Abstract
The plastid rRNA (rrn) operon in chloroplasts of tobacco (Nicotiana tabacum), maize, and pea is transcribed by the plastid-encoded plastid RNA polymerase from a ς70-type promoter (P1). In contrast, the rrn operon in spinach (Spinacia oleracea) and mustard chloroplasts is transcribed from the distinct Pc promoter, probably also by the plastid-encoded plastid RNA polymerase. Primer-extension analysis reported here indicates that in Arabidopsis both promoters may be active. To understand promoter selection in the plastid rrn operon in the different species, we have tested transcription from the spinach rrn promoter in transplastomic tobacco and from the tobacco rrn promoter in transplastomic Arabidopsis. Our data suggest that transcription of the rrn operon depends on species-specific factors that facilitate transcription initiation by the general transcription machinery.
In higher plants the genes for the plastid 16S, 23S, and 5S rRNA are encoded in the plastid genome and are transcribed as a large precursor RNA, which is subsequently processed into the various mature rRNA species. The plastid rrn operon in maize, pea, and tobacco (Nicotiana tabacum) is transcribed from a promoter with two conserved blocks (“-10” and “-35”) of hexameric sequences reminiscent of the Escherichia coli ς70-type promoters (P1 or Nt-Prrn-114; Strittmatter et al., 1985; Sun et al., 1989; Vera and Sugiura, 1995; Allison et al., 1996). The P1 promoter is recognized by the PEP. In addition to P1, the rrn operon in tobacco has a second promoter (P2 or Nt-Prrn-62), which is inactive in chloroplasts and functions only in BY2 tissue culture cells (Vera and Sugiura, 1995) and in plants lacking PEP (Allison et al., 1996). This second promoter is transcribed by the nuclear-encoded plastid RNA polymerase (Allison et al., 1996).
In contrast to maize, tobacco, and pea, the rrn operon in spinach (Spinacia oleracea) and mustard is transcribed from promoters lacking properly spaced -10/-35 elements in vivo (Baeza et al., 1991; Iratni et al., 1997; Pfannschmidt and Link, 1997). In spinach the strong Pc promoter is active in chloroplasts but not in roots. The Pc site appears to be faithfully recognized by the mustard PEP in vitro (Pfannschmidt and Link, 1997). A second weak promoter transcribing the spinach rrn operon is upstream of tRNA(GAC)Val (Iratni et al., 1997). It is not known which of the plastid RNA polymerases is recognizing this second promoter.
Since the rrn operon in the chloroplasts of higher plants is either transcribed from P1 or Pc, promoters that have transcription initiation sites 26 nucleotides apart, it was uncertain whether both promoters may function in the same plastid. Given the potential overlap between Pc and P1, promoter exclusion was proposed as the mechanism to explain transcription from Pc but not from the P1 promoter in spinach (Iratni et al., 1994). Mapping of RNA 5′ ends upstream of rrn in the present study led to the identification of transcripts characteristic of both P1 and Pc in Arabidopsis. Given the confidence that both promoters may be active in the same plastid, we designed transgenic experiments to understand promoter selection in the plastid rrn operon in higher plants. Specifically, we have tested transcription from the spinach rrn promoter in transplastomic tobacco and from the tobacco rrn promoter in transplastomic Arabidopsis. We conclude from this study that tobacco plastids lack the factor required for transcription from Pc, whereas spinach has an intact P1 promoter but lacks the cognate P1 activator. These findings suggest that P1 and Pc activity depends on promoter-specific factors that are essential for the recognition of these promoters by the general transcription machinery.
