). Irp3 has 100EHP102 in the comparable loop (Figures 3B and 5B), the glutamic acid is H-bonded to the carboxyamide of the nicotinamide ring, the proline is cis, and the histidine is 3.5 – 4.0 Å from the carboxyamide of the nicotinamide ring.
Substrate binding
A consensus sequence for substrate binding has been hypothesized for GFOR/AFR/DHDH: GGX3DX3(Y/H) (28, 29), originally proposed from sequence alignment studies of DHDH (41). This sequence is found at the C-terminus of a 28-residue loop and includes the N-terminal two turns of an active site α-helix (Figure 4 lower panels). In GFOR (37), KijD10 (33), and WlbA from Bordetella pertussis (31), which also have the GGX3DX3(Y/H) substrate binding consensus sequence (grey in Figure 5 for KijD10), the aspartic acid is hypothesized to be important for substrate binding. The final amino acid of the sequence has been hypothesized to serve as the general acid/general base for catalysis. Biliverdin reductase does not have the consensus sequence, but the comparable loop/helix is extensive (19 residues in all) and similarly located. Irp3 does not have the consensus sequence nor the loop. Instead, the comparable loop connecting the G strand to the 6 helix is five residues (residues 157-162) and the helix is a turn shorter, effectively removing this potential substrate specificity determinant (cyan in Figure 4). BVR and Irp3 have a serine (166 in Irp3) at the structurally equivalent site to the (Y/H) of the consensus sequence. Irp3 has a tyrosine (128) derived from the linker between the N- and C-terminal domains (loop F-5) that is in a similar three-dimensional location and hydrogen bonded to the carboxyamide of the nicotinamide of NADP+ (Figure 5B). The orange loop of Irp3 in Figures 4 and 5 connects the J and K strands, occupies the space filled in the other proteins by the comparable G-6 loop, and may provide the specificity determinants for substrate binding. Interestingly, the putative substrate binding site in Irp3 is filled in part by the very long loop (residues 250 – 277) which connects the M and N strands from the opposing monomer (M’-N’ blue loop in Figure 5). A portion of the M-N loop is disordered in the structure, the number of disordered residues depending on the monomer (Table 3). This very long loop is unique to Irp3 compared to the others of this structural class, as seen best in Figure 4 (upper panels). The sugar oxidoreductases have a short turn connecting the strands comparable to M and N. Indeed, BVR does not have strands comparable to M and N.
Homology model of PchG
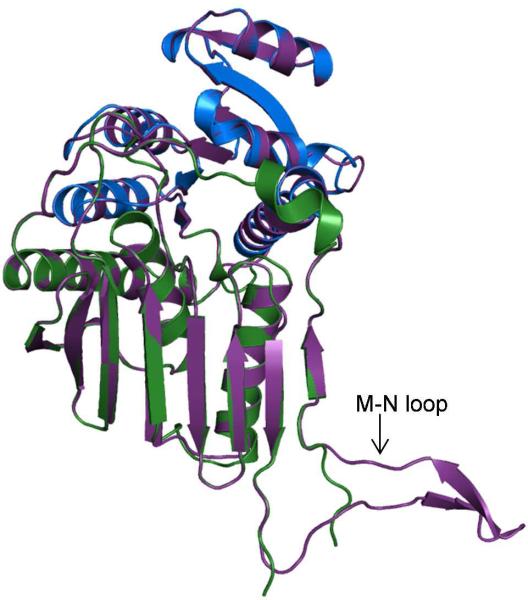
Using the fully automated protein structure homology modeling server SWISS-MODEL (http://swissmodel.expasy.org) (22, 23), and the native Irp3 structure as a template, a model of the thiazolinyl imine reductase involved in pyochelin production from Pseudomonas aeruginosa (PchG) was generated (Figure 6). The sequence identity between the proteins is 26%, with Irp3 comprising 365 residues and PchG containing 349 residues. The PchG model includes residues 4 to 336, with the majority of the unmodeled residues at the C-terminus. The QMEAN score for the PchG model of -0.757 indicates that the model is of good quality (42), and as expected, the overlay of the PchG model with the Irp3 determined structure shows a marked degree of similarity (r.m.s.d. of 0.464 Å for 315 Cα; Figure 6), including a high conservation of active site residues. The notable difference is in the loop connecting strands M and N, which is partially disordered in the Irp3 structure but built in the PchG model and includes two additional short β-strands.
Figure 6. Homology model of PchG from Pseudomonas aeruginosa generated with SWISS-MODEL (22, 23).
