Abstract
Application of the kinase inhibitors in cancer treatment is rapidly growing. However, the methods for monitoring the effectiveness of the inhibitors are still poorly developed and currently rely mainly on tracking changes in the tumor volume, a rather late and relatively insensitive marker of therapeutic response. In contrast, magnetic resonance spectroscopy (MRS) can detect changes in cell metabolism, which has potential for providing early and patient-specific markers of drug activity. Using human B-cell lymphoma models and MRS, we demonstrate that inhibition of the mTOR signaling pathway can be detected in malignant cells in vitro and noninvasively in vivo by measuring lactate levels. An mTOR inhibitor, rapamycin, suppressed lactic acid production in the lymphoma cell line cultures and also diminished steady state lactate levels in xenotransplants. The inhibition was time dependent and first detectable 8 hours after drug administration in cell cultures. In the xenotransplants, two days of rapamycin treatment produced significant changes in lactic acid concentration in the tumor measured in vivo that were followed by tumor growth arrest and tumor volume regression. The rapamycin-induced changes in lactate production strongly correlated with inhibition of expression of hexokinase II, the key enzyme in the glycolytic pathway. These studies suggest that MRS or FDG PET detection of changes in glucose metabolism may be effective noninvasive methods for monitoring mTOR targeting therapy in lymphomas and other malignancies. Furthermore, measuring glucose metabolic inhibition by MRS or by FDG PET imaging may also prove effective in monitoring the efficacy of other kinase inhibitors, given that the rapamycin-sensitive mTOR is down-stream of many oncogenic signaling pathways.
Keywords: mTOR signaling, B-cell lymphoma, Glycolysis, magnetic resonance spectroscopy (MRS), Signal transduction, Lactate
INTRODUCTION
Many different kinase inhibitors are used in cancer therapy and some have already become the drugs of choice in selected malignancies (1–3). The kinase inhibitors target cell-signaling pathways that are persistently activated in the cancer cells and are, therefore, at least partially responsible for the malignant cell transformation. However, despite the rapidly increasing application of signaling inhibitors in various cancer types, monitoring their therapeutic efficacy noninvasively remains underdeveloped. Monitoring response to these agents currently relies primarily on imaging methods that measure changes in tumor volume. This approach has several limitations: foremost that tumor shrinkage tends to occur relatively late after initiation of therapy. Furthermore, kinase inhibitors sometimes display cytostatic rather than cytotoxic properties and, hence, may exert limited effect on tumor volume. Therefore, the availability of reliable metabolic surrogate markers of response capable of direct and precise non-invasive evaluation of the kinase inhibitor-induced suppression of the targeted pathways for individual patients would be of great value in the management of cancer therapy.
mTOR, the mammalian target of rapamycin, is a ubiquitously expressed and highly conserved serine/threonine kinase that associates with either raptor or rictor and other proteins to form the mTORC1 and mTORC2 complexes, respectively. The role of mTORC1 has thus far been elucidated in much greater detail (1–3). mTORC1 affects a number of key cell functions such as cell cycle progression and gene expression and acts by directly activating p70S6kinase 1 (p70S6K1) and inhibiting 4E binding protein 1 (4E-BP1). p70S6K1 is a serine/threonine kinase that phosphorylates an S6 protein of the 40S ribosomal subunit (S6rp, also known as S6 ribosomal protein) at several sites including serines 235 and 236. Rapamycin and its analogs are specific, potent, and relatively non-toxic mTORC1 inhibitors that act by inhibiting the binding of the kinase to FKBP12, a member of the mTORC1 complex. Because persistent activation of mTORC1 occurs in a large spectrum of hematopoietic and non-hematopoietic malignancies, mTORC1 is a very attractive therapeutic target. Indeed, certain types of tumors respond very well to the rapamycin-type mTORC1 inhibitors used as single agents. As a result, two of the rapamycin analogs, CCI-779/temsirolimus and RAD001/everolimus/certican, recently received FDA approval for the treatment of renal cell carcinoma (4,5).
The remarkable propensity of cancer cells to almost universally over-rely on the conversion of glucose to lactic acid to generate ATP and various essential metabolites for cell replication even in the presence of saturating oxygen concentrations (i.e., aerobic glycolysis) has been recognized a long time ago as a hallmark of malignancy and dubbed the “Warburg effect” (reviewed in ref. 6 & 7). Recently, it has been reported that in numerous cancer types the enzyme hexokinase II plays the critical initiating role in aerobic glycolysis (8). This is largely due to its overexpression and binding to the mitochondrial outer membrane where the inhibition by glucose-6-phosphate is prevented and where it has preferred access to mitochondrial ATP generated by ATP synthase located in the inner membrane (8). Magnetic resonance spectroscopy (MRS) is able to monitor cellular metabolic status including measuring the concentration of lactic acid, the end product of glycolysis.
By using the B-cell lymphoma model, we demonstrate in this study that the inhibition of the mTORC1 signaling pathway can be detected in malignant cells in vitro and in vivo by monitoring the lactic acid concentration. The decrease in lactate production is gradual and correlates with the inhibition of hexokinase II expression. These studies suggest that appropriately timed detection of changes in the lactate production rate may permit monitoring of direct or indirect mTORC1 inhibition in patients with lymphomas and other malignancies.
MATERIALS AND METHODS
Cell Lines
The cell lines used in this study have been described in detail before (9). The DLCL2, Ly18 and Val lines were derived from the DLBCL of germinal center origin. The Ramos cell line was obtained from Burkitt Lymphoma. DLCL2 and Ramos cells were cultured in RFPMI1640 (MediaTech) and Ly18 and Val cells in IMDM+GlutaMAX (Invitrogen) supplemented with 10% FBS (Mediatech). Rapamycin was purchased from Cell Signaling Technology.
