Abstract
A ‘shotgun’ lipidomics strategy consisting of sequential functional group selective chemical modification reactions coupled with high-resolution / accurate mass spectrometry and ‘targeted’ tandem mass spectrometry (MS/MS) analysis has been developed and applied toward the comprehensive identification, characterization and quantitative analysis of changes in relative abundances of >600 individual glycerophospholipid, glycerolipid, sphingolipid and sterol lipids between a primary colorectal cancer (CRC) cell line, SW480, and its isogenic lymph node metastasized derivative, SW620. Selective chemical derivatization of glycerophosphoethanolamine and glycerophosphoserine lipids using a ‘fixed charge’ sulfonium ion containing, d6-S,S′-dimethylthiobutanoylhydroxysuccinimide ester (d6-DMBNHS) reagent was used to eliminate the possibility of isobaric mass overlap of these species with the precursor ions of all other lipids in the crude extracts, thereby enabling their unambiguous assignment, while subsequent selective mild acid hydrolysis of plasmenyl (vinyl-ether) containing lipids using formic acid enabled these species to be readily differentiated from isobaric mass plasmanyl (alkyl-ether) containing lipids. Using this approach, statistically significant differences in the abundances of numerous lipid species previously identified as being associated with cancer progression, or that play known roles as mediators in a range of physiological and pathological processes, were observed between the SW480 and SW620 cells. Most notably, these included increased plasmanylcholine and triglyceride lipid levels, decreased plasmenylethanolamine lipids, decreased C-16 containing sphingomyelin and ceramide lipid levels, and a dramatic increase in the abundances of total cholesterol ester and triglyceride lipids in the SW620 cells compared to those in the SW480 cells.
Introduction
Lipids comprise the majority of cellular membranes and are necessary for several cellular functions including structure and signaling1,2. Numerous studies have demonstrated that disruption of lipid metabolism and/or signaling is associated with the onset and progression of several human cancers including lung3, prostate3,4, breast3,5,6, brain7, liver8 and colorectal9–12. Notably, changes in the abundances of ether-linked glycerophosphocholine (PC) and glycerophosphoethanolamine (PE) lipids have been correlated with malignancy and metastatic potential in various cancers3. Specifically for colorectal cancer (CRC), the third leading cause of cancer deaths in the United States13, increased plasmanyl-PC lipid levels and significantly decreased plasmenyl-PE lipids have been reported in malignant colon cancer versus normal tissue11,12. At present, however, little information is available regarding the changes in ‘global’ lipid metabolism profiles that occur between normal and diseased cells, tissues or readily accessible bodily fluids as a function of the onset and progression of CRC. Such information could provide further insights into the biological mechanisms by which lipids are associated with cancer development, or lead to the identification of effective biomarker signatures of the disease, e.g., to differentiate between benign, primary versus metastatic cancers, or serve as targets for therapeutic intervention.
Mass spectrometry (MS) and tandem mass spectrometry (MS/MS) methods are an excellent choice for comprehensive lipidome analysis, providing capabilities for rapid and sensitive monitoring of the molecular compositions and abundances of individual lipid species in complex extracts obtained from limited quantities of tissue, with minimal need for sample preparation14. However, there are several caveats associated with the use of MS methods for lipidome analysis. First and foremost is the structural diversity and complexity of the lipid mixtures that can result from extraction of a given cell or tissue. There are eight general classes of lipids including fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterol lipids, prenol lipids, saccharolipids and polyketides, each with their own subclasses. Glycerophospholipids, for example, may contain any of seven different head groups linked to the sn-3 position of the glycerol backbone, as well as fatty acyl chains with variable carbon lengths and degrees of unsaturation attached to the sn-1 and sn-2 positions. Adding to this diversity, acyl chains typically linked to the sn-1 position may alternatively be substituted with an ether linkage, most commonly consisting of a plasmenyl vinyl-ether (i.e., -O-alk-1′-enyl or plasmalogen) bond, or by an plasmanyl alkyl-ether (i.e., 1-O-alkyl) bond, while hydrolysis of the acyl chain from the sn-2 position results in the formation of a lysophospholipid species. Individual lipids containing different head-group and acyl-chain compositions, but with the same nominal mass, may not be identified on the basis of their m/z values alone when using mass analyzers with only unit mass resolving power (e.g., ion traps or triple quadrupoles). Furthermore, other lipids of the same class but belonging to different subclasses may yield ions with isomeric molecular structures, and therefore the same exact m/z, so cannot be distinguished from one other even when using high resolution/accurate mass instrumentation (e.g., plasmanyl lipids containing one additional site of unsaturation compared to plasmenyl containing lipids, both with the same head group (i.e., [PC(P- 36:0)+H]+ = [PC(O-36:1)+H]+)). Additionally, isomeric lipid species with the same head group but differing in their individual sn-1 and sn-2 acyl chain compositions (e.g., [PC(16:0/22:6)+H]+ and [PC(18:1/20:4) )+H]+), will also be present at the same exact m/z value. Also, lipids that have different molecular elemental compositions may actually yield ions with the same ionic elemental compositions (e.g. [PE(32:0)+H]+ = PC(29:0)+H]+ = [PA(34:1)+NH4]+, and [PS(36:0)+H]+ = PG(36:2)+NH4]+), depending on the ionic adduct that is formed upon ESI-MS. Finally, low abundance lipid species that are present at or below the level of chemical noise present within a mass spectrum are often not identified by MS measurements alone.
Traditionally, to overcome these issues when using direct infusion ‘shotgun’ MS14, identification, structural characterization and quantitative analysis has been accomplished using neutral loss and precursor ion MS/MS scans in triple quadrupole mass spectrometers15,16, or product ion MS/MS scans in an ‘ion mapping’ mode in high resolution quadrupole time-of-flight mass spectrometers17. However, the throughput of these approaches can be limited as individual scans are required for the analysis of each different lipid class or precursor ion m/z value. Alternatively, liquid chromatography (LC) may be employed for the separation of lipids prior to MS and/or MS/MS analysis18, resulting in less complex spectra and potentially enabling the separation of individual lipids with the same nominal or exact m/z values. However, the analysis times associated with these approaches can be relatively slow, and the quantitation of individual lipids can be compromised in complex mixtures19.