The the N-terminal, NADP(H) binding domain of the native Irp3 structure (residues 1-125, 285-310) is blue. The C-terminal substrate binding domain is shown in green (residues 126-284 and 311 – 338). The PchG homology model is shown in purple cartoon. The loop connecting strands M and N, which is partially disordered in the Irp3 structure, contains two additional β-strands in the PchG model.
Discussion
Dimerization may promote binding of substrate
This work provides the first structures of a thiazolinyl imine reductase important in siderophore biosynthesis, including an apo-Irp3 crystal structure, Irp3 with NADP+ bound, and a homology model for PchG generated from the Irp3 structures. An N-terminal Rossmann fold nucleotide binding domain is followed by a C-terminal substrate binding domain, with the active site cleft lying between the two domains. The closest structural homologues are sugar oxidoreductases, along with dihydrodiol dehydrogenase and biliverdin reductase. The structural homologues range in oligomerization state from monomeric to octameric. In solution and in the crystal, Irp3 is dimeric, using the open face of the β-sheet in the C-terminal domain as the most extensive dimeric interface (Figure 2). The question of dimerization in this structural class has been tested in DHDH, where dimer interface mutations were generated and shown to result in active but temperature sensitive monomers, suggesting dimerization promotes stability and thus activity (28). This may also be true in Irp3.
The substrate for Irp3 (depicted in Figure 1) is covalently bound to the peptidyl carrier domain of the very large HMWP1 (350 kDa), consisting of both polyketide synthase and nonribosomal peptide synthetase modules (8, 43). The substrate is bound to the peptidyl carrier domain of the NRPS module (8). Neither the structure nor the oligomerization state of HMWP1 are documented; however, there is evidence for oligomerization of polyketide synthases (44-46) and the predominance of evidence supports monomers for nonribosomal peptide synthetases (47-49). Binding of dimeric Irp3 to HMWP1 (monomeric or dimeric) allows for insertion of the substrate covalently attached to the peptidyl carrier domain of HMWP1 into the active site of Irp3. This binding, which is a catalytic interaction and is most likely transient, may be facilitated by two loops on Irp3. The M-N loop of Irp3 is very long compared to the comparable loop in homologous proteins (~ 25 residues longer). This loop from one monomer projects into the active site of the opposing monomer (Figure 5), with some residues disordered. While the precise placement of the loop may be crystallographic, dimerization orients the M-N loop for participation in binding of HMWP1. If this is true, the C-terminal tail may also be involved in binding to HMWP1, due to proximity to the M-N loop, instead of serving an additional role in Irp3 oligomerization. These extended loops (the M-N loop and the C-terminal tail) are conserved in PchG (homology model depicted in Figure 6). The potential functional role may be conserved as well, with the comparable loops in PchG promoting interaction with PchF, the NRPS to which the PchG substrate is covalently attached. Finally, the G-6 loop is significantly shorter in the thiazolinyl imine reductases relative to the sugar oxidoreductases, preventing this loop from involvement in binding of the substrate. Instead, the J-K loop is extended and occupies a similar location in three dimensions, possibly providing binding residues for the siderophore biosynthetic intermediate (Figures 4 and 5).