Western blot
The washed cells were lysed in radioimmunoprecipitation assay buffer supplemented with 0.5 mM phenylmethylsulfonyl fluoride, phosphatase inhibitor cocktails I and II from Sigma and protease inhibitor cocktail from Roche as described previously (10,11). For normalization of gel loading, the protein extracts were assayed using the Lowry method (Bio-Rad) Dc protein assay). Typically, 20–50 mg of the protein per lane was loaded. To examine protein phosphorylation and expression, the membranes were incubated with antibodies specific for S6rp S235/236, AKT T308, S6rp and hexokinase II (all from Cell Signaling) and Actin (from Santa Cruz Biotechnology). Next, the membranes were incubated with the appropriate secondary, peroxidase-conjugated antibodies. The blots were developed using the ECL Plus System from Amersham.
BrdU
The assay was performed as previously described (9,10). In brief, cell proliferation was evaluated with the bromodeoxyuridine (BrdU) incorporation assay using the Cell Proliferation enzyme-linked immunosorbent assay kit (Roche) according to the manufacturer’s protocol. In brief, cells were seeded in 96-well plates (Corning) at a concentration of 1 × 104 cells/wellin RPMI medium supplemented with 10% fetal bovine serum, cultured for 24 and 48 hr in the presence of 200 nM of rapamycin and labeled with BrdU (Roche) for 4 hr. The amount of incorporated BrdU was determined by incubation with a specific antibody conjugated with peroxidase followed by colorimetric conversion of the substrateand optical density evaluation in the EIA plate reader.
Gene expression analysis
DLCL2 and Ramos cell lines were placed into six-well plates in 10 mL RPMI (10% FBS) for 2 hr followed by addition of 200 nM of rapamycin or medium alone for 6 hr. All experimental groups were examined in triplicate. Cells were harvested, and total RNA was isolated using a TRIZOL and Qiagen RNeasy protocol from the Penn Microarray Facility. RNA integrity and quality were assessed on an Agilent 2100 Bioanalyzer (Agilent). The mRNA was reverse-transcribed using a poly(t)-T7 promoter primer, amplified in a linear fashion by in vitro transcription withbiotin labeling and hybridized to the U133 Plus 2.0 array chips. The chips were washed, stained, and scanned according to the manufacturer's instructions. Normalized expression values were calculated using GCRMA (GeneSpring GX 7.3.1, Agilent). Present/absent/marginal flag calls were calculated using the MAS5 algorithm (GeneChip Operating Software, Affymetrix), and all MAS5 quality control variables were within reference ranges. Genes that appeared with a present flag in all three samples of at least one condition in the experiment were identified for further analysis. Principal component analysis was used to assess variability of data. Differentially expressed genes were identified using ANOVA. A false discovery rate test correction was applied using the Benjamini-Hochberg step-up method (Partek Genomics Suite 6.2, Partek, Inc.). For further analysis, we used gene lists that are estimated to have1% false positive genes (false discovery rate, 0.01).
In vitro measurement of metabolite concentration
Intracellular lactate concentration was measured by 1H MRS of perchloric acid extracts of cells. Cells seeded at 6×105/ml in 75 cm2 flasks were cultured at 37°C and 5% CO2 in either medium or 200 nM rapamycin. At specific time points (2, 4, 8, 24 and 48 hr from treatment), total cell numbers were counted and cells were centrifuged at 2000 rpm for 5 min. The medium was collected and stored on ice for measurement of extracellular glucose and lactate concentrations. The pellets were rinsed twice with ice-cold PBS. Ice-cold 12% perchloric acid was added and the pellets were homogenized with a Dounce homogenizer at 4°C and centrifuged (12000 rpm, 4°C, 30 min). The clear supernatant was neutralized with 3M KOH to pH 7.0 and centrifuged for 5 min at 2000 rpm. The supernatant was stored at −80°C until further processing. The frozen samples were lyophilized, the resulting powder was dissolved in 0.6 ml D2O, and the solution was placed in a 5-mm NMR tube. High-resolution 1H MRS spectra were acquired with a 9.4 T Bruker Avance DMX400 spectrometer (Bruker BioSpin). A capillary containing 0.2 mM sodium 3-(trimethylsilyl)-(2,2,3,3,-2H4)-1-propionate (TSP) was inserted as an external reference. A zgpr pulse sequence (water suppression with presaturation pulses, Bruker) was used with a 45° flip angle, TR = 8.8 s, SW = 6775 Hz, NP = 65536, and NT = 128. Lactate secretion and glucose uptake per cell were calculated by measuring extracellular lactate and glucose concentrations using a YSI Glucose/Lactate analyzer (YSI2300 STAT Plus, YSI) before treatment and at 2, 4, 8, 24 and 48 hr after treatment and dividing the difference at each time point by cell numbers.
In vivo measurement of lactate concentration
1H Magnetic resonance spectroscopy methods were employed: We used the Hadamard-slice encoded selective multiple quantum coherence MRS pulse sequence recently developed by Pickup et al (12). This slice selective sequence enables non-invasive detection of tumor lactate while eliminating overlapping lipid signals with multiple quantum coherence filters. For normalization, a Hadamard-slice encoded spin-echo sequence was employed for measurement of water in the same voxel. We and others have previously demonstrated that normalization of metabolites to water effectively determines the absolute concentration of metabolites within a proportionality factor that depends on the degree of hydration of the tissue (13). Details of the pulse sequence parameters have been previously described (12,14). Two cell lines (DLCL2 and Ramos) were subcutaneously implanted at 107 cells per inoculum into immuno-defficient mice (nude mice for DLCL2 and SCID mice for Ramos). When the tumor volumes reached ~500 mm3, in vivo MRS experiments were started. Animals were anaesthetized with 1% isoflurane and placed in a home-built loop gap resonator (diameter = 13 mm, length = 10 mm) NMR probe. Animal body temperature was kept at 37°C by flowing warm air through the magnet. Ten mg/kg of rapamycin was administered via oral gavage to mice twice a day for the first four days and once a day afterwards.
