Abstract
This paper is aimed at developing and evaluating a content-based retrieval method for contrast-enhanced liver computed tomographic (CT) images using bag-of-visual-words (BoW) representations of single and multiple phases. The BoW histograms are extracted using the raw intensity as local patch descriptor for each enhance phase by densely sampling the image patches within the liver lesion regions. The distance metric learning algorithms are employed to obtain the semantic similarity on the Hellinger kernel feature map of the BoW histograms. The different visual vocabularies for BoW and learned distance metrics are evaluated in a contrast-enhanced CT image dataset comprised of 189 patients with three types of focal liver lesions, including 87 hepatomas, 62 cysts, and 60 hemangiomas. For each single enhance phase, the mean of average precision (mAP) of BoW representations for retrieval can reach above 90 % which is significantly higher than that of intensity histogram and Gabor filters. Furthermore, the combined BoW representations of the three enhance phases can improve mAP to 94.5 %. These preliminary results demonstrate that the BoW representation is effective and feasible for retrieval of liver lesions in contrast-enhanced CT images.
Keywords: Content-based image retrieval, Bag of visual words, Contrast-enhanced CT, Liver lesion, Distance metric learning
Introduction
The medical field is increasingly using digital images for diagnoses. Enabling radiologists to quickly search the images of similar-appearing lesions with accurate diagnosis according to their visual image features would be greatly beneficial for diagnostic decision making, especially when the visual properties play an important role to diagnosis. The task involves finding the visually and semantically similar images in a large image collection based on a given query image. This task can be addressed using content-based image retrieval (CBIR), which is an active research area in the field of medical image analysis [1].
This paper aims to develop a CBIR system to retrieve contrast-enhanced computed tomographic (CT) images that contain similar focal liver lesions and relevant information. The purpose is to aid diagnosis for the given query images, in which the lesion regions are outlined or the regions of interest (ROI) are selected. Contrast-enhanced CT examination is a standard routine for the patients who are suspicious of liver problems. The contrast-enhanced CT examination includes unenhanced, hepatic arterial (HAP), portal venous (PVP), and delayed phases (HDP). Some studies have developed computerized methods to classify and retrieve CT liver lesions [2–6]. However, many existing studies on live CT images only used an unenhanced [3, 5] or PVP image [4, 6] to detect and characterize the liver lesions. Nonetheless, one study used temporal signal tendency to describe the multiphase feature [7]. Liver CT images from each phase may actually contain useful and important diagnostic information [8]. In this paper, the potential ability of single- and multiphase visual information (contents) of contrast-enhanced CT for retrieval of liver lesions is explored.
In general, two key issues are at the forefront of the development of the CBIR system: (1) representing the image contents and (2) defining the similarity between images [9]. The intensity and texture features in liver CT images are considered as the important cues for the computerized detection and classification of liver lesions. First-order statistics [5], gray-level co-occurrence matrix (GLCM) [2, 3], Gabor filters [10, 11], and wavelet transform [12] are the most commonly used methods for describing the intensity and texture features of liver CT images. Although GLCM and wavelet transform can effectively represent texture, recent studies have suggested that the texton and patch exemplar methods are more powerful for texture classification [13, 14]. The patch exemplar and texton representations are closely related to the bag-of-visual-words (BoW) approach [15–20]. BoW is a popular strategy for representing images within the context of image classification and CBIR. Unlike GLCM or Gabor, the BoW approach can provide the way to design task-specific feature representation. In this paper, the BoW approach is used to deal with the first issue mentioned above. However, representing an image (or a region) using the visual features usually results in loss of information. In addition, the appearance of lesions has large variations. Lesions belonging to the same pathological category but coming from a different patient can present diverse appearances in the images. As a result, the extracted visual features may not directly link to the target image category (semantic concept). It is the so-called semantic gap [21]. To address this issue, we use the distance metric learning (DML) methods to obtain the image similarity associated with semantic concepts in the BoW feature space.
Materials and Methods
Image Data
Contrast-enhanced CT was performed using a multidetector row helical scanner at the General Hospital, Tianjin Medical University, China, from 2007 to 2010. CT images of the three types of focal liver lesions, namely, hepatoma, cyst, and hemangioma, were collected. Only one lesion per patient was analyzed, which was manually outlined by three experienced radiologists. For patients with multiple lesions, the dominant lesion (based on size) was analyzed. For each patient, one to five representative slices of each phase were selected by the radiologists to comprise the image dataset. The resulting image dataset consisted of 1,375 CT slices (512 × 512 matrix, 12 bits/pixel) from 189 patients, including 87 hepatomas, 62 cysts, and 60 hemangiomas. Since the clinical settings of patients were different during the imaging procedure, only one to two phases of the CT images from some patients were acquired. Details of the image data are listed in Table 1. Three examples of outlined liver lesions are shown in Fig. 2.
Table 1.
Number of patients in the image dataset
| Phase | Patient number |
|---|---|
| HAP | 118 |
| PVP | 129 |
| HDP | 127 |
| HAP and PVP | 89 |
| HAP and HDP | 79 |
| PVP and HDP | 95 |
| HAP, PVP, and HDP | 68 |
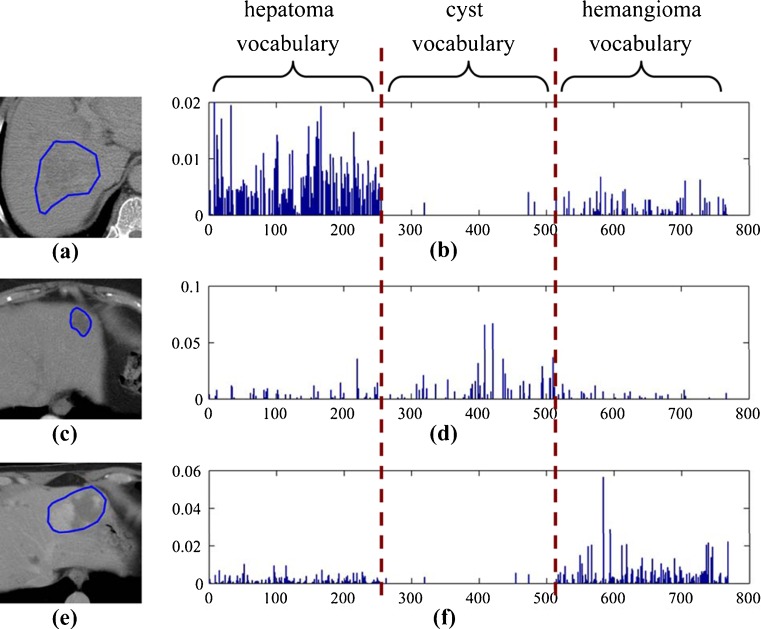
Fig. 2.