MATERIALS AND METHODS
Construction of Vector pPS105
pPS102 is a pBSKS+ plasmid (Stratagene) derivative that contains a chimeric uidA gene as a SacI-HindIII fragment. The chimeric uidA gene consists of the following: between the SacI and NcoI sites, the spinach (Spinacia oleracea) promoter fragment and a ribosome-binding site; between the NcoI and XbaI sites, the uidA-coding region with an N-terminal c-myc tag; and between the XbaI and HindIII sites, the 3′ untranslated region of the rps16 ribosomal protein gene (Trps16; Staub and Maliga, 1994). The SacI-NcoI fragment was obtained by PCR amplification of spinach plastid DNA using the following oligonucleotide primers: O1, ccgagctCCCAACGTCAGTTTTTCT; and O2, ccgctagccatggatccctccctacaacGTAGACAAAGCGGATTC.
In the primer sequences, anchor sequences derived from the plastid genome are designated in uppercase letters; nucleotides added during vector construction are designated in lowercase letters; restriction sites are underlined; and the ribosome-binding site is in italic font. The uidA-coding region was tagged by translationally fusing it with a PCR approach at its N terminus with amino acids 410 to 419 (EQKLISEEDL) of the human c-myc protein (Kolodziej and Young, 1991; L.A. Allison, unpublished data).
Plasmid pPS105 was obtained by cloning the uidA gene as a SacI-HindIII fragment into SacI-HindIII-digested pPRV111A plastid-transformation vector, as described by Zoubenko et al., 1994.
Tobacco Plastid Transformation
For plastid transformation, tungsten particles were coated with DNA and introduced into the leaves of tobacco (Nicotiana tabacum) plants using the Dupont PDS1000He Biolistic gun at 1100 p.s.i. Transgenic shoots were selected aseptically on RMOP medium containing 500 mg/L spectinomycin dihydrochloride (Svab and Maliga, 1993). Transgenic cuttings were rooted and maintained on agar-solidified Murashige-Skoog salts containing 3% Suc (Murashige and Skoog, 1962).
Primer-Extension Analysis
For RNA isolation, wild-type Arabidopsis (RLD ecotype) seeds were germinated and grown in vitro on a medium containing Murashige-Skoog salts (Murashige and Skoog, 1962) and 2% Suc. Cotyledon samples were collected from 10-d-old seedlings. Leaves were harvested from 3-week-old plants. Root tissue was obtained from 2-week-old Arabidopsis cultures grown in liquid ARM medium (Marton and Browse, 1991). The leaves of transgenic pGS31A-16 Arabidopsis plants (Sikdar et al., 1998) were collected from plants maintained on agar-solidified ARM medium. Wild-type and transgenic tobacco leaves were taken from plants grown aseptically on a medium containing Murashige and Skoog salts and 3% Suc (Murashige and Skoog., 1962). Spinach leaves were derived from seedlings grown aseptically on a medium containing Murashige-Skoog salts and 2% Suc.
Total cellular RNA was prepared according to the method of Stiekema et al. (1988). Primer-extension reactions were carried out using 3 μg (wild-type Arabidopsis, tobacco, and spinach leaves and cotyledons) or 10 μg (transgenic tobacco and Arabidopsis leaves; wild-type Arabidopsis roots) of total RNA, as described by Allison and Maliga (1995). The following oligonucleotide primers were used: 16S rRNA: O3, 5′-TTCATAGTTGCATTACTTATAGCTTC-3′; uidA: O4, 5′-GGCCGTCGAGTTTTTTGATTTCACGGGTTGGGG-3′; aadA: O5, 5′-CGCTCGATGACGCCAACTACC-3′.
Sequence ladders generated with the same primers using the Sequenase II kit (United States Biochemical) were used as molecular size markers.
RESULTS
In Arabidopsis Chloroplasts Both P1 and Pc Are Active
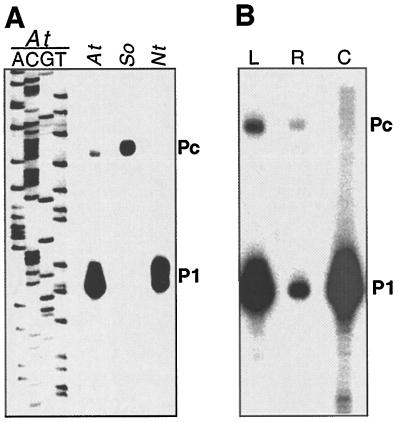
Primer-extension analysis was carried out to identify the plastid rrn operon promoter in Arabidopsis. Two RNA species were identified in leaves with 5′ ends mapping to 111 and 139 nucleotides upstream of 16S rRNA, the first gene of the rrn operon (Fig. 1A). The position of the 5′ end of these transcripts suggests transcription initiation from P1 and Pc promoter homologs (Fig. 2). The ratio of the two transcripts was approximately 10:1.