Statistical analysis
A two-tailed equal variance unpaired Student’s t-test was used to determine p-values between data of vehicle-treated and rapamycin-treated cells. A two-tailed paired Student’s t-test was used to determine p-values between pre-treatment and post-treatment longitudinal animal data measurements. P≤0.05 was considered to be statistically significant.
RESULTS
Rapamycin inhibits mTORC1 signaling and lactate synthesis in B-cell lymphoma cells
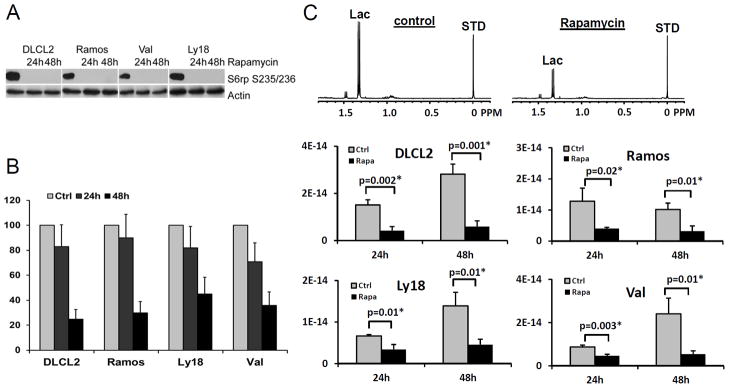
We first examined the effects of rapamycin on mTORC1 signaling and cell growth in four B-cell lymphoma cell lines, sub-classified as either diffuse large cell (DLCL2, Val, and Ly18) or Burkitt (Ramos). As shown in Fig.1A, rapamycin completely suppressed mTORC1 activity in all four cell lines as determined by loss of mTORC1-dependent S6rp phosphorylation. Furthermore, the compound profoundly inhibited cell proliferation as determined by BrdU incorporation (Fig. 1B).
Figure 1.
Rapamycin-induced inhibition of mTORC1 signaling, cell proliferation and glycolysis. The four listed B-cell lymphoma cell lines were treated with 200 nM rapamycin or medium alone for 24 hr and 48 hr and evaluated for inhibition of S6rp phosphorylation (A), BrdU incorporation (B), and intracellular lactic acid concentration (mole/cell) (C). Data are displayed as mean±SD (n=3). P-values were inserted for comparisons of vehicle- and rapamycin-treated groups. * denotes significant values. The upper panel in (C) shows 1H MRS spectra of DLCL2 cells at 24 h (control vs. rapamycin-treated); lactate (1.3 ppm) and the reference TSP peaks (0 ppm, indicated as STD).
Since we anticipated that glycolysis was a key pathway of tumor metabolism under control of mTOR, we monitored the effect of rapamycin on the cellular level of lactate, the end-product of glycolytic metabolism, by 1H MRS. Rapamycin significantly decreased intracellular lactate relative to controls in all four cell lines albeit to various extents (Fig. 1C). The relative reduction in intracellular lactate levels in rapamycin treated cells vs. controls was more pronounced after 48 hr than 24 hr exposure to rapamycin and correlated with the suppression of cell growth.
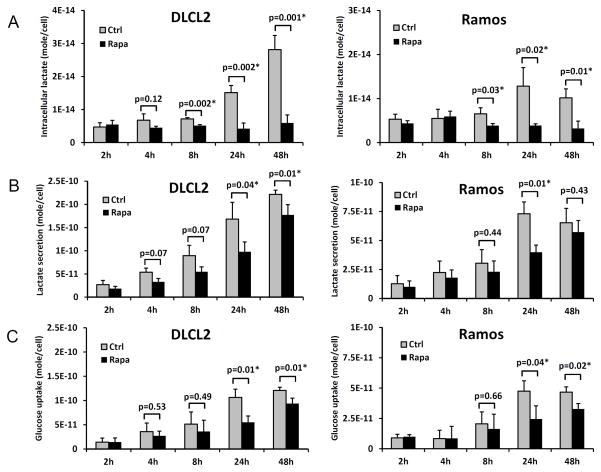
Kinetics of rapamycin-induced suppression of lactic acid synthesis
To determine how early the rapamycin-induced metabolic changes can be detected, we examined the intracellular lactic acid concentration at 2, 4, 8, 24 and 48 hr time points in two B-cell lymphoma cell lines (Fig. 2A). While 2 hr and 4 hr exposure to rapamycin did not have a definitive effect, statistically significant differences between vehicle- and rapamycin-treated cells were noted at 8 hr in both cell lines (see p-values), which increased at 24 hr and 48 hr. Lactate secretion and glucose uptake per cell calculated from changes of lactate and glucose concentrations in the medium manifested diminished glycolysis following rapamycin treatment (Fig. 2B and 2C). In contrast to intracellular lactate, lactate secretion and glucose uptake exhibited maximum differences between vehicle- and rapamycin-treated cell cultures at 24 hr. The amount of secreted lactate per cell was 3–4 orders of magnitude greater than the intracellular lactate levels, which indicates that most of the lactate generated by cellular metabolism was secreted out of the cell. Nonetheless, the level of intracellular lactate was more sensitive for distinguishing metabolic differences between vehicle- and rapamycin-treated cells than secreted lactate levels. This was in part due to the intracellular lactate concentration remaining essentially constant after rapamycin treatment (see Fig 2A).
Figure 2.
Time-course of the rapamycin-induced inhibition of glycolysis. Two B-cell lymphoma cell lines (DLCL2 and Ramos) were exposed to 200 nM of rapamycin or medium alone for the depicted time periods and examined for intracellular lactate concentration (A), lactate secretion (B) and glucose uptake (C). Data are displayed as mean±SD (n=3). * indicates statistically significant p-values.