Typical examples of BoW representation of liver lesions at PVP phase using the category-specific vocabulary. a, c, and e are the CT images of hepatoma, cyst, and hemangioma, respectively. b, d, and f are the corresponding histograms of the lesions in a, c, and e
Bag-of-Visual-Words Representation of Lesions
The bag-of-visual-words image representation is analogous to the bag-of-words representation of text documents, which makes techniques for text retrieval readily applicable to the problem of image retrieval. The BoW model treats an image as a distribution of local descriptors, wherein each descriptor is labeled as a discrete visual prototype. The set containing these prototypes, or visual words, is the so-called visual vocabulary (or dictionary [16], codebook [22]), which is typically obtained by clustering in the feature space of local descriptor. Given a visual vocabulary, an image is represented as a histogram of visual word occurrences on the sampled image patches from the image. The histogram can be considered as a discrete representation of the probability distribution over visual words.
In the BoW framework, the image patches are sampled densely or sparsely by interest point detectors and are depicted by local patch descriptors. The most popular local patch descriptor is SIFT in the computer vision community [23]. Unlike natural images, most of the medical images are taken under the standardized conditions to allow direct comparisons of intensity of the images. There are no very meaningful key points and structures in the liver lesions in CT images, and the intensity is an important cue for diagnosis. Thus, for the current work, we used the raw intensity without normalization as the local patch descriptor [24]. The raw patches were then sampled densely with the stride of one pixel in the liver lesion region to form the BoW representation.
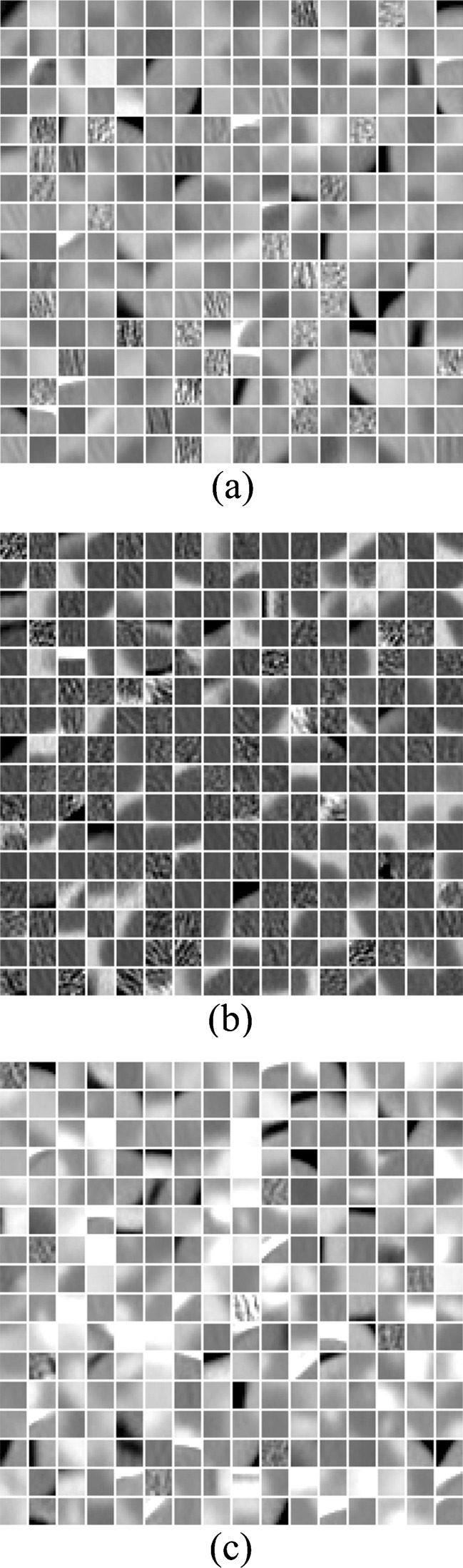
For the BoW method, an important issue is to construct the visual vocabulary which affects the classification and retrieval performance significantly. By clustering algorithms, the found cluster centers or exemplars in the continuous feature space are considered as the visual words. A popular clustering approach for finding visual words is k-means because of its simple and efficient implementation. K-means is an unsupervised clustering algorithm that tries to minimize the variance between k clusters and the training data. Labeling the image patch is equivalent to quantize the high-dimensional local descriptors. Generally, a larger visual vocabulary usually leads to better performance since the quantization error is effectively reduced [25]. A universal vocabulary can be created in the whole feature space by k-means clustering, which may produce more clusters for the high-frequency parts in the feature space, leaving fewer clusters for the remaining parts. However, frequently occurring features are not necessarily informative and discriminative. One way to improve the expressive ability of the visual vocabulary is to incorporate the category information into it. This can be done by constructing a visual vocabulary for each category by clustering (classwise clustering) [25] and then by aggregating them as one overall vocabulary (category-specific vocabulary). We constructed the universal and category-specific vocabularies for each phase for the liver CT images. Figure 1 shows the category-specific vocabulary trained by k-means for the PVP phase. Figure 2 shows the examples of BoW histograms extracted using the category-specific vocabulary.
Fig. 1.
The category-specific visual vocabulary of PVP. The patch size is 11 × 11, and vocabulary size is 256 × 3. a, b, and c are hepatoma, cyst, and hemangioma vocabulary, respectively. The intensity ranges of visual words are adjusted for view
Given a visual vocabulary, the standard BoW approach assigns one visual word to an image patch, namely hard assignment. For each visual word w in a vocabulary V, the BoW model estimates the distribution of visual words in an image (or a ROI) by
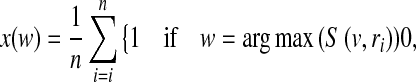 |
where n is the number of sampled regions or patches in the image, ri is an image patch i, and S(w, ri) is the similarity between a word w and a patch ri. Thus, an image is represented by a histogram H of word frequencies that describes the probability distribution over visual words. The similarity S(w, ri) can be defined as a Gaussian kernel 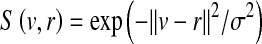 .