The recent development of ultra-high resolution/accurate mass analyzers based on FT-ICR or Orbitrap platforms has opened the door to performing high-throughput direct infusion ‘shotgun’ mass spectrometry for the simultaneous analysis of multiple lipid classes, without need for prior separation and reduced need for extensive MS/MS analysis20. Ultra-high resolution (≥100,000) provides the ability to separate lipid ions with the same nominal m/z values, with substantially improved signal-to-noise and dynamic range for the detection of low abundance lipid species compared to lower resolution instruments. However, the resolution of isobaric mass lipid ions remains an issue. One approach to overcome this issue has been to perform multistep lipid extractions to separate apolar lipids from polar lipids20. However, this requires additional extraction steps which could result in increased experimental error for subsequent quantitative analysis. Negative ion mode MS can be used to eliminate the possibility of m/z overlap for certain lipids. However, the ionization efficiency of many lipid classes in negative ionization mode can be much lower than in positive ionization mode unless different solvent systems are used20, potentially limiting the ability to identify lipids present at trace abundances using this method alone. Furthermore, some lipids that have different molecular elemental compositions can yield ions with the same ionic elemental compositions in negative ionization mode, depending on the ionic adduct that is formed upon ESI-MS (e.g., PC(32:2)+Acetate]− = [PS(36:1)-H]− and [PC(32:2)+Formate]− = [PS(35:1)-H]−). Finally, while differentiation of isobaric mass plasmenyl- and plasmanyl-ether containing glycerophospholipids can be achieved using CID-MS3,21, the practical application of this approach for the analysis of very low abundant species is quite limited. Thus, further developments are required in order for ultra-high resolution/accurate mass spectrometry to become more generally applicable for use in quantitative lipidome profiling.
Materials and Methods
Lipid Extraction
Extraction of lipids from 2×107 SW480 and SW620 lyophilized cells was performed using a modified Folch method, as previously described22,23. Stock solutions were prepared by dissolving crude lipid extracts in 600μL of 4:2:1 IPA/MeOH/CHCl3. Immediately prior to ESI-MS analysis, 5μL of each stock lipid extract, and 10μL each of 10μM PC(14:0/14:0) and PE(14:0/14:0) and 3.25μL PS(14:0/14:0) internal lipid standards were dried under a stream of nitrogen, then re-dissolved in 200μL of 4:2:1 IPA/MeOH/CHCl3 containing either 20mM ammonium acetate or 20mM ammonium formate.
Derivatization of aminophospholipids with d6-S,S′-dimethylthiobutanoylhydroxysuccinimide ester (d6-DMBNHS)
5μL of the stock lipid extract, 10μL each of 10μM PC(14:0/14:0) and PE(14:0/14:0) and 3.25μL PS(14:0/14:0) were dried under a stream of nitrogen, then redissolved in 40μL of 39:1.1 CHCl3 containing 0.0125M TEA and vortexed for 30sec. 1μL of 0.0125M d6-DMBNHS24 in DMF was then added to the lipid mixture and vortexed for 30min. The reaction was quenched by drying under a stream of nitrogen then re-dissolved in 200μL 4:2:1 IPA/MeOH/CHCl3 containing 20mM ammonium formate for immediate analysis by ESI-MS.
Mild acid hydrolysis of plasmenyl-ether containing lipids
Following derivatization with d6-DMBNHS as described above, then drying under N2, 40μL of 80% formic acid in 4:2:1 IPA/MeOH/CHCl3 was added to the sample and allowed to react for 1min. The solvent was then immediately evaporated under reduced pressure and samples were re-dissolved in 200μL 4:2:1 IPA/MeOH/CHCl3 containing 20mM ammonium formate for immediate analysis by ESI-MS.
Mass Spectrometry Analysis
Samples (n=5 replicates) were introduced into a high resolution / accurate mass Thermo Scientific model LTQ Orbitrap Velos mass spectrometer (San Jose, CA) using an Advion Triversa Nanomate nano-electrospray ionization (nESI) source (Advion Ithaca, NY). Mass spectra were acquired in positive and negative ionization modes using the FT analyzer operating at 100,000 resolving power. Assignment of the molecular lipid compositions25 of selected ions from the underivatized sample was achieved using Higher-Energy Collision Induced Dissociation (HCD-MS/MS) and/or ion trap CID-MS/MS and -MSn. Further details are available in the attached Supplemental Information.
Data Analysis
Lipid identification (i.e., assignment of the lipid headgroup, the nature of the linkage of the hydrophobic tails (i.e., diacyl versus alkyl or alkenyl) and the total number of carbons and degree of unsaturation) and relative quantification was performed using the Lipid Mass Spectrum Analysis (LIMSA) v.1.0 software linear fit isotope correction algorithm26, in conjunction with an ‘in-house’ developed database of hypothetical lipid compounds for automated peak finding and correction of 13C isotope effects. Further details are available in the attached Supplemental Information.
Results
‘Shotgun’ high resolution/accurate mass spectrometry and functional group specific chemical modification for comprehensive lipid identification
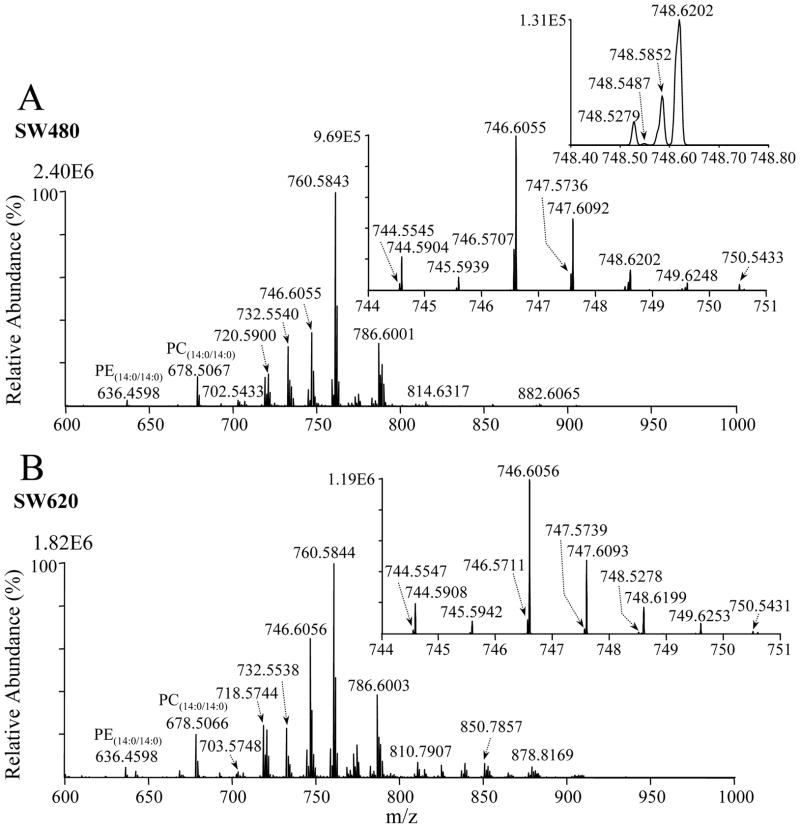
The mass spectra (m/z 600–1000) obtained following positive ionization mode ESI-MS analysis of the crude lipid extracts from the primary CRC cell line, SW480, and its isogenic metastatic derivative, SW620, are shown in Figures 1A and 1B, respectively. The addition of 20 mM ammonium formate to the spray solution was found to provide an approximately two-fold increase in ion abundance in positive ion mode compared to the use of 20 mM ammonium acetate (data not shown); therefore, ammonium formate was employed for all the results shown herein. A number of qualitative differences in the presence and/or abundance of several ions between the SW480 and SW620 cell lipid extracts are immediately apparent. For example, the ion at m/z 746.6055 and numerous ions within the m/z range of 800–920 have all increased in abundance between the SW480 and SW620 extracts, relative to that of the internal standard PC(14:0/14:0) lipid, while the ion at m/z 760.5861 has decreased in abundance.