Implications for catalysis
A mechanism has evolved in the literature for the sugar oxidoreductases that includes a general acid/base residue for donation or abstraction of a proton from substrate and donation of a hydride to/from the nicotinamide cofactor to the substrate, depending on whether oxidation or reduction of a carbon-oxygen bond is being undertaken (25, 27, 31, 33). A similar mechanism is proposed for reducing the carbon-carbon bond in biliverdin, with a proton donated from a tyrosine general acid and a hydride donated from the C4 of the nicotinamide (35). The residue hypothesized to serve as the general acid/base has been hypothesized to be to be 1) the middle residue of the EKP consensus sequence (31, 33, 35) (tyrosine in BVR) or 2) the final residue of the GGX3DX3(Y/H) consensus sequence (25, 27-29). These hypotheses have been tested by mutagenesis. In AFR, the K94G-variant (EKP) and the H180A-variant (GGX3DX3(Y/H)) produced similar changes in kinetic parameters leading the authors to suggest that these residues were important in substrate binding and catalysis (27). In KijD10, a battery of K102-variants (EKP) either had no activity, or in the case of K102E, a reduction of four orders of magnitude in kcat/Km, whereas the Y186F-variant (GGX3DX3(Y/H)) showed mostly unchanged kinetic parameters. The authors propose the lysine of EKP to be the general acid/base with the glutamic acid of the K102E-variant serving the role ineffectively (33). In BVR, the tyrosine that corresponds to the lysine of EKP has been mutated to phenylalanine, producing the Y97F-variant, which showed 50% of the activity of wildtype. The authors conclude that Y97 is the general acid, but that the hydride transfer from NADPH was the dominant process in catalysis (35). BVR does not contain the GGX3DX3(Y/H) consensus sequence, and neither do Irp3 or PchG. However, there is a structurally equivalent residue at the (Y/H) site, which is serine (a possible general acid) in BVR and Irp3 and alanine (incapable of serving as a general acid) in PchG. Taken together, the middle residue at the EKP sequence is the more likely the general acid/base, especially considering the following conserved cis-proline for orientation of the acid/base in the proper conformation. Therefore, by analogy, histidine-101 of Irp3 (EHP) is the general acid required for catalysis, which is conserved in PchG (histidine-97). However, we cannot rule out that tyrosine-128 of Irp3 that is derived from loop connecting strand F on the N-terminal domain to helix 5 of the C-terminal domain which is in a similar location to the final residue of the GGX3DX3(Y/H) in the sugar oxidoreductases may also serve as a general acid (Figure 5B). This residue is conserved in PchG (tyrosine-124), but not in the other proteins under discussion.
Conclusions
Thiazolinyl imine reductases (Irp3 and PchG) are dimeric enzymes with structural similarity to sugar oxidoreductase such as GFOR, AFR, KijD10 and WlbA and to biliverdin reductase. Irp3 and PchG may interact with the nonribosomal peptide synthetase modules to which their substrates are attached using loops that are unique from their structural homologues. The reduction of the C-N double bond of the substrate thiazoline ring is hypothesized to be carried out by proton donation from a general acid, most likely histidine-101 or alternatively tyrosine-128, and hydride transfer from C4 of the nicotinamide of the NADPH cofactor.
Acknowledgements
We are grateful to A. S. Chilton for technical assistance. Diffraction data were collected at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. We would like to thank the staff at the Stanford Synchrotron Radiation Laboratory for their support and assistance.
This publication was made possible by funds from the NIH grants numbered R01 AI77725 and K02 AI093675 from the National Institute for Allergy and Infectious Disease (A.L.L), and by the Graduate Training Program in Dynamic Aspects of Chemical Biology NIH grant number T32 GM08545 (K.M.M.).
Abbreviations
- AFR
1,5-anhydro-D-fructose reductase from Sinorhizobium morelense
- BVR
rat biliverdin reductase
- DHDH
monkey dimeric dihydrodiol dehydrogenase
- GFOR
glucose-fructose oxidoreductase from Zymomonas mobilis
- HMWP1
high molecular weight protein 1, a nonribosomal peptide synthetase from Yersinia enterocolitica
- Irp3
thiazolinyl imine reductase from Yersinia enterocolitica
- Irp3-his6-C
Irp3 with a C-terminal histidine purification tag
- KijD10
a ketoreductase from Actinomadura kijaniata
- N-his6-Irp3
Irp3 with an N-terminal histidine purification tag
- NRPS
nonribosomal peptide synthetase
- Ppant
phosphopantetheine
- PchD
salicylate adenylase from Pseudomonas aeruginosa
- PchE
nonribosomal peptide synthetase from Pseudomonas aeruginosa
- PchF
nonribosomal peptide synthetase from Pseudomonas aeruginosa
- PchG
thiazolinyl imine reductase from Pseudomonas aeruginosa
- r.m.s.d.