.
Labeling an image patch with the single best visual word (hard assignment) ignores all ambiguity regarding the meaning of the image patch. Unlike hard assignment, assigning a degree of similarity to an image patch (soft assignment) can help in modeling the inherent uncertainty of the image patch while considering the continuous nature of image patches [22]. Soft assignment can be easily incorporated in the BOW model by
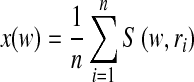 |
Soft assignment weighs each word based on the similarity of an image region to the visual word to model the uncertainty of the meaning of an image patch. The sparse coding method on a visual dictionary, such as locality constrained linear coding (LLC) [26], is another alternative of hard assignment.
Learning Distance Metrics Between BoW Histograms
Using visual features to represent an image (e.g., BoW histogram in the present paper) usually results in loss of information. Using a common distance metric, such as Euclidean distance and χ2 distance between histograms as the similarity (dissimilarity) measure, a CBIR system cannot achieve the expected performance and capture the important information for diagnosis. A CBIR system, therefore, should be able to reduce the semantic gap between the low-level visual features and the image category. DML methods can be used to find a linear transformation that projects the image features to a new meaningful feature space to reduce this semantic gap. Previous work showed that appropriately designed distance metrics could improve CBIR performance compared with Euclidean distance [27]. For the BoW model, the semantic meaning of visual words is ambiguous; thus, the retrieval performance can be improved by embedding the semantic information to BoW representation by supervised DML.
Let L be a d × D matrix and W = LTL. The (squared) Mahalanobis distance between image representations xi and xj is:
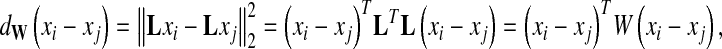 |
where T denotes the transposition of a matrix or vector. The aim of distance metric learning is to find an optimal projection L or a metric W to minimize an objective function. Most existing algorithms obtain a metric either by dimensionality reduction (subspace learning) such as principal components analysis, linear discriminant analysis (LDA), and local Fisher discriminant analysis (LFDA) [28], or by explicit metric learning, such as Xing's method [29], and large margin nearest neighbor (LMNN) [30], close-form metric learning (CFML) [31]. The intuitive goal of metric learning is keeping all intraclass data points close, while separating all interclass data points as far as possible in the subspace projected by L. Among DML methods, some require semidefinite programming to obtain solutions and are computationally expensive, and some are formulated as trace ratio and have close-form solutions.
LDA is a powerful approach to learn a subspace that preserves the variance between class labels. Suppose we have a set of n samples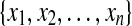 belonging to c classes. Since there are many variants of LDA [32], a regularized form of LDA (rLDA) is used in this paper. The objective of rLDA to search a transformation matrix L* is as follows:
belonging to c classes. Since there are many variants of LDA [32], a regularized form of LDA (rLDA) is used in this paper. The objective of rLDA to search a transformation matrix L* is as follows:
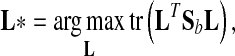 |
1 |
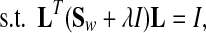 |
where
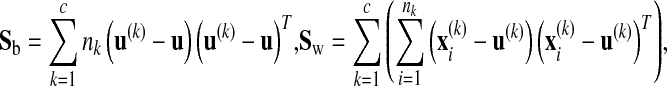 |
tr() denotes matrix trace, u is the total sample mean vector, nk is the number of samples in the kth class, u(k) is the average vector of the kth class, and 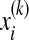 is the ith sample in the kth class. Sw is the within-class scatter matrix and Sb is the between-class scatter matrix. The optimization problem in Eq. (1) is equivalent to finding the eigenvectors associated with maximum eigenvalues of a generalized eigenproblem. The rank of Sb is upper bounded by c–1(<<D). The regularization parameter λ(λ > 0) enforces an isotropic Gaussian prior on Sw, which ensures that the eigenvalue problem is well conditioned when Sw is low rank.
is the ith sample in the kth class. Sw is the within-class scatter matrix and Sb is the between-class scatter matrix. The optimization problem in Eq. (1) is equivalent to finding the eigenvectors associated with maximum eigenvalues of a generalized eigenproblem. The rank of Sb is upper bounded by c–1(<<D). The regularization parameter λ(λ > 0) enforces an isotropic Gaussian prior on Sw, which ensures that the eigenvalue problem is well conditioned when Sw is low rank.
CFML, a close-form DML algorithm proposed by Alipanahi [31], is closely related to LDA. We assume that the images and lesions are labeled, i.e., whether or not two image representations are in the same category or not. Below, we refer to the image representations in the same and different categories as similar and dissimilar, respectively. Let the set of similar pairs be denoted by
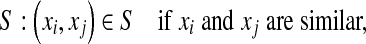 |
and the set of dissimilar pairs be denoted by
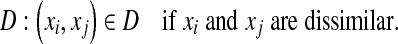 |
The objective of CFML to search the d × D transformation matrix L* is as follows
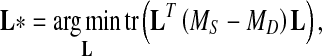 |
2 |
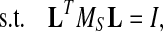 |
where
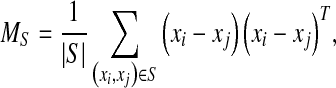 |
and
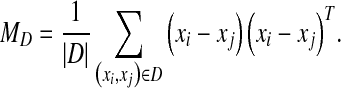 |
CFML attempts to minimize the squared Mahalanobis distance between similar points, while maximizing the squared Mahalanobis distance between dissimilar points. The solution of the optimization problem in Eq. (2) is provided by the matrix of eigenvectors corresponding to the largest eigenvalues of the matrix 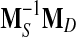 . In this paper, a regularization form of CFML is implemented by substituting LTMSL =
I with LT(MS + λI)L =
I.
. In this paper, a regularization form of CFML is implemented by substituting LTMSL =
I with LT(MS + λI)L =
I.