Figure 1.
Positive ion mode ESI high-resolution mass spectrometric analysis of crude lipid extracts from the human adenocarcinoma cell lines (A) SW480 and (B) SW620. An internal standard lipid PC(14:0/14:0) was included to enable relative quantification of the observed lipid species, while the internal standard lipids PE(14:0/14:0) and PS(14:0/14:0) (not labeled) were included to allow completion of subsequent reaction with d6-DMBNHS to be monitored.
Based on the high resolution performance and accurate mass values obtained using the Orbitrap, the sum compositions25 of the glycerolipid, sphingolipid, cholesterol, cholesterol ester and PI lipid ions, including (i) assignment of the head group identities, (ii) the nature of their hydrophobic chain linkages (i.e., diacyl versus plasmanyl- or plasmenyl-ether) and (iii) the total number of carbons and double bonds that were present within their hydrophobic chains, could be automatically assigned by in silico comparison with the calculated masses of a database of hypothetical lipid compounds, using the LIMSA software (Table 1 and Supplemental Tables 1–15). For example, m/z 850.7857 in Figure 1A was assigned as the ammonium ion adduct of a TG(50:1) lipid (calculated m/z 850.7858), with the experimental and calculated m/z values differing by only 0.0001 Da (0.08 ppm). For saturated plasmanyl-ether containing lipids, assignment of the lipid identity was also straightforward, while for ether linked lipids containing at least one site of unsaturation, assignment of lipid identity was more complicated as these could overlap in exact mass with plasmenyl-ether containing lipid ions with the same total number of carbons but one less site of unsaturation.
Table 1.
Summary of the number of assigned lipid ions, identified lipids, and total lipid species from crude lipid extracts derived from the SW480 and SW620 CRC cell lines.
| SW480 | SW620 | |||||
|---|---|---|---|---|---|---|
| Lipid Class | Assigned Lipid Ions1 | Identified Lipids2 | Total Lipids3 | Assigned Lipid Ions1 | Identified Lipids2 | Total Lipids3 |
| PC | 85 | 93 | 130 | 80 | 125 | 154 |
| PE | 77 | 154 | 164 | 60 | 149 | 158 |
| PA | 15 | 30 | 30 | 12 | 26 | 26 |
| PS | 25 | 62 | 62 | 27 | 74 | 74 |
| PI | 24 | 33 | 39 | 19 | 32 | 36 |
| PG | 7 | 9 | 9 | 7 | 9 | 9 |
| LPC | 4 | 4 | 4 | 5 | 5 | 5 |
| LPE | 15 | 18 | 18 | 15 | 19 | 19 |
| LPS | 5 | 5 | 5 | 5 | 5 | 5 |
| SM | 17 | 1 | 17 | 17 | 1 | 17 |
| Cer | 6 | 0 | 6 | 6 | 0 | 6 |
| TG | 30 | 60 | 68 | 59 | 96 | 133 |
| DG | 24 | 0 | 27 | 26 | 0 | 30 |
| MG | 14 | 15 | 15 | 14 | 16 | 16 |
| Chol | 1 | 1 | 1 | 1 | 1 | 1 |
| Chol ester | 5 | 5 | 5 | 15 | 15 | 15 |
| Total | 354 | 490 | 600 | 368 | 573 | 694 |
Total number of peaks identified for each lipid sub-class, for which the sum composition (i.e., the identity of the individual headgroup and the combined total number of carbons and double bonds within the acyl, alkyl and/or alkenyl chains) could be assigned from the mass spectra.
Identified lipids categorizes those species whose molecular lipid identities of their individual fatty acyl, alkyl and/or alkenyl chains could be determined by MS/MS.
Total lipids includes the combined number of lipids whose identities of their individual fatty acyl, alkyl and/or alkenyl chains could be determined by MS/MS, as well as those lipids where only the total number of carbons and double bonds within the acyl, alkyl and/or alkenyl chains could be assigned from the mass spectra.
The sum compositions of PC, PE, PA, lipid species could also not be assigned directly from their accurate mass values alone, due to the possibility of exact mass overlap between protonated PE lipid ions with the ammonium ion adducts of PA lipids containing 2 additional carbons and 1 additional double bond, as well as with odd-numbered chain length protonated PC ions containing 3 additional carbons. (e.g. m/z 744.5545 in Figure 1A, could be [PC(33:2)+H]+ and/or [PE(36:2)+H]+ and/or [PA(38:3)+NH4]+; calculated exact m/z=744.5537). Similarly, protonated PS lipids can also overlap in exact mass with the ammonium ion adducts of PG lipids containing 2 additional double bonds.
The presence of unsaturated PC, PE and PA, or PS and PG phospholipids containing plasmanyl- or plasmenyl-ether linkages introduces even greater complexity. For example, m/z 744.5904 in the inset to Figure 1A could be assigned as [PC(P-34:1) and/or PC(O-34:2)+H]+ and/or [PE(P-37:1) and/or PE(O-37:2)+H]+ and/or [PA(P-39:2) and/or PA(O-39:3)+NH4]+, while m/z 748.5487 could be assigned as [PS(P-34:0) and/or PS(O-34:1)+H]+ and/or [PG(P-34:2) and/or PG(O-34:3)+NH4]+. Note that shoulders can be seen on the low m/z side of the ions at m/z 748.5852 and 748.6202, which correspond to the M+2 isotope peaks of the m/z 746.5707 and 746.6055 ions, respectively. Although these ions cannot be fully resolved at the 100,000 mass analyzer resolving power available on the mass spectrometer employed in this study, their presence does not impede either the assignment of lipid identities, or their quantification using the LIMSA software.
To overcome the limitations associated with the identification of isobaric PC, PE and PA, or PS and PG lipids, as well as to enable the differentiation of plasmanyl- and plasmenyl-ether lipid containing ions, a sequential chemical modification strategy involving selective amino-functional group derivatization of PE and PS lipids, followed by selective mild formic acid hydrolysis of plasmenyl-ether containing lipids, was developed. A particular advantage of this approach is that no additional sample extraction steps are required, with only a minimal increase in sample handling, thereby minimizing the possibility of increased sample handling related variances for subsequent quantitative analysis. Derivatization with the ‘fixed’ charge sulfonium ion containing reagent, d6-DMNBHS, shifts the m/z values of the amino functional group containing PE and PS ions by 136.0829 mass units, thereby separating them from PC or PA and PG lipid ions, respectively, allowing the headgroups and total number of carbons and double bonds within the ions from each lipid class to be individually assigned and quantified. A range of derivatization reagents and techniques have previously been employed to improve ionization efficiency, facilitate quantitation, or enhance the structural characterization of lipids using ESI-MS and -MS/MS strategies27–32. For example, Han et al described a method involving 9-fluorenylmethoxylcarbonylchloride (Fmoc-Cl) derivatization of PE and lysoPE lipids in extracts from mouse retinas to provide enhanced sensitivity for their identification and quantification in negative ion mode MS and MS/MS31. Zemski Berry et al. reported a similar strategy involving isotopically labeled N-methylpiperazine acetic acid NHS ester29 or 4-(dimethylamino)benzoic acid NHS ester30 reagents for the quantitative analysis of diacyl, ether, and plasmalogen PE lipids in a RAW 264.7 macrophage cell line. Of particular relevance to the work described herein, Han et al. noted an additional significant benefit of derivatization, i.e., that PE species are shifted in mass from the region of the mass spectrum where they could potentially overlap in nominal mass with other lipid classes (e.g., phosphatidylinositol), thereby facilitating the quantification of both species31.