Figure 1.
Identification of promoters for the plastid rrn operon in Arabidopsis. A, Primer-extension analysis to identify RNA 5′ ends upstream of the rrn operon in Arabidopsis (At), spinach (So), and tobacco (Nt) leaves. The Arabidopsis rrn promoter sequence obtained with the same primer is shown for comparison. Nucleotides at which P1 and Pc transcription initiates are marked in Figure 2. B, Pc is active in Arabidopsis leaves (L) and roots (R) but not in cotyledons (C).
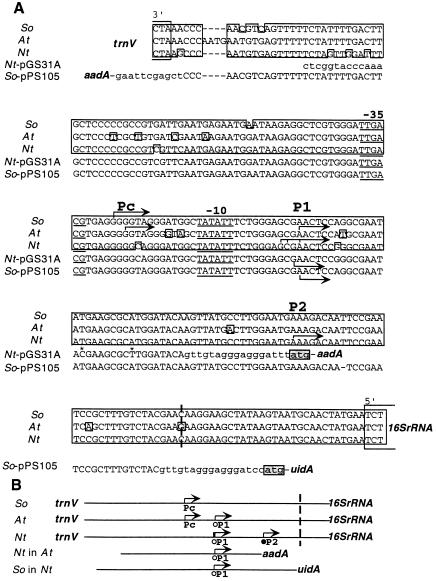
Figure 2.
Promoters for the rrn operon in spinach (So), Arabidopsis (At), and tobacco (Nt). A, Alignment of DNA sequences between trnV and 16SrDNA. DNA sequences of the tobacco rrn promoter fragment in transplastomic Arabidopsis from plasmid pGS31A and of the spinach rrn promoter fragment in transplastomic tobacco from plasmid pPS105 are also shown. Nucleotides derived from the plastid genome are shown in uppercase letters, whereas those added during vector construction are shown in lowercase letters. Conserved sequences in the plastid genomes are boxed. Transcription-initiation sites are marked with horizontal arrows. The RNA-processing site is marked with a dashed vertical line. Stars in pGS31A denote mutations introduced to eliminate translation-initiation codons. B, Schematic representation of transcription-initiation sites in A. PEP and nuclear-encoded plastid RNA polymerase promoters are marked with open and filled circles, respectively.
Both P1 and Pc are active in roots and in leaves (Fig. 1B). However, no −139-transcript was detectable in cotyledons, suggesting organ-specific utilization of Pc (Fig. 1B).
Testing Transcription from the Spinach rrn Promoter in Tobacco Chloroplasts
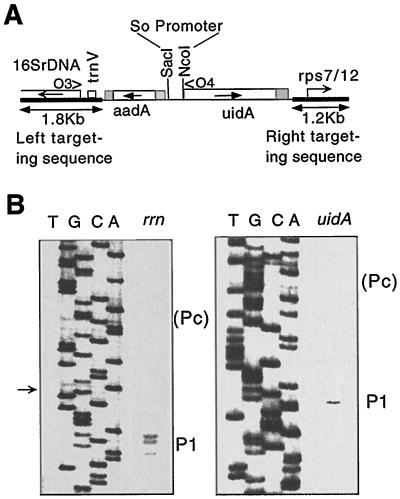
To test whether sequence differences between the spinach and tobacco rrn operon promoters are responsible for promoter choice, transcription from the spinach promoter was tested in transgenic tobacco plants. For this, the spinach rrn operon promoter was PCR amplified as a 292-bp DNA fragment containing the region between trnV and the rrn operon-processing site (Fig. 2). This promoter fragment was cloned upstream of a uidA reporter gene, which was subsequently linked to a spectinomycin-resistance (aadA) gene in plastid vector pPRV111A (Fig. 3A). The resulting plasmid, pPS105, was introduced into tobacco chloroplasts by particle bombardment, where the linked aadA and uidA genes integrated into the plastid genome via the plastid-targeting sequences (Fig. 3A). Plastid transformants were selected by spectinomycin resistance.