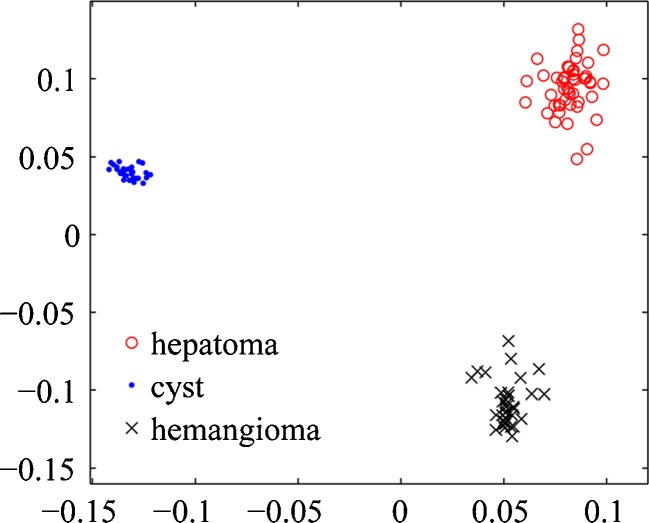
For high-dimensional BoW representations, we find that the explicit regularization is essential for good performance even when Sw or MS is full rank. The number of available samples in this paper is less than 200, while the BoW histograms have a large number of bins. This is a typical high-dimensional small-sample problem. As shown in Fig. 3, the 768-dimensional BoW histograms of liver CT images are embedded into 2-dimensional feature space by rLDA. This scatter plot demonstrates the powerfully discriminative ability of the BoW representation. In the training sample set, three types of liver lesions were linearly separable in the subspace learned by rLDA.
Fig. 3.
The embedding space of BoW representations by rLDA
Since the BoW histograms can be regarded as the discrete probability distributions, they do not perform well for retrieval and classification in Euclidean space. In comparison, histogram intersection (which is equivalent to L1 distance), χ2 distance, and Hellinger distance have consistently been found to perform well in applications for the histogram-based representations. Particularly, the squared Hellinger distance between two distributions, P and Q, can be expressed as
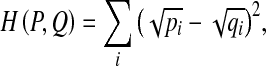 |
which has a similar form to Euclidean distance. Analogous to Mahalanobis distance, we define the generalized Hellinger distance
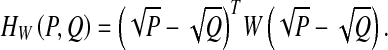 |
To learn a generalized Hellinger distance, the square-rooting operation on the elements of the BoW vector is the only requirement. Actually, the square-rooting BoW histogram is the feature map of Hellinger kernel 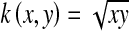 , which can improve the retrieval performance [33, 34].
, which can improve the retrieval performance [33, 34].
To develop the CBIR system of contrast-enhanced liver CT images, the visual vocabularies of each phase and the distance metrics were learned offline for each single- or multiphase CT image. These were then restored in the database. The BoW histograms of each patient at each phase were also computed offline and restored in the database for indexing the patients. Figure 4 presents the architecture of the retrieval system based on the BoW representations and the learned distance metrics. All of the acquired slices at the single or multiple phases of a patient are fed to the system as a query. The BoW histograms of a query were computed using the stored visual vocabularies for each phase online and then compared with the BoW histograms stored in the database using the learned distance metrics for single or multiple phases. The images and the known diagnostic information of patients corresponding to the most similar BoW histograms were retrieved from the database and were then presented to the user.
Fig. 4.
Architecture of the CBIR system of multiphase liver CT images
Retrieval Evaluation Measures
To evaluate the CBIR system, several performance evaluation measures have been proposed based on precision and recall:
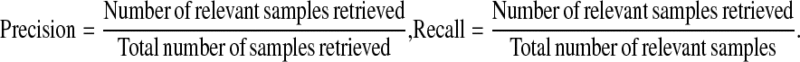 |
Precision and recall values are usually represented in a precision–recall curve, which summarizes the precision–recall pairs for varying numbers of retrieved samples. The most common way to summarize this precision–recall curve into one value is the mean of average precision (mAP). More precisely, precision at the top k retrieved samples (Prec@k in short) is defined as the proportion of relevant samples up to position k:
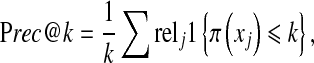 |
where 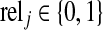 is the relevance label of xj for a given query xq (1 for relevant, and 0 for irrelevant), π(xj) is the position or rank of the jth sample in the ranking list, and 1{} is the indicator function. Average precision (AP) is the average of the precisions at the positions where there is a relevant sample:
is the relevance label of xj for a given query xq (1 for relevant, and 0 for irrelevant), π(xj) is the position or rank of the jth sample in the ranking list, and 1{} is the indicator function. Average precision (AP) is the average of the precisions at the positions where there is a relevant sample:
 |
where N+ is the number of relevant samples. mAP is the mean of AP over all queries.
Results
Experimental Settings
The performance of BoW representations for retrieval of liver lesions was evaluated on single- and multiphase CT images. The patients were partitioned by tenfold cross-validation method. The slices of patients in the training set were used as the dataset images. The slices of each patient in the test set were used as a query to retrieve the training set to report performance. The visual vocabulary for each enhance phase was constructed by k-means clustering. For each patient, the BoW histogram was computed using all of the slices at each phase.
The shapes of image patches were set to be blocks with the sizes of 7 × 7, 9 × 9, 11 × 11, and 13 × 13 pixels, respectively. The image patches were sampled densely on grids with step of one pixel to form the BoW histograms. For each phase, the universal vocabulary and category-specific vocabulary were constructed separately. For the category-specific vocabulary, the sizes of vocabulary (number of visual words) for each type of liver lesion were set to 64, 128, 256, and 512. For fair comparison, the sizes of universal vocabulary were set to 192, 378, 768, and 1,536.
In this paper, two patients with the slices at single (or multiple) phase(s) containing lesions in the same category were defined as relevant (similar); otherwise, they were considered as irrelevant (dissimilar). For the DML algorithms, the optimal regularization parameters and embedding dimensionality were estimated by cross-validation.