Rapamycin-induced lactate concentration decrease correlates with inhibition of lymphoma growth in vivo
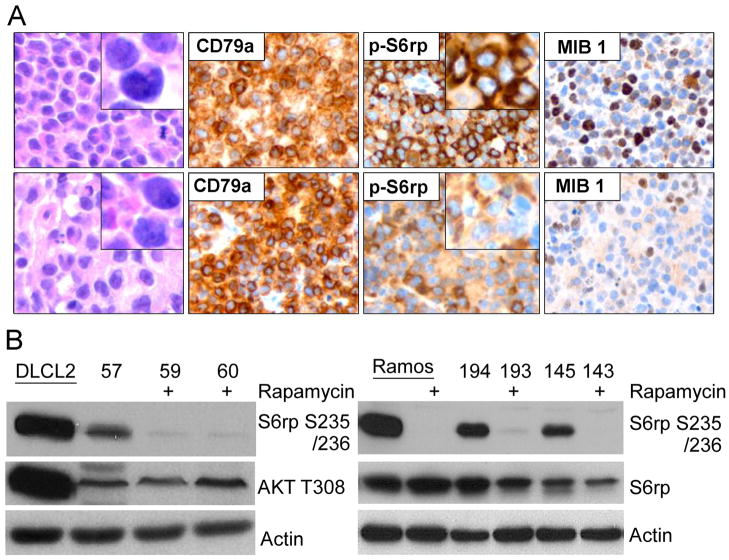
To determine if the rapamycin-induced suppression of lactate concentration can also be detected in vivo, we evaluated xenotransplanted lymphoma models, the DLCL2 and Ramos B-cell lines studied in greatest detail in vitro, as described above. The tumors displayed the appropriate transformed lymphoid cell morphology and human B-cell phenotype (Fig. 3A, left panels) confirming that they originated from the implanted lymphoma cell lines.
Figure 3.
Rapamycin-induced inhibition of mTORC1 signaling in vivo. The mice with established xenotransplants of DLCL2 and Ramos B-cell lymphoma cell lines were treated twice daily with 10 mg/kg of rapamycin for the first four days then once daily for 10 days (DLCL2) and 8 days (Ramos). A: the DLCL2 tumors were harvested at 8 hr after the last dose and microscopically evaluated for cell morphology, expression of the human B-cell markers (CD79), mTORC1 activation (p-S6rp), and proliferative rate (mib-1 expression). B: the DLCL2 and Ramos tumors (labeled with numbers) were examined for mTORC1 activation by Western blotting. In vitro cultured cell lines, either untreated (DLCL2) or untreated and rapamycin-treated (Ramos), served as controls.
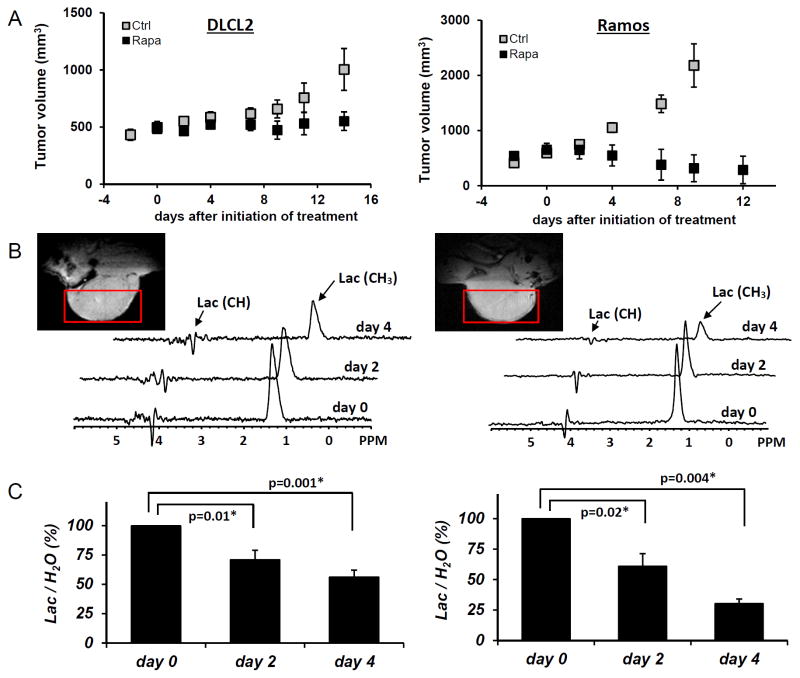
Treatment of the xenotransplanted mice with rapamycin inhibited mTORC1 signaling and decreased the proliferative rate of the lymphoma cells as determined by suppression of S6rp phosphorylation and decreased expression of the cell cycle-associated marker mib1, respectively (Fig. 3A, right panels). Effective and specific mTORC1 inhibition was confirmed in Western blots, which revealed greatly diminished S6rp phosphorylation and intact phosphorylation of Akt (Fig. 3B). Extended treatment with rapamycin resulted in the tumor growth arrest for the DLCL2 cell line and a marked decrease in tumor volume of the Ramos cell line (Fig. 4A). Note that these effects were preceded by decreases in tumor lactic acid concentrations measured by the in vivo MRS method (see Materials and Methods section). The decrease was already statistically significant at 48 hr following initiation of rapamycin treatment and increased at 96 hr (Fig. 4B and 4C). Note that at 2 days after initiation of treatment, the effect of rapamycin on tumor volume was not noticeable (see Fig. 4A) while tumoral lactate had already significantly decreased. The result strongly suggests that in vivo measurement of lactate originating from the tumor provides an early marker of the action of the mTOR inhibitor rapamycin that correlates with the tumor volume responses that are observable at later time points.
Figure 4.
Rapamycin-induced inhibition of tumor growth and glucose metabolism in vivo. Xenotransplants of DLCL2 cells (left panel) and Ramos cells (right panel). Drug treatments are in Materials and Methods. A: tumor growth delay curve with the data representing mean±SD (n≥5 in each group). B: Times course in vivo localized MRS of tumor with rapamycin treatment. Tumor MRI images and the selected voxels for MRS are also displayed. C: Lac/H2O ratios normalized to the pretreatment values for the rapamycin-treated tumors are presented as mean±SE (n=5). Statistically significant changes in Lac/H2O were observed at 48 hr and after from initiation of rapamycin treatment in both xenografts (P values are displayed. * denotes statistical significance) while control tumors (n=5) did not have significant changes in Lac/H2O until 96 hr.