Figure 3.
Transcription initiation in the spinach rrn promoter fragment in tobacco chloroplasts. A, Plastid-targeting region of plasmid pPS105. Shown are: aadA-selectable marker gene, uidA reporter gene, and SacI and NcoI sites for insertion of spinach rrn promoter. Direction of transcription is shown by horizontal arrows. O3 and O4 mark the position of oligonucleotides used for primer extension. B, Primer-extension analysis to map RNA 5′ ends in transplastomic Nt-pPS105 tobacco plants. Data are shown for the native rrn operon using primer O3 and for the uidA transgene driven by the spinach rrn promoter region using primer O4. No signal was detected at the Pc position, even when the film was overexposed. DNA sequences obtained with the same primers are shown alongside primer extension. Sequences above the mark for rrn were derived from cloning vector.
Promoter utilization in the uidA gene was tested in the leaves of transgenic Nt-pPS105–1 tobacco plants. Primer-extension analysis identified a single 5′ end mapping to the P1 site within the spinach promoter fragment (Fig. 3B). This finding indicates that all sequence information required for transcription from the P1 promoter is present in spinach. Since there is no transcript initiating from the Pc site, even when the blots are overexposed, tobacco chloroplasts cannot recognize Pc sequences.
Testing Transcription from the Tobacco rrn Promoter in Arabidopsis Chloroplasts
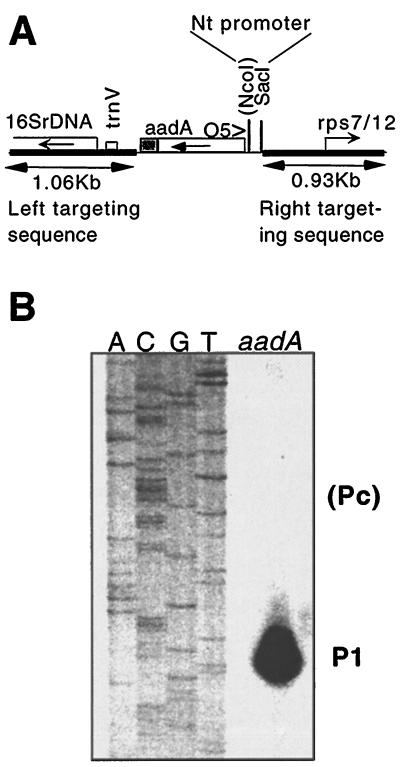
Because tobacco chloroplasts lack the ability to initiate transcription from Pc, we were interested in determining whether sequences for Pc function are present. Spinach plastid transformation was not available; therefore, we could not test transcription initiation from the tobacco rrn operon promoter in spinach. However, an Arabidopsis plant expressing aadA from the tobacco Prrn promoter was available (Sikdar et al., 1998; Fig. 2). Therefore, we could test transcription from the tobacco rrn promoter in the At-pGS31A transplastomic plants in which both Pc and P1 are active upstream of the native rrn operon. In the transplastomic plants, primer-extension analysis identified only one RNA 5′ end upstream of aadA. This 5′ end maps to the P1 site (Figs. 2 and 4B). Since in the transgenic Arabidopsis plant no transcript initiates at Pc within the tobacco rrn promoter fragment, tobacco plastids apparently lack functional Pc promoter sequences.
Figure 4.