Comparison of Different Distances
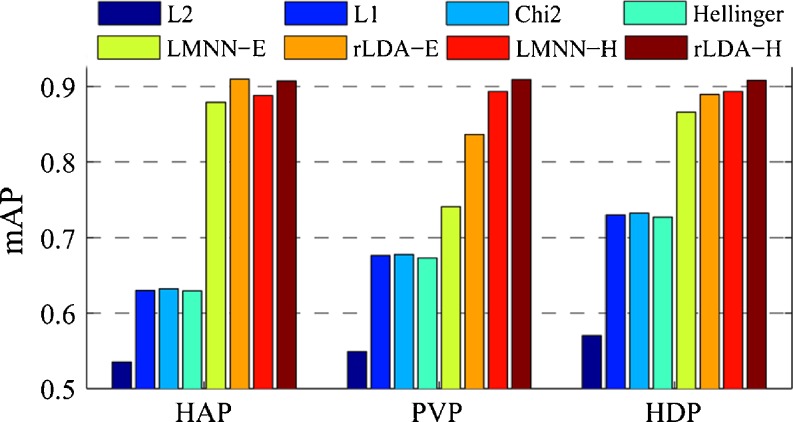
First, the sizes of the image patch and visual vocabulary were fixed, after which the retrieval performances of the different distances on the BoW representations in terms of mAP were summarized. Figure 5 shows the mAP values of different distance metrics for retrieval of CT images at each single phase using category-specific vocabulary with the size of 256 × 3 and the image patch size of 11 × 11. It can be seen that the mAP values of Euclidean (L2), L1, χ2 (chi2), and Hellinger distance are significantly lower than those of the learned distance metrics. Using the feature map of Hellinger kernel of BoW histograms, the distance metrics learned by LMNN and rLDA (denoted as LMNN-H and rLDA-H) achieved slightly better retrieval performance in terms of mAP than the original histograms (denoted as LMNN-E and rLDA-E). We found that the feature map of Hellinger kernel of BoW histogram was an effective way to improve the retrieval performance in the experiments. Only the results on the feature map of Hellinger kernel are reported in the rest of this paper.
Fig. 5.
Retrieval performance of different distance metrics on BoW representations
Figure 6 shows the precision–recall curve and the precision–scope curve of different distance metrics for retrieval of the CT images at PVP phase using the BoW representations. We can see that the precision of χ2 and Hellinger distance is rather high when recall or scope is small, even the mAP of χ2 and Hellinger distance is relatively small. It is verified that the visual similarity induced by the BoW histogram is related to the semantic similarity. However, with respect to semantic meaning, the importance of visual word is ambiguous and different. Directly using χ2 and Hellinger distance cannot efficiently express well the discriminant information, and the precision drops dramatically when scope increases. By DML, the discriminant ability of BoW representation was explored. As shown in Fig. 6b, the high precision is kept up when the scope increases up to 30.
Fig. 6.
Precision–scope curves of different distances
Impact of Vocabulary Size and Patch Size
Previous studies suggested that the vocabulary size and the patch size were important factors to the performance of image categorization and retrieval [24, 25]. We varied the image patch size and the vocabulary size. Using the category-specific vocabulary, we reported the retrieval performance of the learned distance metrics.
The image patch size was set to 9 × 9 to assess the impact of the vocabulary size on the retrieval performance. Figure 7 shows the mAP values of the distance metrics learned by rLDA, CFML, LFDA, and LMNN using the BoW histograms with different vocabulary sizes at each phase. For all the three phases and the learned distance metrics, larger vocabulary led to higher mAP. It is interesting that rLDA outperforms the state-of-the-art metric learning algorithm LMNN in some cases. It is interesting to note that, in some cases, rLDA outperformed the state-of-the-art metric learning algorithm LMNN. However, a large vocabulary requires higher computational cost. Since the gain on mAP is not significant when the vocabulary size is greater than 768, this vocabulary size is an appropriate value for the category-specific vocabulary size in balancing the computational cost and the retrieval performance.
Fig. 7.
Performance of BoW representations with different vocabulary sizes at single phase: a HAP, b PVP, c HDP
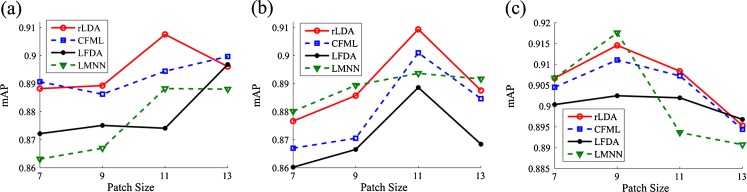
The vocabulary was set to 768 to assess the impact of the image patch size on the retrieval performance. The category-specific vocabularies were used to obtain the BoW histograms of liver lesion at each phase. The patch size is a critical factor for the local image feature. A larger patch size can achieve higher discriminative power of a local feature. However, a larger feature is less repeatable and more sensitive to image variations. Usually, the medium size of a patch is a good choice as the tradeoff between discriminative power and generalization ability of the local feature.
The medium size of the image patch led to better retrieval performance in terms of mAP at a single phase (Fig. 8). For the HAP and PVP phases, the BoW representations on 11 × 11 image patches outperformed the other image patch sizes using the different DML algorithms. For the HDP phase, the best choice of image patch size was 9 × 9.
Fig. 8.
Performance of BoW representations with different patch size at single phase: a HAP, b PVP, and c HDP
Universal Vocabulary vs. Category-Specific Vocabulary
This section presents a comparison between the two types of visual vocabularies. For a fair comparison, the universal vocabulary and the category-specific vocabulary were trained by k-means on the same image patch set for each phase. Their sizes were also set to the same value. The category-specific vocabulary outperformed the universal vocabulary when the vocabulary size was relatively small (Fig. 9). When the vocabulary size was large enough, the performance of the universal vocabulary and the category-specific vocabulary tended to be identical for the learned distance metrics. In addition, rLDA outperformed LMNN in most of the cases for the category-specific vocabulary.
Fig. 9.
Performance of the universal vocabulary (UV in short) and category-specific vocabulary (CSV in short) at single phase: a HAP, b PVP, c HDP
Comparison of BoW Representation and the Other Image Descriptors
Gabor filters, a widely used approach for texture classification, had been employed for the retrieval of liver CT images [10]. The Gabor filter bank and descriptors proposed by Manjunath were implemented in the experiments [35]. Table 2 lists the retrieval performance in terms of mAP and precision at the top 10 and 20 retrieved patients for each single phase. The retrieval performance of the intensity histogram was also reported. For the intensity histogram, the CT values were quantized to 16, 32, 64,128, and 256 levels. The intensity histogram was actually equivalent to the BoW histogram of 1 × 1 image patches. The intensity histogram of 128 bins achieved the best performance. For the BoW histograms, the category-specific vocabularies of 11 × 11 image patches were used. From Table 2, we can see that Gabor vector performed worst. For each single phase, mAP of the BoW histograms with the learned distance metric can achieve more than 90 % which is significantly higher than those of the intensity histogram and Gabor vector.