Rapamycin inhibits expression of hexokinase II
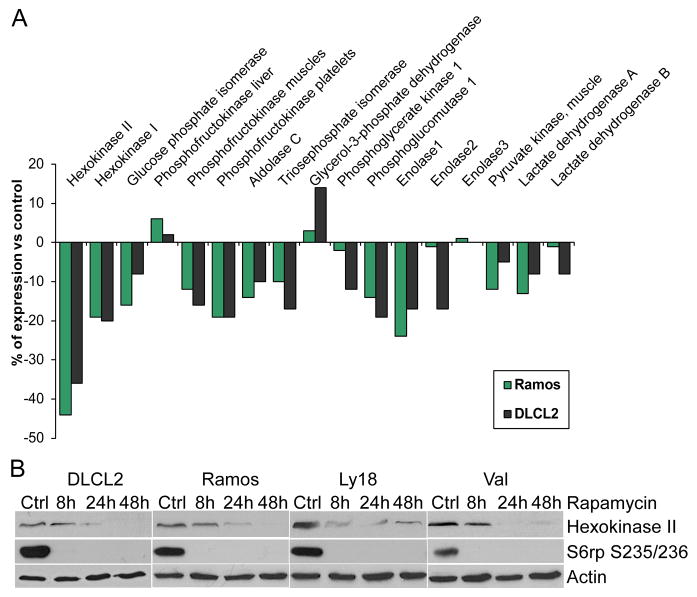
While inhibition of mTORC1 signaling by rapamycin reaches an optimum within minutes after drug administration (9,10,11), the drug’s effects on the lactic acid concentration were not detectable until 8 hr and became more pronounced at 24 hr and 48 hr in cultured cells (Fig. 2A). This marked timing difference indicates that the inhibition of glycolysis does not directly result from inhibition of the mTORC1 signaling but most likely originates from diminished expression of enzymes involved in the glycolytic pathway. To test this assumption, we performed gene expression profiling of the DLCL2 and Ramos cell lines treated with rapamycin or medium alone. The only component of the glycolytic pathway whose gene expression was significantly inhibited by the rapamycin treatment was hexokinase II, although some other glycolytic genes showed similar tendencies (Fig. 5A). The steady loss of hexokinase II expression (Fig. 5B) that paralleled the inhibition of lactate production (Fig. 2A and 2B) was identified in all four B-cell lines at the protein level.
Figure 5.
Rapamycin-induced inhibition of hexokinase II expression. A: Expression of the glycolysis-related genes in the depicted cell lines treated for 6 hr with rapamycin or medium alone. The results are presented as a percentage of the rapamycin-induced change in the gene expression with the medium-treated cells serving as the baseline value. B: Kinetics of the rapamycin-induced changes in hexokinase II protein expression.
DISCUSSION
Despite the large number of studies of kinase inhibitors as approved or potential therapeutic agents for cancer therapy, the standard methods currently used to monitor the effects of these inhibitors in vivo have a number of serious limitations (15). The most broadly used radiological methods for evaluating tumor response to therapy typically measure changes in tumor volume. However, these changes occur relatively late and can be quite subtle for some of the kinase inhibitors that often exert a cytostatic rather than cytotoxic effect. An alternative approach of obtaining tumor biopsy samples for immunohistochemical or Western blot evaluation may not be applicable to poorly accessible tumors, and the need for multiple measurements during the course of therapy is very difficult to satisfy given their invasive nature (16). Another approach is to measure tumor-associated biomarkers present in the patient’s peripheral blood. However, this method suffers from low sensitivity of detection and is prone to the technical artifacts related to sample handling (17). Finally, the analysis of the surrogate normal tissues or cells such as the blood lymphocytes to determine the therapeutic impact of the inhibitors often does not correspond to the inhibitor’s effect on the actual malignant cells.
MRS offers a non-invasive tool to repeatedly measure the metabolic status of tumors in cancer patients and animal models (15,18,19). As we have demonstrated, inhibition of the mTOR signaling pathway can be detected by 1H MRS in B-cell lymphoma cells by measuring changes in lactate concentration. Rapamycin, the inhibitor of the mTORC1 complex, decreased lactic acid production (intra- plus extracellular, Fig. 2A and 2B) in lymphoma cell line cultures or the steady state lactic acid concentration in mouse xenotransplants (Fig. 4). The inhibition of lactate production was progressive, detectable as early as 8 hr after initiation of the therapy in cell cultures, and correlated with inhibition of the lymphoma cell growth.
In the kinetic study (Fig. 2), there were time-related differences between the changes in intracellular and extracellular lactate concentration. First, intracellular and extracellular lactate of untreated DLCL2 cells increased until 48 hr. The same is true for the Ramos cells except that maximal intracellular and extracellular lactate levels were observed at 24 hr. Following treatment the intracellular lactate level of both cell lines remained essentially constant for 48 hr, while extracellular lactate increased monotonically but at a slower rate compared to controls. Tumor cells have evolved to maintain intracellular pH homeostasis despite high levels of aerobic lactic acid production as a result of the Warburg effect. This is largely accomplished through the expression of high levels of monocarboxylate transporters that facilitate the export of stoichiometric equivalents of lactate and protons. This mechanism appears to be partially overwhelmed in the absence of rapamycin as indicated by the rise in intracellular lactate in the untreated control cells. However, addition of rapamycin leads to a decrease in lactate production rates that permits re-establishment of lactate homeostasis, hence explaining the post-treatment constant level of intracellular lactate in DLCL2 and Ramos cells.