Transcription initiation in the tobacco rrn promoter fragment in Arabidopsis chloroplasts. A, Plastid-targeting region of plasmid pGS31A with the spectinomcyin-resistance (aadA) gene. The position of primer O5 is shown. Direction of transcription is marked by horizontal arrows. B, Primer-extension analysis to map RNA 5′ ends. The primer-extension product is run alongside the DNA sequence obtained with the same primer.
DISCUSSION
We conclude from our studies that transcription initiation from the P1 and Pc promoters depends on promoter-specific transcription factors that are necessary for transcription initiation by the general transcription machinery. Spinach apparently has the P1 promoter sequence, as evidenced by transcription initiation from the spinach rrn promoter in tobacco at the P1 site. Although other genes, e.g. rbcL, atpB, and psbB, are transcribed by PEP from ς70-type promoters (Mullet et al., 1985; Westhoff, 1985), the P1-PEP promoter is silent in spinach. Therefore, the lack of P1 activity in spinach is probably due to the lack of a functional P1-specific activating factor. This conclusion is strengthened by the observation that P1 is silent in spinach roots where Pc is inactive, indicating that transcription from Pc is not the reason for the lack of P1 activity in chloroplasts (Iratni et al., 1997). Tobacco chloroplasts apparently lack the factor required for Pc recognition, otherwise transcription from the spinach rrn promoter region in tobacco would initiate at both the P1 and the Pc site. Since tobacco does not have a Pc factor, it is not surprising that it does not have a functional Pc promoter sequence, as suggested by the lack of transcription initiation at Pc from the tobacco rrn promoter in Arabidopsis plastids. Relevant in this regard could be point mutations in tobacco compared with spinach in the rrn upstream region (Fig. 2A).
Our data can be best explained by the requirement for a positive regulatory factor(s) for rrn transcription. A good candidate for the Pc-activating factor is the protein CDF2, the binding of which is correlated with transcription from Pc in spinach (Iratni et al., 1997). The target site of the rrn P1 activator in pea seems to be sequences upstream of the −35 hexamer required for specific transcription initiation (Sun et al., 1989). This site coincides with the CDF2-binding site (Baeza et al., 1991). The proposed plastid transcriptional activators would have a function analogous to the Fis protein, for which there are multiple binding sites upstream of the P1 ribosomal operon promoter in E. coli (Ross et al., 1990, 1993; Condon et al., 1992).
Selective activation of plastid housekeeping genes early during chloroplast development is an important regulatory step (Baumgartner et al., 1993; DuBell and Mullet, 1995). In the case of the rRNA operon, this may be accomplished by a gene-specific transcriptional activator that allows selective control of plastid gene transcription by the nucleus.
ACKNOWLEDGMENTS
We thank Samir Sikdar for the leaf sample of transgenic At-pGS31A-16 plants, German Serino for plasmid pGS31A, and Lori Allison for the c-myc-tagged uidA.
Abbreviation:
- PEP
plastid-encoded plastid RNA polymerase
Footnotes
This research was supported by the National Science Foundation (grant no. MCB 96-30763) and the Monsanto Company. P.S. was the recipient of a Johanna and Charles Busch Predoctoral Fellowship Award.