Table 2.
Retrieval performance of different image descriptors (mean ± standard deviation) (in percent)
| Phase | Descriptor | Distance | mAP | Prec@10 | Prec@20 |
|---|---|---|---|---|---|
| HAP | Intensity histogram (128 bins) | χ2 | 58.0 ± 5.0 | 61.1 ± 7.0 | 57.1 ± 6.7 |
| rLDA | 70.8 ± 7.7 | 67.8 ± 10.3 | 67.6 ± 9.9 | ||
| Gabor vector (24 D) | L1 | 47.5 ± 4.4 | 50.1 ± 7.5 | 45.7 ± 6.8 | |
| rLDA | 55.0 ± 7.8 | 52.5 ± 10.9 | 51.7 ± 10.3 | ||
| BoW histogram (256 × 3 bins) | χ2 | 63.2 ± 5.7 | 70.9 ± 9.2 | 63.7 ± 6.9 | |
| rLDA | 90.8 ± 6.0 | 86.9 ± 8.0 | 86.9 ± 8.0 | ||
| PVP | Intensity histogram (128 bins) | χ2 | 60.2 ± 6.5 | 64.6 ± 11.0 | 62.9 ± 9.5 |
| rLDA | 71.4 ± 8.4 | 68.7 ± 12.6 | 68.1 ± 11.6 | ||
| Gabor vector (24 D) | L1 | 55.2 ± 5.5 | 62.6 ± 9.0 | 56.8 ± 7.7 | |
| rLDA | 63.0 ± 4.3 | 65.4 ± 7.2 | 62.5 ± 5.3 | ||
| BoW histogram (256 × 3 bins) | χ2 | 67.8 ± 5.8 | 73.1 ± 8.6 | 70.2 ± 8.5 | |
| rLDA | 90.9 ± 6.3 | 87.5 ± 8.9 | 87.6 ± 8.8 | ||
| HDP | Intensity histogram (128 bins) | χ2 | 70.1 ± 5.6 | 76.5 ± 8.0 | 72.6 ± 7.0 |
| rLDA | 78.2 ± 9.3 | 79.8 ± 13.0 | 78.3 ± 11.1 | ||
| Gabor vector (24 D) | L1 | 53.6 ± 4.0 | 59.0 ± 5.5 | 54.4 ± 5.6 | |
| rLDA | 60.6 ± 6.7 | 63.0 ± 9.6 | 60.0 ± 8.7 | ||
| BoW histogram (256 × 3 bins) | χ2 | 73.2 ± 5.7 | 83.1 ± 6.2 | 78.0 ± 7.3 | |
| rLDA | 90.8 ± 6.5 | 89.2 ± 8.3 | 89.2 ± 8.2 |
Retrieval Using Multiphase BoW Representations
For some patients, more than one phase contrast-enhanced CT images were acquired. The retrieval performance may be improved by combining multiphase information. For multiphase retrieval, the BoW histograms of each single phase were merged into one vector to represent the multiphase contents. The settings of BoW representations were the same as in the previous section. Table 3 lists mAP and precision of the multiphase BoW histograms. By combining the information of multiple phases, mAP of χ2 distance was consistently improved. However, Prec@10 and Prec@20 of χ2 distance achieved the highest values for the HDP phase; the mAP values of the two-phase BoW histograms with the distance metrics learned by rLDA were slightly higher or at times lower than single-phase BoW histograms. Combining the three-phase BoW histograms resulted in the highest mAP (94.5 %) and Prec@10 (92.1 %) for the learned distance metric by rLDA.
Table 3.
Retrieval performance of BoW histograms using single- and multiphase representations (mean ± standard deviation) (in press)
| Phase | χ2 | rLDA | ||||
|---|---|---|---|---|---|---|
| mAP | Prec@10 | Prec@20 | mAP | Prec@10 | Prec@20 | |
| HAP | 63.2 ± 5.7 | 70.9 ± 9.2 | 63.7 ± 6.9 | 90.8 ± 6.0 | 86.9 ± 8.0 | 86.9 ± 8.0 |
| PVP | 67.8 ± 5.8 | 73.1 ± 8.6 | 70.2 ± 8.5 | 90.9 ± 6.3 | 87.5 ± 8.9 | 87.6 ± 8.8 |
| HDP | 73.2 ± 5.7 | 83.1 ± 6.2 | 78.0 ± 7.3 | 90.8 ± 6.5 | 89.2 ± 8.3 | 89.2 ± 8.2 |
| HAP + PVP | 68.5 ± 13.1 | 72.4 ± 11.1 | 64.9 ± 7.7 | 88.8 ± 5.4 | 84.2 ± 8.0 | 84.2 ± 8.0 |
| PVP + HDP | 81.5 ± 13.6 | 78.9 ± 8.9 | 71.7 ± 8.2 | 93.1 ± 8.3 | 91.4 ± 10.4 | 91.2 ± 10.7 |
| HAP + HDP | 84.0 ± 5.0 | 77.5 ± 9.3 | 66.9 ± 7.4 | 91.4 ± 8.6 | 88.8 ± 11.2 | 86.3 ± 12.2 |
| HAP + PVP + HDP | 84.1 ± 5.0 | 77.5 ± 8.5 | 63.7 ± 7.8 | 94.5 ± 8.3 | 92.1 ± 12.1 | 87.0 ± 12.2 |
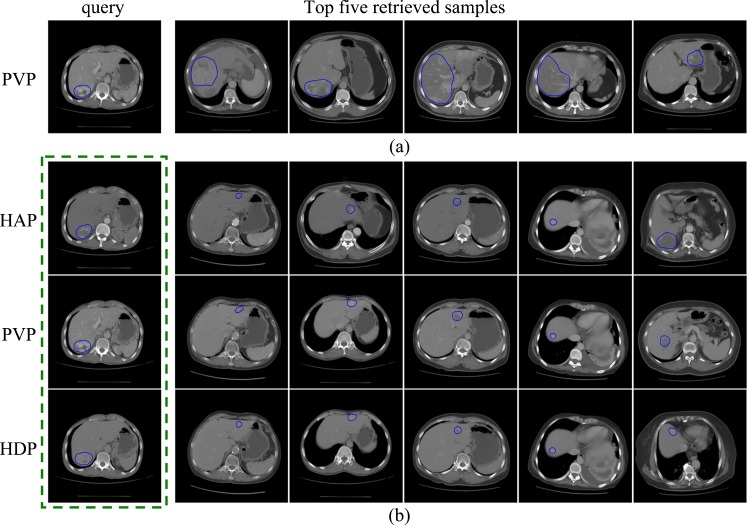
Retrieval Examples
Figure 10 presents the retrieval examples using BoW representations and the distance metrics learned by rLDA. For BoW, the category-specific vocabularies (patch size 11 × 11) of size 768 for each phase were used. Only one representative slice of each retrieved patient was displayed. For multiphase retrieval, all slices of the query patient at three phases are fed to the retrieval system as one query. The images in each column are the slices of one patient at three phases in Fig. 10b. In Fig. 10, all of the query lesions and the top five retrieved lesions are hemangiomas. From Fig. 10a, we can see that the query lesion and the retrieved lesions have very similar appearance; the visual similarity between the query lesion and the retrieved lesions at one or two phases can also be observed in Fig. 10b.