It is noteworthy that while glycolysis is expected to be the major pathway of lactate production, there are other pathways by which this metabolite is produced including glutaminolysis followed by malic enzyme or pyruvate carboxylase activlty, transamination reaction converting alanine to lactate, ets. For this reason, we have avoided referring to glycolysis as the exclusive source of lactic acid. Complete analysis of the other pathways will require 13C MRS studies of labeled substrates with appropriate metabolic network analysis (20). Such studies are planned for the future.
The diminished production of lactic acid also correlated with inhibition of the expression of hexokinase II, the critical enzyme in the glycolytic pathway. These results indicate that measuring the impact of mTOR inhibition on the metabolic status of malignant cells represents an effective and sensitive method for determining the drug’s effect and suggests that this monitoring approach should be applicable to cancer patients. Rather unexpectedly in this context, the recent evaluation of patients with several different malignancies and of the few related animal models failed to demonstrate the correlation between mTOR inhibition as detected by the positron emission tomography (PET) and the treatment outcome (21). Of note, the study did not include any of the malignancies known to respond well to an mTOR inhibitor monotherapy. In fact, none of the evaluated patients experienced an objective response. Whether this lack of the therapeutic effect resulted from overall insufficient mTOR inhibition, functional independence of the evaluated tumors from mTOR control, or other factors remains to be determined. In contrast to that study, a pre-clinical examination of glioblastoma cell lines found good correlation between the PET-detected mTOR signaling inhibition and tumor volume (22).
It is interesting to note that mTOR controls glucose metabolism at least partially by inducing the expression of hexokinase II (Fig. 5). Although it is well established that mTOR activity is controlled by the availability of glucose and other nutrients (23), this observation indicates that mTOR also actively regulates a key pathway of metabolic activity of the cancer cells. Hexokinase II is highly specific for glucose and binds the sugar with high affinity (8). Since it is bound to the outer surface of the mitochondrial membrane in close proximity to ATP synthase, hexokinase II is strategically located to access ATP as it is produced and to utilize it to phosphorylate glucose and initiate glycolytic metabolism. Therefore, by regulating its expression, mTOR effectively controls the entire glycolytic pathway and, hence, a key portion of the critical ATP-based cellular energy supply. Because hexokinase II expression rather than its enzymatic activity is affected, this control is relatively delayed leading to the detectable decrease in the lactic acid concentration after several hours and even more profoundly after a few days (Fig. 2). Taking into account the distinct kinetics of the decrease should facilitate optimal design of imaging protocols for monitoring the mTOR inhibitor therapy in cancer patients.
Accumulating evidence indicates that mTORC1 can be activated by a variety of cell-surface receptor complexes. While the original studies implicated the activation of receptors for the insulin-family growth factors (1–3), observations made by some of us (9–11) and others (24–29) demonstrated that a large spectrum of receptors and their ligands triggered mTORC1 activation. These include interleukin-2, anaplastic lymphoma kinase, CD40 ligand, notch, thyroid-stimulating hormone, fibroblast growth factor-9, polycystein-1, and prostaglandin F2a. These diverse ligand-receptor complexes activate mTORC1 through the signaling pathways PI3K/Akt (30–32), MEK/ERK (33) and possibly Syk (34). Therefore, the inhibition of these pathways should also be amenable to monitoring changes in glycolysis. Given that these pathways are frequently dysregulated in malignant cells and clinical-grade inhibitors either are already available or should soon become available, evaluation of glucose metabolism as a surrogate for monitoring mTOR activity may prove useful in the management of cancer patients treated with a variety of kinase inhibitors. Indeed, inhibition of the oncogenic BCR/ABL tyrosine kinase in the inhibitor-responsive leukemic cells led to decreased glucose uptake and lactate production (35,36). Although in those studies no signaling pathways down-stream of BCR/ABL were implicated in modulation of the glucose metabolism, in the light of our current findings, it is all but certain that mTORC1 was involved, given the capacity of BCR/ABL to activate mTORC1 (37).
Both MRS-based and PET methods should be able to detect decreases in glycolytic metabolism associated with decreased expression of hexokinase II. PET typically measures uptake of the radioactive tracer 18F fluorodeoxyglucose (FDG) by cancer cells via the GLUT1 transporter. FDG is phosphorylated by hexokinase II and trapped inside the cell until it decays or is eliminated by interaction with glucose-6-phosphatases. Whereas FDG-PET measures the initial step of glycolysis, 1H lactate MRS measures the end product of glycolysis. The major strength of FDG-PET is its high sensitivity and ability to simultaneously detect tumors in all regions of the body. The MRS methods are much less sensitive than the PET techniques, but 1H MRS lactate detection is the most sensitive of the MRS methods for detection of glycolytic activity, and it does not require a potentially harmful radioactive probe as used in PET. Moreover, 1H MRS detects metabolism along the entire glycolytic pathway, and this imaging method can be implemented on all existing MRI systems. 13C MRS is another way of detecting glycolysis but requires infusion of 13C-labeled glucose into blood, modification of the NMR instrument for 13C detection and 1H-decoupling and has significantly lower signal to noise ratio than 1H MRS; consequently, the method requires a prolonged scan time. 1H MRS lactate imaging method has recently been translated onto a clinical 3T instrument by some of us (18), A 70% decrease of lactate in the tumor of a non-Hodgkin's lymphoma patient was detected two days following initiation of multi-agent immunochemotherapy (38). Based on the results of the current study, we anticipate that 1H MRS lactate imaging could also monitor therapeutic response in patients treated with rapamycin or other signal transduction inhibitors non-invasively and early after treatment.
Acknowledgments
Supported in part by the NCI grants R01-CA89194, R01-CA96856, RO1-CA101700 and U24CA08315.
This study was supported in part by the Institute for Translational Medicine and Therapeutics' (ITMAT) Transdisciplinary Program in Translational Medicine and Therapeutics. The animal imaging studies were performed at the Small Animal Imaging Facility of the Department of Radiology of the University of Pennsylvania and the Abramson Cancer Center.