root mean squared deviation
- SeMet
selenomethionine
- WlbA
from Bordetella pertussis, Chromobacterium violaceum and Pseudomonas aeruginosa, enzymes involved in 2,3-diacetamido-2,3-dideoxy-D-manuronic acid production
- YbtU
thiazolinyl imine reductase from Yersinia pestis
Reference
- 1.Vasil ML, Ochsner UA. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 1999;34:399–413. doi: 10.1046/j.1365-2958.1999.01586.x. [DOI] [PubMed] [Google Scholar]
- 2.Cornelis P, Matthijs S. Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environ Microbiol. 2002;4:787–798. doi: 10.1046/j.1462-2920.2002.00369.x. [DOI] [PubMed] [Google Scholar]
- 3.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marahiel MA, Stachelhaus T, Mootz HD. Modular Peptide Synthetases Involved in Nonribosomal Peptide Synthesis. Chem Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 5.Patel HM, Walsh CT. In vitro reconstitution of the Pseudomonas aeruginosa nonribosomal peptide synthesis of pyochelin: characterization of backbone tailoring thiazoline reductase and N-methyltransferase activities. Biochemistry. 2001;40:9023–9031. doi: 10.1021/bi010519n. [DOI] [PubMed] [Google Scholar]
- 6.Geoffroy VA, Fetherston JD, Perry RD. Yersinia pestis YbtU and YbtT are involved in synthesis of the siderophore yersiniabactin but have different effects on regulation. Infect. Immun. 2000;68:4452–4461. doi: 10.1128/iai.68.8.4452-4461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reimmann C, Patel HM, Serino L, Barone M, Walsh CT, Haas D. Essential PchG-dependent reduction in pyochelin biosynthesis of Pseudomonas aeruginosa. J. Bacteriol. 2001;183:813–820. doi: 10.1128/JB.183.3.813-820.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller DA, Luo L, Hillson N, Keating TA, Walsh CT. Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem. Biol. 2002;9:333–344. doi: 10.1016/s1074-5521(02)00115-1. [DOI] [PubMed] [Google Scholar]
- 9.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 10.Kabsch W. XDS. Acta Crystallogr. 2010;D66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leslie AGW, Powell HR. Processing diffraction data with Mosflm. Evolv. Meth. Macromol. Cryst. 2007;245:41–51. [Google Scholar]
- 13.Collaborative Computational Project, N The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 14.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 15.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 16.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. 2005;D61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 17.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 18.Kleywegt GJ, Jones TA. Detecting folding motifs and similarities in protein structures. Methods Enzymol. 1997;277:525–545. doi: 10.1016/s0076-6879(97)77029-0. [DOI] [PubMed] [Google Scholar]
- 19.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. 2004;D60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 20.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 21.DeLano W. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. www.pymol.org. [Google Scholar]
- 22.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinf. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 23.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucl Acids Res. 2009;37:D387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buehner M, Ford GC, Moras D, Olsen KW, Rossmann MG. Structure determination of crystalline lobster D-glyceraldehyde-3-phosphate dehydrogenase. Journal of molecular biology. 1974;82:563–585. doi: 10.1016/0022-2836(74)90249-6. [DOI] [PubMed] [Google Scholar]
- 25.Kingston RL, Scopes RK, Baker EN. The structure of glucose-fructose oxidoreductase from Zymomonas mobilis: an osmoprotective periplasmic enzyme containing non-dissociable NADP. Structure. 1996;4:1413–1428. doi: 10.1016/s0969-2126(96)00149-9. [DOI] [PubMed] [Google Scholar]
- 26.Lott JS, Halbig D, Baker HM, Hardman MJ, Sprenger GA, Baker EN. Crystal structure of a truncated mutant of glucose-fructose oxidoreductase shows that an N-terminal arm controls tetramer formation. J. Mol. Biol. 2000;304:575–584. doi: 10.1006/jmbi.2000.4245. [DOI] [PubMed] [Google Scholar]
- 27.Dambe TR, Kuhn AM, Brossette T, Giffhorn F, Scheidig AJ. Crystal structure of NADP(H)-dependent 1,5-anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 A resolution: construction of a NADH-accepting mutant and its application in rare sugar synthesis. Biochemistry. 2006;45:10030–10042. doi: 10.1021/bi052589q. [DOI] [PubMed] [Google Scholar]
- 28.Carbone V, Hara A, El-Kabbani O. Structural and functional features of dimeric dihydrodiol dehydrogenase. Cell Mol Life Sci. 2008;65:1464–1474. doi: 10.1007/s00018-008-7508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carbone V, Endo S, Sumii R, Chung RP, Matsunaga T, Hara A, El-Kabbani O. Structures of dimeric dihydrodiol dehydrogenase apoenzyme and inhibitor complex: probing the subunit interface with site-directed mutagenesis. Proteins. 2008;70:176–187. doi: 10.1002/prot.21566. [DOI] [PubMed] [Google Scholar]