Fig. 10.
The retrieval results of a hemangioma query: a using a BoW representation of the PVP phase only and b using BoW representations of the three phases
Discussion
From the experimental results, the BoW representations are effective for retrieval of contrast-enhanced CT images of liver lesions. Unlike Gabor filters [10] and the other hand-drift filter banks [13], the task-specific and subtle representation can be learned in the BoW framework within the specific image domain, e.g., liver CT images in this paper. In general, the Gabor filter bank and the other filter bank can be more suitable for classification of periodic and structural textures which appear less in liver CT images. The large visual vocabulary of BoW and the learned distance metrics served as factors that facilitated good retrieval performance. Although the larger visual vocabulary requires more computational cost and storage, the distance metric learning algorithms can be employed to reduce the dimensionality of feature vectors and to speed up the retrieval procedure. In addition, incorporating the category information as the category-specific vocabulary is an effective way to construct a relatively compact vocabulary.
Each phase of contrast-enhanced liver CT images are effective and useful for the retrieval task. From the experimental results, the retrieval performance of BoW representations for each enhance phase is close. This implies that the retrieval system can work well even if only one phase of contrast-enhanced liver CT images is acquired. The retrieved results can be more accurate if all three phases of liver CT imaging of a patient are simultaneously fed into the retrieval system. The reason may be that more information of lesion is provided by multiphase CT images. However, more phases can increase patient exposure to ionizing radiation and lead to higher CT examination cost.
In this paper, the relevant patients are defined as the patients who have the same category lesions. It is expected that the relevant or categorical similar CT images to the query images could help the diagnosis. A more proper approach to define this relevance is by annotating the patients and the CT images by medical experts, as discussed in a previous study [6]. We plan to construct a liver CT image dataset with more types of liver lesions that are manually annotated by radiologists. We also intend to verify the clinical usefulness of the CBIR system.
The focal liver lesions in the CT images need to be outlined manually in the current retrieval method. It is not convenient for radiologists to use the CBIR system. Moreover, there are interobservation variations in the outlined lesion regions. To address these problems, the existing methods of automatic detection and segmentation of liver lesion in CT images can be integrated to the CBIR system. It is interesting to develop a retrieval system in a simpler and more efficient way, in which the lesion or an ROI only requires to be bounded by a box or an ellipse.
Aside from the standard BoW with hard assignment procedure, there are also other BoW variants. The state-of-the-art methods include soft assignment, LLC, etc. We also tested experimentally the retrieval performance of these methods. Their performance, however, was similar to hard assignment, and no significant improvement was found.
Conclusions
We presented a CBIR method for the retrieval of categorically similar focal liver lesions in the contrast-enhanced CT images. The BoW histograms represented the contents of the liver lesions at each phase. Raw intensity was used as the local descriptor of image patch, and the dense sampling method was employed to form the BoW representations. We used the distance metric learning algorithms in obtaining the semantic similarity between the CT images. Both single-phase and multiple-phase contrast-enhanced CT images were evaluated in the retrieval performance of the BoW representation. Preliminary results demonstrated that the BoW representation of the single phase achieved mAP greater than 90 %. Additionally, combining the BoW representations of three phases could improve mAP to 94.5 %. These encouraging results suggest that it is feasible to retrieve similar lesions in the contrast-enhanced CT image via BoW representation. In future works, we hope to extend the BoW approach on a liver CT image dataset composed of more types of liver lesions, in which the similarity is manually annotated. We also intend to develop a practical system for clinical decision making and medical education.
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (973 Program) (no. 2010CB732505) and National Natural Science Funds of China (no. 81101109, no. 30900380 and no. 31000450).