References
- 1.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 2.Petroulakis E, Mamane Y, Le Bacquer O, Shahbazian D, Sonenberg N. mTOR signaling: implications for cancer and anticancer therapy. Br J Cancer. 2006;94:195–199. doi: 10.1038/sj.bjc.6602902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corradettia MN, Guan KL. Upstream of the mammalian target of rapamycin: do all roads pass through mTOR? Oncogene. 2006;25:6347–6360. doi: 10.1038/sj.onc.1209885. [DOI] [PubMed] [Google Scholar]
- 4.Hudes G, Carducci M, Tomczak P, Dutcher J, Figlin R, Kapoor A, Staroslawska E, Sosman J, McDermott D, Bodrogi I, Kovacevic Z, Lesovoy V, Schmidt-Wolf IG, Barbarash O, Gokmen E, O'Toole T, Lustgarten S, Moore L, Motzer RJ Global ARCC Trial. Global ARCC Trial. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N Engl J Med. 2007;356:2271–2281. doi: 10.1056/NEJMoa066838. [DOI] [PubMed] [Google Scholar]
- 5.Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, Grünwald V, Thompson JA, Figlin RA, Hollaender N, Urbanowitz G, Berg WJ, Kay A, Lebwohl D, Ravaud A RECORD-1 Study Group. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372:449–456. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 6.Kim JW, Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 7.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell 2008. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 8.Mathupala SP, Ko YH, Pedersen PL. Hexokinase-2 bound to mitochondria: cancer's stygian link to the “Warburg Effect” and a pivotal target for effective therapy. Semin Cancer Biol. 2008;19:17–24. doi: 10.1016/j.semcancer.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wlodarski P, Kasprzycka M, Liu X, Marzec M, Robertson ES, Slupianek A, Wasik MA. Activation of mTOR in transformed B lymphocytes is nutrient-dependent but independent of Akt, MEK, IGF-I, and serum. Cancer Res. 2005;65:7800–7808. doi: 10.1158/0008-5472.CAN-04-4180. [DOI] [PubMed] [Google Scholar]
- 10.Marzec M, Kasprzycka M, Liu X, El-Salem M, Halasa K, Raghunath PN, Bucki R, Wlodarski P, Wasik MA. Oncogenic tyrosine kinase NPM/ALK induces activation of the rapamycin-sensitive mTOR signaling pathway. Oncogene. 2007;26:5606–5614. doi: 10.1038/sj.onc.1210346. [DOI] [PubMed] [Google Scholar]
- 11.Marzec M, Liu X, Kasprzycka M, Witkiewicz A, Raghunath PN, El-Salem M, Robertson E, Odum N, Wasik MA. IL-2- and IL-15-induced activation of the rapamycin-sensitive mTORC1 pathway in malignant CD4+ T lymphocytes. Blood. 2008;111:2181–2189. doi: 10.1182/blood-2007-06-095182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pickup S, Lee SC, Mancuso A, Glickson JD. Lactate imaging with Hadamard-encoded slice-selective multiple quantum coherence chemical-shift imaging. Magn Reson Med. 2008;60:299–305. doi: 10.1002/mrm.21659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shungu DC, Bhujwalla ZM, Li SJ, Rose LM, Wehrle JP, Glickson JD. Determination of absolute phosphate metabolite concentrations in RIF-1 tumors in vivo by 31P-1H-2H NMR spectroscopy using water as an internal intensity reference. Magn Reson Med. 1992;28:105–121. doi: 10.1002/mrm.1910280111. [DOI] [PubMed] [Google Scholar]
- 14.Lee SC, Poptani H, Pickup S, Jenkins WT, Kim S, Koch CJ, Delikatny EJ, Glickson JD. Early detection of radiation therapy response in non-Hodgkin's lymphoma xenografts by in vivo 1H magnetic resonance spectroscopy and imaging. NMR Biomed. 2010;23:624–632. doi: 10.1002/nbm.1505. [DOI] [PubMed] [Google Scholar]
- 15.Belouche-Barbari M, Chung YL, Al-Saffar NM, Falck-Miniotis M, Leach MO. Metabolic assessment of the action of targeted cancer therapeutics using magnetic resonance spectroscopy. Br J Cancer. 2009;102:1–7. doi: 10.1038/sj.bjc.6605457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Workman P, Aboagye EO, Chung YL, Griffiths JR, Hart R, Leach MO, Maxwell RJ, McSheehy PM, Price PM, Zweit J Cancer Research UK Pharmacodynamic/Pharmacokinetic Technologies Advisory Committee. Minimally invasive pharmacokinetic and pharmacodynamic technologies in hypothesis-testing clinical trials of innovative therapies. J Natl Cancer Inst. 2006;98:580–598. doi: 10.1093/jnci/djj162. [DOI] [PubMed] [Google Scholar]
- 17.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 18.Mellon EA, Lee SC, Pickup S, Kim S, Goldstein SC, Floyd TF, Poptani H, Delikatny EJ, Reddy R, Glickson JD. Detection of lactate with a hadamard slice selected, selective multiple quantum coherence, chemical shift imaging sequence (HDMD-SelMQC-CSI) on a clinical MRI scanner: Application to tumors and muscle ischemia. Magn Reson Med. 2009;62:1404–1413. doi: 10.1002/mrm.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SC, Arias-Mendoza F, Poptani H, Delikatny EJ, Wasik M, Marzec M, Schuster SJ, Nasta SD, Svoboda J, Goldstein SC, Arias-Mendoza F, Glickson JD. Molecular imaging of cancer: Prediction and early detection of response by NMR spectroscopy and imaging. International Drug Discovery. 2011 Aug-Sep;:20–23. doi: 10.1016/j.cpet.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shestov AA, Mancuso A, Leeper DB, Glickson JD. Metabolic network analysis of melanoma: How much energy comes from aerobic glycolysis?. Proceedings of the 39th annual meeting of the International Society on Oxygen Transport to Tissue; Washington, D.C., USA. 2011. p. 101. [Google Scholar]