The completeness of the d6-DMNBHS derivatization reaction performed here was demonstrated to be greater than 95%, by monitoring for the disappearance of the MS precursor ion abundances from the PE(14:0/14:0) and PS(14:0/14:0) internal standards. An overall uniform decrease in absolute abundance of non-derivatized lipids of approximately 30% was observed after the d6-DMNBHS derivatization reaction, presumably due to the presence of the excess derivatization reagent causing some ionization suppression in the sample. The d6-containing DMBNHS reagent was used in order to minimize the possibility of overlap with other ions that may be present within the m/z range where the derivatized PE and PS lipids. However, at the 100,000 mass resolving power available using the Orbitrap, the m/z of some low mass d6-DMBNHS derivatized PE lipids can partially overlap with the m/z values of high mass PC lipids, such that the LIMSA data analysis software is unable to differentiate between these ions. In these instances, the PC ion abundances determined from the underivatized sample were manually subtracted from the d6-DMBNHS derivatized ion abundances prior to quantification of the PE lipids. A13C-labeled DMBNHS reagent, for which no overlap can occur, is planned for use in future studies to fully overcome this issue.
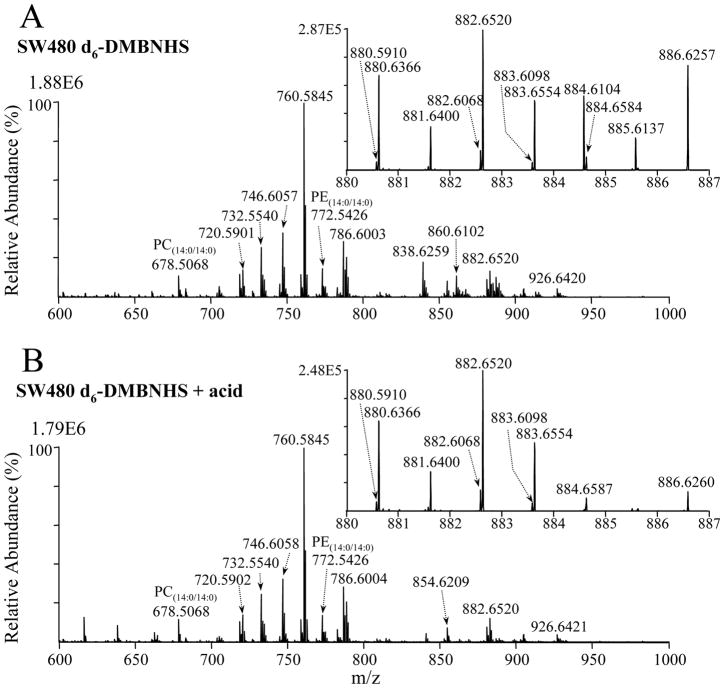
The mass spectrum obtained following d6-DMNBHS derivatization of the SW480 cell line crude lipid extract is shown in Figure 2A. It can be seen that several ions have either disappeared or reduced in abundance, or have appeared in the higher m/z range compared to the underivatized spectrum in Figure 1A, indicative of the presence of derivatized PE and PS lipids.
Figure 2.
Positive ion mode ESI high-resolution mass spectrometric analysis of a crude lipid extract from the SW480 human adenocarcinoma cell line, obtained (A) after reaction with the amine-specific derivatization reagent, d6-DMBNHS, and (B) after reaction with d6-DMBNHS followed by mild formic acid hydrolysis.
For example, the ions at m/z 748.5279 and m/z 748.5487 in Figure 1A are not present in Figure 2A (see an expanded m/z range of the spectrum in Supplemental Figure S1A). This, together with the appearance of the corresponding d6-DMBNHS derivatized ions at m/z 884.6104 and 884.6316, respectively (compare the inset to Figure 2A with the expanded m/z range of the underivatized spectrum in Supplemental Figure S2) allows these two ions to be assigned exclusively as containing [PE(P-38:6) and/or PE(O-38:7)+H]+ and [PS(P-34:0) and/or PS(O-34:1)+H]+ lipids, respectively. Similarly, m/z 744.5545, that was observed to almost (but not completely) disappear upon derivatization, and the derivatized ion appearing at m/z 880.6366, indicated the presence of a major [PE(36:2)+H]+ lipid (along with a minor [PC(33:2)+H]+ and/or [PA(38:3)+NH4]+ lipid). In contrast, m/z 746.5707 was observed to decrease in relative abundance by only approximately 30% upon derivatization (Supplemental Figure S1A), with a corresponding derivatized ion appearing at m/z 882.6520, indicating the presence of both [PE(36:1)+H]+ and the isobaric mass [PC(33:1)+H]+ and/or [PA(38:2)+NH4]+ lipids at this m/z value.
Typically, odd-numbered chain length fatty acyl-containing lipids in mammalian systems are considered to be relatively low in concentration compared to even chain fatty acyls33. Note, however, that low concentration odd-numbered-chain length protonated PC lipids could result in the formation of highly abundant ions at the same m/z as even chain length ammonium adducts of PA lipids, due to the substantially different electrospray ionization potentials for these species in positive ionization mode. Furthermore, the presence of abundant odd-numbered carbon fatty acyl-containing phospholipids have recently been reported in lipidomic analyses of macrophages and other human tissue and cell extracts34. Here, the presence of odd-numbered chain length PC lipids was first confirmed by ESI-MS HCD-MS/MS in positive and negative ion modes. Then, to determine the relative contributions of overlapping even chain PA ions with odd-numbered chain length PC ions at a given m/z value in positive ion mode, the lipid extracts were also analyzed by negative ionization mode high resolution/accurate mass ESI-MS, where no overlap of PA lipids with other lipid species can occur. By measuring the ratio of the positive and negative ion mode abundances for a series of PA lipid standards, an appropriate correction factor could be applied to the positive ion mode abundances. After performing this correction, with the exception of four cases in which both even-numbered chain length PA and odd-numbered chain length PC ions were found to be present (and when overlap was possible), only the odd-numbered chain length PC lipids were observed. For example, m/z 748.5853 in Supplemental Figure S1A, could be exclusively assigned as containing [PC(33:0)+H]+ rather than [PA(38:1)+NH4]+.