LITERATURE CITED
- Allison LA, Maliga P. Light-responsive and transcription-enhancing elements regulate the plastid psbD core promoter. EMBO J. 1995;14:3721–3730. doi: 10.1002/j.1460-2075.1995.tb00042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison LA, Simon LD, Maliga P. Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 1996;15:2802–2809. [PMC free article] [PubMed] [Google Scholar]
- Baeza L, Bertrand A, Mache R, Lerbs-Mache S. Characterization of a protein binding sequence in the promoter region of the 16S rRNA gene of the spinach chloroplast genome. Nucleic Acids Res. 1991;19:3577–3581. doi: 10.1093/nar/19.13.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner BJ, Rapp JC, Mullet JE. Plastid genes encoding the transcription/translation apparatus are differentially transcribed early in barley (Hordeum vulgare) chloroplast development. Plant Physiol. 1993;101:781–791. doi: 10.1104/pp.101.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, Philips J, Fu ZY, Squires C, Squires CL. Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. EMBO J. 1992;11:4175–4185. doi: 10.1002/j.1460-2075.1992.tb05511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBell AN, Mullet JE. Differential transcription of pea chloroplast genes during light-induced leaf development. Plant Physiol. 1995;109:105–112. doi: 10.1104/pp.109.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iratni R, Baeza L, Andreeva A, Mache R, Lerbs-Mache S. Regulation of rDNA transcription in chloroplasts: promoter exclusion by constitutive repression. Genes Dev. 1994;8:2928–2938. doi: 10.1101/gad.8.23.2928. [DOI] [PubMed] [Google Scholar]
- Iratni R, Diederich L, Harrak H, Bligny M, Lerbs-Mache S. Organ-specific transcription of the rrn operon in spinach plastids. J Biol Chem. 1997;272:13676–13682. doi: 10.1074/jbc.272.21.13676. [DOI] [PubMed] [Google Scholar]
- Kolodziej PA, Young RA. Epitope tagging and protein surveillance. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- Marton L, Browse J. Facile transformation of Arabidopsis thaliana. Plant Cell Rep. 1991;10:235–239. doi: 10.1007/BF00232565. [DOI] [PubMed] [Google Scholar]
- Mullet JE, Orozco EM, Chua NH. Multiple transcripts for higher plant rbcL and atpB genes and localization of the transcription initiation site of the rbcL gene. Plant Mol Biol. 1985;4:39–54. doi: 10.1007/BF02498714. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Pfannschmidt T, Link G. The A and B forms of plastid DNA-dependent RNA polymerase from mustard (Sinapis alba L.) transcribe the same genes in a different developmental context. Mol Gen Genet. 1997;257:35–44. doi: 10.1007/s004380050621. [DOI] [PubMed] [Google Scholar]
- Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severionov K, Gourse RL. The third recognition element in bacterial promoters: DNA binding by the α subunit of the RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- Ross W, Thompson JF, Newlands JT, Gourse RL. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikdar SR, Serino G, Chaudhuri S, Maliga P (1998) Plastid transformation in Arabidopsis thaliana. Plant Cell Rep (in press)
- Staub JM, Maliga P. Translation of psbA mRNA is regulated by light via the 5′-untranslated region in tobacco plastids. Plant J. 1994;6:547–553. doi: 10.1046/j.1365-313x.1994.6040547.x. [DOI] [PubMed] [Google Scholar]
- Stiekema WJ, Heidekamp F, Dirkse WG, van Beckum J, de Haan P, ten Bosch C, Louwerse JD. Molecular cloning and analysis of four potato tuber mRNAs. Plant Mol Biol. 1988;11:255–269. doi: 10.1007/BF00027383. [DOI] [PubMed] [Google Scholar]
- Strittmatter G, Godzicka-Josefiak A, Kössel H. Identification of an rRNA operon promoter from Zea mays chloroplast which excludes the proximal tRNAval from the primary transcript. EMBO J. 1985;4:599–604. doi: 10.1002/j.1460-2075.1985.tb03672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun E, Wu BW, Tewari KK. In vitro analysis of the pea chloroplast 16S rRNA gene promoter. Mol Cell Biol. 1989;9:5650–5659. doi: 10.1128/mcb.9.12.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera A, Sugiura M. Chloroplast rRNA transcription from structurally different tandem promoters: an additional novel-type promoter. Curr Genet. 1995;27:280–284. doi: 10.1007/BF00326161. [DOI] [PubMed] [Google Scholar]
- Westhoff P. Transcription of the gene encoding the 51 kd chlorophyll a-apoprotein of the photosystem II reaction centre from spinach. Mol Gen Genet. 1985;201:115–123. [Google Scholar]
- Zoubenko OV, Allison LA, Svab Z, Maliga P. Efficient targeting of foreign genes into the tobacco plastid genome. Nucleic Acids Res. 1994;22:3819–3824. doi: 10.1093/nar/22.19.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]