References
- 1.Müller H, Michoux N, Bandon D, Geissbuhler A. A review of content-based image retrieval systems in medical applications–clinical benefits and future directions. Int J Med Informatics. 2004;73(1):1–23. doi: 10.1016/j.ijmedinf.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 2.Chen EL, Chung PC, Chen CL, Tsai HM, Chang CI. An automatic diagnostic system for CT liver image classification. IEEE Trans Biomed Eng. 1998;45(6):783–794. doi: 10.1109/10.678613. [DOI] [PubMed] [Google Scholar]
- 3.Gletsos M, Mougiakakou SG, Matsopoulos GK, Nikita KS, Nikita AS, Kelekis D. A computer-aided diagnostic system to characterize CT focal liver lesions: design and optimization of a neural network classifier. IEEE Trans Info Tech Biomed. 2003;7(3):153–162. doi: 10.1109/TITB.2003.813793. [DOI] [PubMed] [Google Scholar]
- 4.Bilello M, Gokturk SB, Desser T, Napel S, Jeffrey RB, Jr, Beaulieu CF. Automatic detection and classification of hypodense hepatic lesions on contrast-enhanced venous-phase CT. Med Phys. 2004;31(9):2584–2593. doi: 10.1118/1.1782674. [DOI] [PubMed] [Google Scholar]
- 5.Mougiakakou SG, Valavanis IK, Nikita A, Nikita KS. Differential diagnosis of CT focal liver lesions using texture features, feature selection and ensemble driven classifiers. Artif Intell Med. 2007;41(1):25–37. doi: 10.1016/j.artmed.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Napel SA, et al. Automated retrieval of CT images of liver lesions on the basis of image similarity: method and preliminary results. Radiology. 2010;256(1):243–252. doi: 10.1148/radiol.10091694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye J, Sun Y, Wang S, Gu L, Qian L, Xu J: Multi-phase CT image based hepatic lesion diagnosis by SVM. In the 2nd International Conference on Biomedical Engineering and Informatics 2009
- 8.Nino-Murcia M, Olcott EW, Jeffrey RB, Lamm RL, Beaulieu CF, Jain KA. Focal liver lesions: pattern-based classification scheme for enhancement at arterial phase CT. Radiology. 2000;215(3):746–751. doi: 10.1148/radiology.215.3.r00jn03746. [DOI] [PubMed] [Google Scholar]
- 9.Akgül C, Rubin D, Napel S, Beaulieu C, Greenspan H, Acar B. Content-based image retrieval in radiology: current status and future directions. J Digit Imaging. 2011;24(2):202–222. doi: 10.1007/s10278-010-9290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao CG, Cheng HY, Huo YL, Zhuang TG: Liver CT-image retrieval based on Gabor texture. In EMBS, 2004 [DOI] [PubMed]
- 11.Lee C-C, Chen S-H, Tsai H-M, Chung P-C, Chiang Y-C: Discrimination of liver diseases from CT images based on Gabor filters. In the 19th IEEE Symposium on Computer-Based Medical Systems, 2006
- 12.El-Gendy MM, El-Zahraa Bou-Chadi F: An automated system for classifying computed tomographic liver images. In National Radio Science Conference, 2009
- 13.Varma M, Zisserman A. A statistical approach to texture classification from single images. Int J Comput Vis. 2005;62(1):61–81. [Google Scholar]
- 14.Manik V, Andrew Z. A statistical approach to material classification using image patch exemplars. IEEE Trans Pattern Anal Mach Intell. 2009;31(11):2032–2047. doi: 10.1109/TPAMI.2008.182. [DOI] [PubMed] [Google Scholar]
- 15.Li F-F, Perona P: A bayesian hierarchical model for learning natural scene categories. In IEEE Conference on Computer Vision and Pattern Recognition, 2005
- 16.Winn J, Criminisi A, Minka T: Object categorization by learned universal visual dictionary. In International Conference on Computer Vision, 2005
- 17.Florent P. Universal and adapted vocabularies for generic visual categorization. IEEE Trans Pattern Anal Mach Intell. 2008;30(7):1243–1256. doi: 10.1109/TPAMI.2007.70755. [DOI] [PubMed] [Google Scholar]
- 18.Gemert JC, Snoek CGM, Veenman CJ, Smeulders AWM, Geusebroek J-M. Comparing compact codebooks for visual categorization. Computer Vision and Image Understanding. 2010;114(4):450–462. doi: 10.1016/j.cviu.2009.08.004. [DOI] [Google Scholar]
- 19.Jégou H, Douze M, Schmid C. Improving bag-of-features for large scale image search. Int J Comput Vis. 2011;87(3):316–336. doi: 10.1007/s11263-009-0285-2. [DOI] [Google Scholar]
- 20.Avni U, Greenspan H, Sharon M, Konen E, Goldberger J: X-ray image categorization and retrieval using patch-based visual words representation. In the Sixth IEEE International Conference on Symposium on Biomedical Imaging, 2009
- 21.Deserno T, Antani S, Long R. Ontology of gaps in content-based image retrieval. J Digit Imaging. 2009;22(2):202–215. doi: 10.1007/s10278-007-9092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gemert JC, Veenman CJ, Smeulders AWM, Geusebroek J-M. Visual Word Ambiguity. IEEE Trans Pattern Anal Mach Intell. 2009;32(7):1271–1283. doi: 10.1109/TPAMI.2009.132. [DOI] [PubMed] [Google Scholar]
- 23.Lowe DG. Distinctive image features from scale-Invariant keypoints. Int J Comput Vis. 2004;60(2):91–110. doi: 10.1023/B:VISI.0000029664.99615.94. [DOI] [Google Scholar]
- 24.Coates A, Lee H, Ng AY: An analysis of single-layer networks in unsupervised feature learning. In the 14th International Conference on Artificial Intelligence and Statistics (AISTATS), 2011
- 25.Wojcikiewicz W, Binder A, Kawanabe M: Enhancing image classification with class-wise clustered vocabularies. In the 20th International Conference on Pattern Recognition, 2010
- 26.Wang J, Yang J, Yu K, Lv F, Huang T, Gong Y: Locality-constrained Linear Coding for Image Classification. In IEEE Conference on Computer Vision and Pattern Recognition, 2010
- 27.Chang H, Yeung D-Y. Kernel-based distance metric learning for content-based image retrieval. Image and Vision Computing. 2007;25(5):695–703. doi: 10.1016/j.imavis.2006.05.013. [DOI] [Google Scholar]
- 28.Masashi S. Dimensionality reduction of multimodal labeled data by local Fisher discriminant analysis. J Mach Learn Res. 2007;8:1027–1061. [Google Scholar]
- 29.Xing EP, Ng AY, Jordan MI, Russell SJ: Distance metric learning with application to clustering with side-information. In Conference on Neural Information Processing Systems (NIPS), 2002
- 30.Weinberger KQ, Saul LK. Distance metric learning for large margin nearest neighbor classification. J Mach Learn Res. 2009;10:207–244. [Google Scholar]
- 31.Alipanahi B, Biggs M, Ghodsi A: Distance metric learning vs. Fisher discriminant analysis. In the 23rd national conference on Artificial intelligence, 2008
- 32.Zhang Z, Dai G, Xu C, Jordan MI. Regularized discriminant analysis, ridge regression and beyond. J Mach Learn Res. 2010;11(3):2199–2228. [Google Scholar]
- 33.Perronnin F, Senchez J, Xerox Y: Large-scale image categorization with explicit data embedding. In IEEE Conference on Computer Vision and Pattern Recognition, 2010
- 34.Vedaldi A, Zisserman A: Efficient additive kernels via explicit feature maps. In IEEE Conference on Computer Vision and Pattern Recognition, 2010 [DOI] [PubMed]
- 35.Manjunath BS, Ma WY. Texture features for browsing and retrieval of image data. IEEE Trans Pattern Anal Mach Intell. 1996;18(8):837–842. doi: 10.1109/34.531803. [DOI] [Google Scholar]