Although the d6-DMNBHS derivatization reaction allowed the PE lipid containing ions to be effectively resolved from PC and/or PA, and the PS lipid containing ions to be resolved from PG, the differentiation of isobaric unsaturated plasmanyl-ether and plasmenyl-ether containing lipids could not be achieved using this strategy alone. Murphy et al. previously demonstrated that plasmenyl lipids could be selectively acid hydrolyzed by extraction in a 0.2N HCl solution35. However, this requires an additional liquid-liquid extraction step prior to analysis, which may result in increased sample loss or variance for subsequent quantitative analysis. In contrast, we determined here that 80% formic acid can also be used to selectively and completely hydrolyze plasmenyl-ether containing lipids, without degradation of diacyl or plasmanyl-ether lipid containing species, and without requirement for additional extraction steps prior to analysis. The mass spectrum obtained following formic acid hydrolysis of the d6-DMNBHS derivatized SW480 extract is shown in Figure 2B. The ion at m/z 746.6057 in Figure 2A, potentially containing [PC(O-34:1)+H]+ and/or [PC(P-34:0)+H]+ lipids, underwent essentially no change in abundance after formic acid treatment, and therefore could be assigned as exclusively containing a [PC(O-34:1)+H]+ plasmanyl-ether lipid. In contrast, m/z 884.6104 in Figure 2A, potentially containing [PE(O-38:7)+d6-DMBNHS+H]+ and/or [PC(P-38:6)+d6-DMBNHS+H]+ lipids, had completely disappeared upon hydrolysis, and could therefore be assigned as containing the PE(P-38:6) plasmanyl-ether lipid.
Enhanced PE and PS lipid ionization sensitivity upon derivatization with d6-DMNBHS
Along with the ability to resolve PE lipids from PC and PA, and PS lipids from PG, derivatization with the d6-DMBNHS reagent provides an additional benefit of significantly increasing PE and PS lipid ionization efficiencies, thereby enhancing the dynamic range for their detection and quantification. For example, m/z 748.5487 ([PS(O-34:1)+H]+) had an intensity of 1.26E3 in the underivatized spectrum in Figure 1A, (note that this ion is present at a relative abundance of only 0.05% compared to the most abundant ion), but yielded a derivatized ion (m/z 884.6316) in Figure 2A with an intensity of 2.99E3. Overall, the average fold change increase in intensity after derivatization was found to be 3.31 ± 0.37 for the PS lipids and 6.08 ± 0.39 for the PE lipids. These results are similar to those previously reported using other aminophospholipid derivatization strategies30–32, as well as to those results recently reported from use of the DMBNHS reagent to enhance the ionization sensitivity of derivatized phosphopeptides during RP-HPLC-ESI-MS36. Notably, the increased ion abundances observed from the d6-DMBNHS derivatized reaction allowed numerous PE and PS lipids to be identified that otherwise would have been too low in abundance to assign from the underivatized spectrum.
Quantification of Changes in Lipid Abundances between SW480 and SW620 cell lines
The combination of high-resolution/accurate mass, d6-DMBNHS derivatization and formic acid treatment resulted in assignment of the sum compositions for 354 and 368 lipid containing ions from the SW480 and SW620 cell lipid extracts, respectively (Table 1), in terms of their individual headgroup identities and the total number of carbons and double bonds within the acyl, plasmanyl and/or plasmenyl chains. The abundances of the ions corresponding to each assigned lipid were then expressed as a percentage of the total ion abundance for a given lipid class, to allow quantitative comparison between the SW480 and SW620 cell lines.
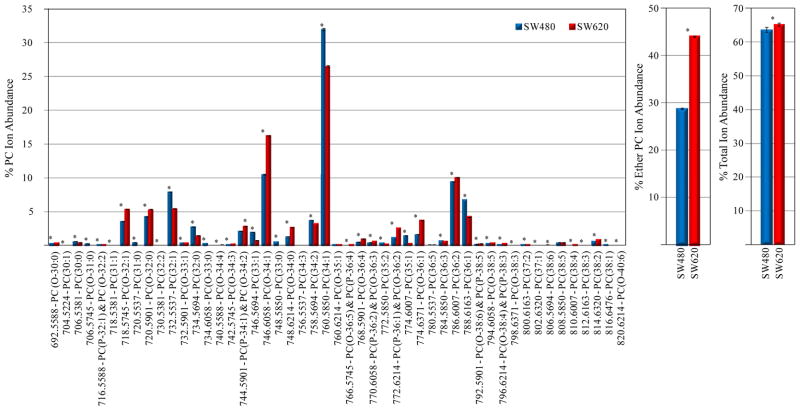
Figures 3 and 4 show the changes observed in individual PC and PE lipid % ion abundances, respectively, between the d6-DMBNHS derivatized SW480 and SW620 cell lipid extracts, for all ions whose abundances were found to be greater than 0.1% of the total ion abundance in each lipid class. Numerous statistically significant changes (p<0.01) were observed between the two cell lines, most notably with respect to increases in the abundances of plasmanyl-ether containing PC lipids between the SW620 and SW480 cell crude lipid extracts, and a corresponding decrease in the abundance of their diacyl counterparts (e.g., PC(O-34:1) at m/z 746.6058 versus PC(34:1) at m/z 760.5850) (Figure 3A). The % total ether-linked PC ion abundance (corresponding to approximately 30% of the total PC lipid ion abundance) increased by approximately 50% percent (Figure 3B), whereas the % total PC ion abundance compared to the total ion abundance for all identified lipids increased to only a small extent (Figure 3C). A similar significant increase was observed in the abundance of the major plasmanyl-LPC lipid present in the lipid extracts i.e., LPC(O-16:0)), along with an overall increase in the % total LPC ion abundance between the SW620 and SW480 cells (Supplemental Figure S3). Note that while we compare here the % total ion abundances for the various lipid classes compared to the total ion abundance for all identified lipids, these do not reflect the actual lipid concentrations as the ionization efficiency of individual lipid classes are very different from each other. However, it is useful to include these values in order to demonstrate the wide dynamic range of the Orbitrap mass spectrometer for the detection of lipid classes observed at very low ion abundances (see below).
Figure 3.
Quantification of PC lipids from the d6-DMBNHS derivatized SW480 and SW620 cell crude lipid extracts. (A) Percent individual PC ion abundances compared to the total ion abundance for all PC lipids. (B) Percent total ether-linked PC ion abundance compared to the total PC lipid ion abundance. (C) Percent total PC ion abundance compared to the total ion abundance for all identified lipids. n=5, *=p<0.01.