- 30.Carbone V, Sumii R, Ishikura S, Asada Y, Hara A, El-Kabbani O. Structure of monkey dimeric dihydrodiol dehydrogenase in complex with isoascorbic acid. Acta Crystallogr. 2008;D64:532–542. doi: 10.1107/S0907444908004538. [DOI] [PubMed] [Google Scholar]
- 31.Thoden JB, Holden HM. Biochemical and structural characterization of WlbA from Bordetella pertussis and Chromobacterium violaceum: enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-D-mannuronic acid. Biochemistry. 2011;50:1483–1491. doi: 10.1021/bi101871f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thoden JB, Holden HM. Structural and functional studies of WlbA: A dehydrogenase involved in the biosynthesis of 2,3-diacetamido-2,3-dideoxy-D-mannuronic acid. Biochemistry. 2010;49:7939–7948. doi: 10.1021/bi101103s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubiak RL, Holden HM. Combined structural and functional investigation of a C-3″-ketoreductase involved in the biosynthesis of dTDP-L-digitoxose. Biochemistry. 2011;50:5905–5917. doi: 10.1021/bi200514b. [DOI] [PubMed] [Google Scholar]
- 34.Kikuchi A, Park SY, Miyatake H, Sun D, Sato M, Yoshida T, Shiro Y. Crystal structure of rat biliverdin reductase. Nat. Struct. Biol. 2001;8:221–225. doi: 10.1038/84955. [DOI] [PubMed] [Google Scholar]
- 35.Whitby FG, Phillips JD, Hill CP, McCoubrey W, Maines MD. Crystal structure of a biliverdin IXalpha reductase enzyme-cofactor complex. J. Mol. Biol. 2002;319:1199–1210. doi: 10.1016/S0022-2836(02)00383-2. [DOI] [PubMed] [Google Scholar]
- 36.Perez-Miller S, Zou Q, Novotny MV, Hurley TD. High resolution X-ray structures of mouse major urinary protein nasal isoform in complex with pheromones. Protein Sci. 2010;19:1469–1479. doi: 10.1002/pro.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nurizzo D, Halbig D, Sprenger GA, Baker EN. Crystal structures of the precursor form of glucose-fructose oxidoreductase from Zymomonas mobilis and its complexes with bound ligands. Biochemistry. 2001;40:13857–13867. doi: 10.1021/bi011355d. [DOI] [PubMed] [Google Scholar]
- 38.Bellamacina CR. The nicotinamide dinucleotide binding motif: a comparison of nucleotide binding proteins. Faseb J. 1996;10:1257–1269. doi: 10.1096/fasebj.10.11.8836039. [DOI] [PubMed] [Google Scholar]
- 39.Hanukoglu I, Gutfinger T. cDNA sequence of adrenodoxin reductase. Identification of NADP-binding sites in oxidoreductases. Eur. J. Biochem. 1989;180:479–484. doi: 10.1111/j.1432-1033.1989.tb14671.x. [DOI] [PubMed] [Google Scholar]
- 40.Jornvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 41.Arimitsu E, Aoki S, Ishikura S, Nakanishi K, Matsuura K, Hara A. Cloning and sequencing of the cDNA species for mammalian dimeric dihydrodiol dehydrogenases. Biochem J. 1999;342(Pt 3):721–728. [PMC free article] [PubMed] [Google Scholar]
- 42.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinf. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suo Z, Tseng CC, Walsh CT. Purification, priming, and catalytic acylation of carrier protein domains in the polyketide synthase and nonribosomal peptidyl synthetase modules of the HMWP1 subunit of yersiniabactin synthetase. Proc. Natl. Acad. Sci. USA. 2001;98:99–104. doi: 10.1073/pnas.021537498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leibundgut M, Maier T, Jenni S, Ban N. The multienzyme architecture of eukaryotic fatty acid synthases. Curr. Opin. Struct. Biol. 2008;18:714–725. doi: 10.1016/j.sbi.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 45.Maier T, Leibundgut M, Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008;321:1315–1322. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- 46.Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc. Natl. Acad. Sci. USA. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell CA, Shi C, Aldrich CC, Gulick AM. Structure of PA1221, a nonribosomal peptide synthetase containing adenylation and peptidyl carrier protein domains. Biochemistry. 2012;51:3252–3263. doi: 10.1021/bi300112e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sieber SA, Linne U, Hillson NJ, Roche E, Walsh CT, Marahiel MA. Evidence for a monomeric structure of nonribosomal Peptide synthetases. Chem. Biol. 2002;9:997–1008. doi: 10.1016/s1074-5521(02)00214-4. [DOI] [PubMed] [Google Scholar]
- 49.Sundlov JA, Shi C, Wilson DJ, Aldrich CC, Gulick AM. Structural and functional investigation of the intermolecular interaction between NRPS adenylation and carrier protein domains. Chem. Biol. 2012;19:188–198. doi: 10.1016/j.chembiol.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]