- 21.Ma WW, Jacene H, Song D, Vilardell F, Messersmith WA, Laheru D, Wahl R, Endres C, Jimeno A, Pomper MG, Hidalgo M. (18F) fluorodeoxyglucose positron emission tomography correlates with Akt pathway activity but is not predictive of clinical outcome during mTOR inhibitor therapy. J Clin Oncol. 2009;27:2697–2704. doi: 10.1200/JCO.2008.18.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei LH, Su H, Hildebrandt IJ, Phelps ME, Czernin J, Weber WA. Changes in tumor metabolism as readout for Mammalian target of rapamycin kinase inhibition by rapamycin in glioblastoma. Clin Cancer Res. 2008;14:3416–3426. doi: 10.1158/1078-0432.CCR-07-1824. [DOI] [PubMed] [Google Scholar]
- 23.Peng T, Golub TR, Sabatini DM. The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol Cell Biol. 2002;22:5575–5584. doi: 10.1128/MCB.22.15.5575-5584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakata A, Kuwahara K, Ohmura T, Inui S, Sakaguchi N. Involvement of a rapamycin-sensitive pathway in CD40-mediated activation of murine B cells in vitro. Immunol Lett. 1999;68:301–309. doi: 10.1016/s0165-2478(99)00053-x. [DOI] [PubMed] [Google Scholar]
- 25.Chan SM, Weng AP, Tibshirani R, Aster JC, Utz PJ. Notch Signals Positively Regulate Activity of the mTOR Pathway in T Cell Acute Lymphoblastic Leukemia. Blood. 2007;110:278–286. doi: 10.1182/blood-2006-08-039883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suh JM, Song JH, Kim DW, Kim H, Chung HK, Hwang JH, Kim JM, Hwang ES, Chung J, Han JH, Cho BY, Ro HK, Shong M. Regulation of the phosphatidylinositol 3-kinase, Akt/protein kinase B, FRAP/mammalian target of rapamycin, and ribosomal S6 kinase 1 signaling pathways by thyroid-stimulating hormone (TSH) and stimulating type TSH receptor antibodies in the thyroid gland. J Biol Chem. 2003;278:21960–21971. doi: 10.1074/jbc.M300805200. [DOI] [PubMed] [Google Scholar]
- 27.Wing L-Y, Chen H-M, Chuang P-C, Wu M-H, Tsai S-J. The mTOR-S6K1 but not PI3K-Akt signaling is responsible for fibroblast growth factor-9-induced cell proliferation. J Biol Chem 2005. 2005;280:19937–19947. doi: 10.1074/jbc.M411865200. [DOI] [PubMed] [Google Scholar]
- 28.Shillingford JM, Murchid NS, Larson CH. The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc Natl Acad Sci USA. 2006;103:5466–5471. doi: 10.1073/pnas.0509694103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arvisais EM, Romanelli A, Hou X, Davis JS. AKT-independent phosphorylation of TSC2 and activation of mTOR and ribosomal protein S6 kinase signaling by prostaglandin F2alpha. J Biol Chem. 2006;281:26904–26913. doi: 10.1074/jbc.M605371200. [DOI] [PubMed] [Google Scholar]
- 30.Navé BT, Ouwens M, Withers DJ, Alessi DR, Shepherd PR. Mammalian target of rapamycin is a direct target for protein kinase B: Identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem J. 1999;344:427–431. [PMC free article] [PubMed] [Google Scholar]
- 31.Sekulić A, Hudson CC, Homme JL, Yin P, Otterness DM, Karnitz LM, Abraham RT. A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells. Cancer Res. 2000;60:3504–3513. [PubMed] [Google Scholar]
- 32.Tee AR, Anjum R, Blenis J. Inactivation of the tuberous sclerosis complex-1 and -2 gene products occurs by phosphoinositide 3-kinase/Akt-dependent and -independent phosphorylation of tuberin. J Biol Chem. 2003;278:37288–37296. doi: 10.1074/jbc.M303257200. [DOI] [PubMed] [Google Scholar]
- 33.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 34.Leseux L, Hamdi SM, Al Saati T, Capilla F, Recher C, Laurent G, Bezombes C. Syk-dependent mTOR activation in follicular lymphoma cells. Blood. 2006;108:4156–4162. doi: 10.1182/blood-2006-05-026203. [DOI] [PubMed] [Google Scholar]
- 35.Gottschalk S, Anderson N, Hainz C, Eckhardt SG, Serkova NJ. Imatinib (STI571)-mediated changes in glucose metabolism in human leukemia BCR-ABL-positive cells. Clin Cancer Res. 2004;10:6661–6668. doi: 10.1158/1078-0432.CCR-04-0039. [DOI] [PubMed] [Google Scholar]
- 36.Kominsky DJ, Klawitter J, Brown JL, Boros LG, Melo JV, Eckhardt SG, Serkova NJ. Abnormalities in glucose uptake and metabolism in imatinib-resistant human BCR-ABL-positive cells. Clin Cancer Res. 2009;15:3442–3450. doi: 10.1158/1078-0432.CCR-08-3291. [DOI] [PubMed] [Google Scholar]
- 37.Carayol N, Katsoulidis E, Sassano A, Altman JK, Druker BJ, Platanias LC. Suppression of programmed cell death 4 (PDCD4) protein expression by BCR-ABL-regulated engagement of the mTOR/p70 S6 kinase pathway. J Biol Chem. 2008;283:8601–8610. doi: 10.1074/jbc.M707934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee SC, Poptani H, Delikatny EJ, Pickup S, Nelson DS, Schuster SJ, Nasta SD, Svoboda J, Goldstein SC, Wallace SG, Loevner LA, Mellon EA, Reddy R, Glickson JD. NMR metabolic and physiological markers of therapeutic response. Adv Exp Med Biol. 2011;701:129–135. doi: 10.1007/978-1-4419-7756-4_18. [DOI] [PubMed] [Google Scholar]