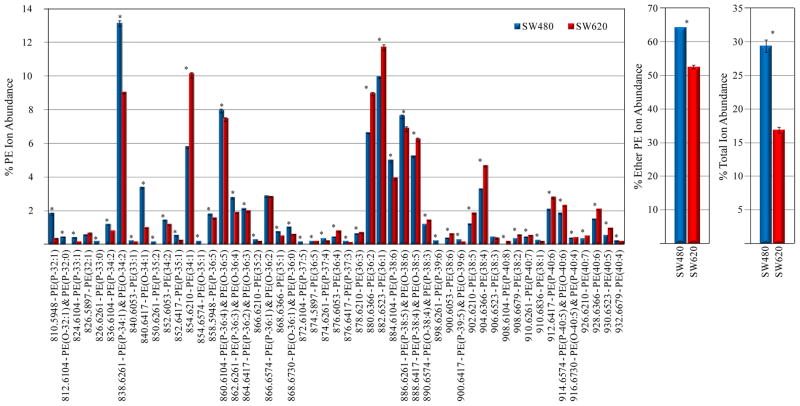
Figure 4.
Quantification of PE lipids from the d6-DMBNHS derivatized SW480 and SW620 cell crude lipid extracts. (A) Percent individual PE ion abundances compared to the total ion abundance for all PE lipids. (B) Percent total ether-linked PE ion abundance compared to the total PE lipid ion abundance. (C) Percent total PE ion abundance compared to the total ion abundance for all identified lipids. n=5, *=p<0.01.
Numerous statistically significant changes in PE lipid ion abundances were also observed between the d6-DMBNHS derivatized SW480 and SW620 cells (Figure 4A). In contrast to the PC lipids, an increase in the % abundance of diacyl-PE lipid containing ions was generally observed, with a corresponding decrease in the % ion abundance of plasmenyl- and plasmanyl-ether containing PE lipids, particularly for shorter chain length species (e.g., the d6-DMBNHS derivatized PE(P-34:1) and PE(O-34:2) lipids at m/z 838.6261, PE(P-32:1) at m/z 810.5948 and PE(O-34:1) at m/z 840.6417). Similar changes were also observed for the LPE lipids (Supplemental Figure S4). Decreases in the % total ether-linked PE ion abundance compared to the total PE lipid ion abundance (Figure 4B), as well as the % total PE ion abundance compared to the total ion abundance for all identified lipids (Figure 4C), were also observed. Finally, the abundance of odd-numbered chain length PC and PE lipid species (diacyl, plasmanyl and plasmenyl), as well as odd-numbered chain length LPE lipid species, were all observed to significantly decrease in abundance between the SW480 and SW620 cells.
It has been known for some time that elevated levels of glycerol alkyl ethers and alkyl-phosphoglycerides are present in neoplastic human tissues when compared to normal tissues37. Howard et al. have suggested that a correlation exists between the rate of tumor growth and the glyceryl-ether content in phospholipids, and ether-lipid levels have been shown to be at higher concentrations in cells that are more rapidly growing than other cells observed38. This, along with previous reports of a correlation between increased plasmanyl-PC and decreased plasmenyl-PE lipid levels with malignancy and metastatic potential in various other cancers3,5,6,11,12, are consistent with our findings, given that the SW620 cells were derived from a lymph node metastatic tumor, which is more proliferative than the primary adenocarcinoma SW480 cell line. Although the functional role of changes in ether-lipid metabolism, and particularly plasmanyl-lipids, in malignancy and metastasis are still poorly understood, it is well established that changes in the physical properties of the cellular membrane as a function of its plasmalogen lipid composition may play an important role in regulating cell growth, fusion, intracellular transport and signal transduction, and may also influence the sensitivity of cells to certain forms of therapy39.
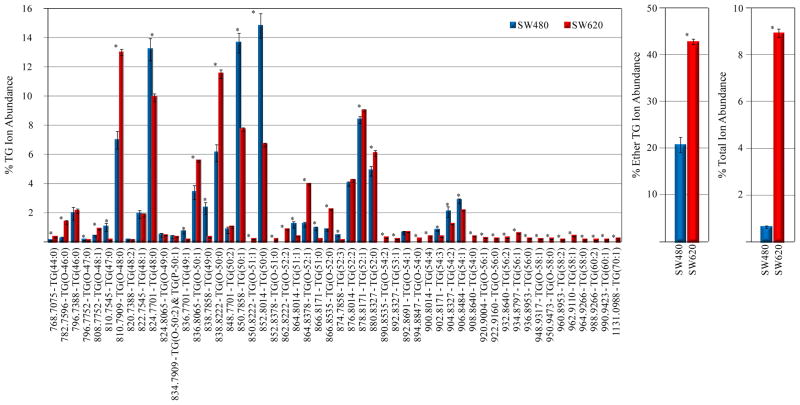
Quantitative analysis of the TG lipids (Figure 6) revealed a dramatic increase in the total % TG ion abundance in the SW620 cells, along with similar trends in changes to the % ion abundances of individual monoether-linked (primarily plasmanyl) and triacyl-TG lipids as those observed for the PC lipids (e.g., increased TG(O-48:0), TG(O-50:1) and TG(O-50:0) and correspondingly decreased TG(48:0), TG(50:1) and TG(50:0)). Notably, numerous long chain length TG lipids (>56 total carbons) were observed exclusively in the SW620 cell lipid extract. Consistent with the other lipid classes (except PS), odd-numbered chain length TG lipids decreased in abundance in the SW620 cells. The increase in overall TG species (including ether-linked TG lipids that are the likely source of the increase in ether-linked phospholipids in the SW620 cells) is likely due to increased de novo fatty acid synthesis in malignant cells which is used to supply the membrane with fatty acids for increased proliferation40. Increases in ether-linked TG content has previously been correlated with malignant and metastatic rat liver41 and mammary tissue42 and in malignant hepatocellular carcinoma tissues compared to normal human liver43.
The results obtained for the PA, PS, LPS, PG and PI lipids are shown in Supplemental Figures S5–S9, respectively. An overall significant increase in the % total ether-linked (primarily plasmanyl) PA ion abundance compared to the total PA lipid ion abundance was observed between the SW480 and SW620 cells, whereas the % total PA ion abundance underwent a significant decrease. There was an increase in the % total ion abundance of both PS and PG lipids in the SW620 compared to the SW480 cells, as well as an increase in % total ether-linked PS ion abundance. No ether-linked or odd-numbered chain length species were observed for the PG lipids. The most abundant LPS lipid in the SW480 cells (d6-DMBNHS derivatized LPS(19:0), m/z 676.4125) underwent a significant decrease in abundance, following the trend observed for the other phospholipids. However, significant increases in the % ion abundance of multiple odd-numbered chain length PS species were observed in the SW620 cells compared to the SW480 cell extract, in contrast to the other phospholipids and glycerolipids determined here. Interestingly, ether-linked PI lipids were only observed in the SW480 cells.
The results obtained from quantitative analysis of the DG and MG lipids are shown in Supplemental Figures S10 and S11. Small increases in total DG and MG lipid ion abundances were observed between the SW620 and SW480 cells. For individual DG lipids, the most obvious difference arose from a decrease in DG(32:0) and an increase in DG(34:0).
Significantly decreased SM(42:1) and Cer(42:1) lipid abundances (i.e., SM(d18:1/16:0) and Cer(d18:1/16:0)) were observed in the SW620 cells, despite an overall increase in SM and Cer % total ion abundances (Supplemental Figures S12 and S14, respectively). Importantly, decreased C16-ceramide levels have previously been associated with the resistance of cells toward activation of apoptotic signaling9. No overall change was found in the level of Chol (Supplemental Figure S14). However, a dramatic (approximately 80 fold) increase was observed in the % lipid abundance of Chol esters, which has previously also been reported in human glioblastomas7.
MS/MS and MS3 structural characterization of individual lipid ions
The above differential quantification was based on the 354 and 368 lipid ions identified from the SW480 and SW620 cell lipid extracts, respectively, that were initially assigned by LIMSA in terms of their headgroup and total number of carbons and double bonds present in the lipid fatty acyl chains. In order to further confirm these assignments, as well as to identify the individual acyl, plasmanyl and/or plasmenyl chain constituents, a series of CID-MS/MS experiments were performed. For the lysophospholipids, cholesterol ester and MG lipids, no further characterization was required, as only one acyl, plasmanyl or plasmenyl chain can be present (note that we did not attempt to assign the SN-linkage position for these species). For the remaining lipid classes containing two or more acyl and/or alkyl chain substituents, and where various structural isomers in terms of the combinations of carbon lengths and number of double bonds within each chain may be present (and when the individual precursor ions were observed at sufficient abundance to allow their isolation), product ion MS/MS or MS3 spectra were acquired to obtain the additional information required to assign the molecular lipid species that were present44–46 (Supplemental Tables 1–16). Upon performing HCD-MS/MS and/or CID/HCD-MS3 analysis, characterization of the head group identity and acyl and/or alkyl chain lengths and degree of unsaturation for 490 lipids in the SW480 cell line and 573 lipids in the SW620 cell line was achieved (summarized in Table 1 and shown in detail in Supplemental Tables 1–16). Note that complete structurally defined molecular lipid characterization was not achieved here, as no information was obtained using these conventional MS/MS approaches to assign the position(s) of double bond(s) within the fatty acyl and/or alkyl chains. For lipids that were positively identified from the high-resolution/accurate mass MS spectra (in conjunction with the additional information provided from the d6-DMBNHS derivatization and formic acid treatment), but whose intensities were too low to allow for the acquisition of MS/MS and/or MS3 spectra to assign the molecular lipid species, the identities are shown simply as the total number of carbons and double bonds, and are included in the column ‘Total Lipids’ in Table 1; 110 ions for the SW480 and 121 ions for the SW620 extracts fall into this category. Therefore, upon combining both the number of lipids that could be assigned as molecular lipid species by MS/MS, as well as those that could only be assigned in terms of their sum compositions, a total of 600 and 694 lipids from the SW480 and SW620 cell extracts, respectively, were identified from this study.
Although we report here only qualitative identification and quantitative differences in relative abundances between the SW480 and SW620 cell lines (i.e., we did not attempt to perform absolute quantification of individual molecular lipid species), the number of lipids that could be identified using this approach compares very favorably to prior reports in the literature describing comprehensive cellular lipidomic analysis strategies20,47,48. For example, global analysis of the Saccharomyces cerevisiae lipidome by quantitative ‘shotgun’ mass spectrometry resulted in the absolute quantification of 250 molecular lipid species20, while the use of LC-MS based approaches have reported the identification and quantification of over 500 distinct molecular lipid species distributed among the main lipid categories from human plasma47, and over 400 major lipid molecular species that showed dynamic changes from mouse macrophage cells in response to an inflammatory response and/or a statin drug48.
Conclusions
We have demonstrated here that selective functional group derivatization reactions combined with high resolution / accurate mass spectrometry can serve as an effective ‘chemical’ separation strategy to resolve isobaric mass lipids from within complex mixtures, as well as to enhance the ionization sensitivity and detection of low abundance lipid species, without requirement for chromatographic fractionation or differential extraction steps prior to analysis. The application of this novel ‘shotgun’ lipidomics approach to identify and quantify differences in the abundances of >600 lipids in 4 major lipid classes and 36 lipid subclasses from the isogenic SW480 and SW620 CRC cell lines, represents the most comprehensive ‘global’ lipidome analysis performed to date to monitor differences between isogenic primary and metastatic CRC cells. The results obtained may provide important new insights into the role of aberrant lipid metabolism in the onset and progression of CRC.
Supplementary Material
Figure 5.
Quantification of TG lipids from the d6-DMBNHS derivatized SW480 and SW620 cell crude lipid extracts. (A) Percent individual TG ion abundances compared to the total ion abundance for all TG lipids. (B) Percent total ether-linked TG ion abundance compared to the total TG lipid ion abundance. (C) Percent total TG ion abundance compared to the total ion abundance for all identified lipids. n=5, *=p<0.01.
Acknowledgments
Support for this work was provided by funding from the National Institutes of Health (GM103508) to GER, and, in part, from the National Health and Medical Research Council of Australia (Program grant #487922) to RJS and HJ.
Footnotes
Additional information as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Green DE, Tzagoloff A. J Lipid Res. 1966;7:587–602. [PubMed] [Google Scholar]
- 2.van Meer G, Voelker DR, Feigenson GW. Nature Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith RE, Lespi P, Di Luca M, Busos C, Marra FA, de Alaniz MJT, Marra CA. Lipids. 2008;43:79–89. doi: 10.1007/s11745-007-3133-6. [DOI] [PubMed] [Google Scholar]
- 4.Min HK, Lim S, Chung BC, Moon MH. Anal Bioanal Chem. 2011;399:823–830. doi: 10.1007/s00216-010-4290-7. [DOI] [PubMed] [Google Scholar]
- 5.Dória ML, Cotrim Z, Macedo B, Simões C, Domingues P, Helguero L, Domingues MR. Breast Cancer Res Treat. 2012;133:635–648. doi: 10.1007/s10549-011-1823-5. [DOI] [PubMed] [Google Scholar]
- 6.Katz-Brull R, Seger D, Rivenson-Segal D, Rushkin E, Degani H. Cancer Res. 2002;62:1966–1970. [PubMed] [Google Scholar]
- 7.Slagel DE, Dittmer JC, Wilson CB. J Neurochem. 1967;14:789–798. doi: 10.1111/j.1471-4159.1967.tb10314.x. [DOI] [PubMed] [Google Scholar]
- 8.Griffitts J, Saunders D, Tesiram YA, Reid GE, Salih A, Liu S, Lydic TA, Busik JV. Biochim Biophys Acta. 2010;1801:1133–1144. doi: 10.1016/j.bbalip.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White-Gilbertson S, Mullen T, Senkal C, Lu P, Ogretmen B, Obeid L, Voelkel-Johnson C. Oncogene. 2009;28:1132–1141. doi: 10.1038/onc.2008.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schonberg SA, Lundemo AG, Fladvad T, Holmgren K, Bremseth H, Nilsen A, Gederaas O, Tvedt KE, Egeberg KW, Krokan HE. FEBS J. 2006;273:2749–2765. doi: 10.1111/j.1742-4658.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 11.Merchant TE, Kasimos JN, de Graaf PW, Minsky BD, Gierke LW, Glonek T. Int J Colorectal Dis. 1991;6:121–126. doi: 10.1007/BF00300208. [DOI] [PubMed] [Google Scholar]
- 12.Dueck D, Chan M, Tran K, Wong JT, Jay FT, Littman C, Stimpson R, Choy PC. Mol Cell Biochem. 1996;162:97–103. doi: 10.1007/BF00227535. [DOI] [PubMed] [Google Scholar]
- 13.Siegel R, Jemal A. Colorectal Cancer Facts & Figures 2011–2013. American Cancer Society; 2011. [Google Scholar]
- 14.Han X, Yang K, Gross RW. Mass Spectrom Rev. 2012;31:134–178. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang K, Cheng H, Gross RW, Han X. Anal Chem. 2009;81:4356–4368. doi: 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busik JV, Reid GE, Lydic TA. Meth Mol Biol. 2009;579:33–70. doi: 10.1007/978-1-60761-322-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ståhlman M, Ejsing CS, Tarasov K, Perman J, Borén J, Ekroos K. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:2664–2672. doi: 10.1016/j.jchromb.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 18.Houjou T, Yamatani K, Imagawa M, Shimizu T, Taguchi R. Rapid Commun Mass Spectrom. 2005;19:654–666. doi: 10.1002/rcm.1836. [DOI] [PubMed] [Google Scholar]
- 19.Han X, Gross RW. Mass Spectrom Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 20.Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A. Proc Natl Acad SciUSA. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu F, Turk J. J Am Soc Mass Spectrom. 2007;18:2065–2073. doi: 10.1016/j.jasms.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Folch J, Lees M, Sloane Stanley GH. J Biol Chem. 1956;226:497–509. [PubMed] [Google Scholar]
- 23.Lydic TA, Busik JV, Esselman WJ, Reid GE. Anal Bioanal Chem. 2009;394:267–275. doi: 10.1007/s00216-009-2717-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X, Lu Y, Wang W, Borhan B, Reid GE. J Am Soc Mass Spectrom. 2010;21:1339–1351. doi: 10.1016/j.jasms.2010.03.047. [DOI] [PubMed] [Google Scholar]
- 25.Ekroos K. Lipidomics. Chpt 1. Wiley-VCH; 2012. [Google Scholar]
- 26.Haimi P, Uphoff A, Hermansson M, Somerharju P. Anal Chem. 2006;78:8324–8331. doi: 10.1021/ac061390w. [DOI] [PubMed] [Google Scholar]
- 27.Sandhoff R, Brügger B, Jeckel D, Lehmann WD, Wieland FT. J Lipid Res. 1999;40:126–132. [PubMed] [Google Scholar]
- 28.Li YL, Su X, Stahl PD, Gross ML. Anal Chem. 2007;79:1569–1574. doi: 10.1021/ac0615910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang W, Adamec J, Regnier FE. Anal Chem. 2007;79:5150–5157. doi: 10.1021/ac070311t. [DOI] [PubMed] [Google Scholar]
- 30.Zemski Berry KA, Murphy RC. Anal Biochem. 2006;349:118–128. doi: 10.1016/j.ab.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 31.Zemski Berry KA, Turner WW, VanNieuwenhze MS, Murphy RC. Eur J Mass Spectrom. 2010;16:463–470. doi: 10.1255/ejms.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han X, Yang K, Cheng H, Fikes KN, Gross RW. J Lipid Res. 2005;46:1548–1560. doi: 10.1194/jlr.D500007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horning MG, Martin DB, Karmen A, Vagelos PR. J Biol Chem. 1961;236:669–672. [PubMed] [Google Scholar]
- 34.Ivanova PT, Milne SB, Brown HA. J Lipid Res. 2010;51:1581–1590. doi: 10.1194/jlr.D003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy EJ, Stephens R, Jurkowitz-Alexander M, Horrocks LA. Lipids. 1993;28:565–568. doi: 10.1007/BF02536090. [DOI] [PubMed] [Google Scholar]
- 36.Lu Y, Zhou X, Stemmer PM, Reid GE. J Am Soc Mass Spectrom. 2012;23:577–593. doi: 10.1007/s13361-011-0190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snyder F, Wood R. Cancer Res. 1969;29:251–257. [PubMed] [Google Scholar]
- 38.Howard BV, Morris HP, Bailey JM. Cancer Res. 1972;32:1533–1538. [PubMed] [Google Scholar]
- 39.Braverman NE, Moser AB. Biochim Biophys Acta. 2012;1822:1442–1452. doi: 10.1016/j.bbadis.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Menendez JA, Lupu R. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 41.Snyder F, Blank ML, Morris HP. Biochim Biophys Acta. 1969;176:502–510. doi: 10.1016/0005-2760(69)90217-3. [DOI] [PubMed] [Google Scholar]
- 42.Friedberg SJ, Smajdek J, Anderson K. Cancer Res. 1986;46:845–849. [PubMed] [Google Scholar]
- 43.Lin HJ, Wu PC, Ho JCI. Brit J Cancer. 1980;41:320–324. doi: 10.1038/bjc.1980.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsu F, Turk J. J Chromatogr B. 2009;877:2673–2695. doi: 10.1016/j.jchromb.2009.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McAnoy AM, Wu CC, Murphy RC. J Am Soc Mass Spectrom. 2005;16:1498–1509. doi: 10.1016/j.jasms.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 46.Valsecchi M, Mauri L, Casellato R, Prioni S, Loberto N, Prinetti A, Chigorno V, Sonnino S. J Lipid Res. 2007;48:417–424. doi: 10.1194/jlr.M600344-JLR200. [DOI] [PubMed] [Google Scholar]
- 47.Quehenberger O, Armando AM, Brown AH, Milne SB, Meyers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, Sullards CM, Wang E, Murphy RC, Barkley RM, Leiker TJ, Raetz CRH, Guan Z, Laird GM, Six DA, Russell DW, McDonald JG, Subramaniam S, Fahy E, Dennis EA. J Lipid Res. 2010;51:3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dennis EA, Deems RA, Harkewicz R, Quehenberger O, Brown HA, Milne SB, Myers DS, Glass CK, Hardiman G, Reichart D, Merrill AH, Jr, Sullards MC, Wang E, Murphy RC, Raetz CRH, Garrett TA, Guan Z, Ryan AC, Russell DW, McDonald JG, Thompson BM, Shaw WA, Sud M, Zhao Y, Gupta S, Maurya MR, Fahy E, Subramaniam S. J Biol Chem. 2010;285:39976–39985. doi: 10.1074/jbc.M110.182915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.