Abstract
Background
DNA methylation of gene promoters is associated with transcriptional inactivation. Changes in DNA methylation can lead to differences in gene expression levels and thereby influence disease development. We hypothesized that epigenetics underlies the pathogenesis of minimal change nephrotic syndrome (MCNS).
Methods
Genome-wide DNA methylation changes between relapse and remission in monocytes (n = 6) and naive T helper cells (Th0s) (n = 4) isolated from patients with MCNS were investigated using the microarray-based integrated analysis of methylation by isochizomers (MIAMI) method. We confirmed the MIAMI results using bisulfite-pyrosequencing analysis. Expression analysis was performed using quantitative real-time PCR.
Results
Three gene loci (GATA2, PBX4, and NYX) were significantly less methylated in Th0s during relapse than in remission, compared to none in monocytes. In addition, the distance distribution from the regression line of all probes in MIAMI was significantly different between monocytes and Th0s. The mRNA levels of the three genes in Th0s were not significantly different between relapse and remission.
Conclusions
Our results demonstrate that the change in DNA methylation patterns from remission to relapse in MCNS occurs predominantly in Th0s rather than in monocytes and suggest that epigenetic regulation in Th0s underlies the pathogenesis of MCNS.
Electronic supplementary material
The online version of this article (doi:10.1007/s00467-012-2248-z) contains supplementary material, which is available to authorized users.
Keywords: DNA methylation, Nephrotic syndrome, Monocytes, Naive T helper cells, Microarray-based integrated analysis of methylation by isoschizomers (MIAMI) method, Genome-wide, Children
Introduction
Epigenetics is the study of mitotically heritable changes in gene expression that occur without direct DNA sequence alterations. DNA methylation, one of the principal epigenetic mechanisms in mammals, involves the covalent addition of a methyl group to a cytosine residue that is followed by a guanine (CpG) [1]. DNA methylation regulates gene expression and is essential for differentiation, embryonic development [2], genomic imprinting [3], and X-chromosome inactivation [4]. DNA methylation within the promoter region of a gene is commonly associated with transcriptional inactivation, whereas demethylation contributes to transcriptional activation. Changes in the DNA methylation profile can also lead to differences in gene expression patterns and thereby influence the development of diseases, such as cancer [5].
Minimal change nephrotic syndrome (MCNS) is the most common cause of nephrotic syndrome in children and is characterized by massive proteinuria and hypoalbuminemia in a relapse/remission course without histological evidence of immune-mediated inflammatory damage. These manifestations are typically reversible with the use of corticosteroid therapy. Although the pathogenesis of MCNS remains to be elucidated, immunological disruption has been implicated in this disease [6] as T cell-derived vascular permeability factors have been shown to be responsible for alterations in glomerular permeability [7–9]. The incidence of MCNS in childhood is twofold higher in boys, with a prevalence that is inversely proportional to age, and recurrent relapse tends to lessen after adolescence [10, 11]. Since the characteristic features of MCNS include (1) a recurrent relapse/remission course, (2) gender preference, (3) age preference of onset and relapse, and (4) steroid response in most patients, a genetic defect cannot explain the pathogenesis of this disease; however, epigenetic alterations may occur without a direct change in the genetic sequence. DNA methylation changes with age and environmental factors, even in the same individual, and is involved with X-chromosome inactivation.
Audard et al. reported that NFRKB (nuclear factor related to kappaB binding protein) was highly expressed in the nuclear compartment during relapse and that NFRKB promotes hypomethylation of genomic DNA, suggesting epigenetic involvement in the pathogenesis of MCNS [12]. The epigenotype is influenced by the environment and alters the regulation of gene expression, leading Elie et al. to suggest a probable impact of epigenetic modifications in infected cells since MCNS relapses are frequently triggered by external or internal environmental factors, including viral infection [13]. Zhang et al. reported significant differences in histone H3 lysine 4 tri-methylation of peripheral blood mononuclear cells (PBMCs) from adult patients with MCNS compared with those from healthy subjects. Their results indicate that alterations in epigenotype are associated with the pathogenesis of MCNS [14].
The aim of this study is to elucidate whether the DNA methylation profile changes between relapse and remission in MCNS cases and whether this process is immune-competent cell-type-specific. Ultimately, we wished to determine whether epigenetics underlies the pathogenesis of MCNS.
Patients and methods
Patients
Samples for microarray-based integrated analysis of methylation by isochizomers (MIAMI) analysis were obtained from six male patients with MCNS (Table 1), while samples for quantitative real-time PCR (qRT-PCR) were obtained from an additional seven patients with MCNS (3 boys, 4 girls) (Table 2) at relapse and also following complete remission. All patients were diagnosed according to the criteria of the International Study of Kidney Disease in Children [15] and had developed nephrotic syndrome prior to 16 years of age. Informed consent was obtained from the parents of each child and from older children/adolescents as necessary. This study was approved by the Ethics Committee of Gunma University Graduate School of Medicine, Japan (Receipt Number 89).
Table 1.
Characteristics of patients with minimal change nephrotic syndrome at sampling whose DNA was isolated for methylation analysis
| Patient no. | Sex | Age at onset | Total follow-up perioda (months) | Frequent relapser | Biopsy | Sampling age at relapse | Relapse times at sampling | Intervalsb (weeks) | PSL at relapse (mg/day) | PLS at remission (mg/day) | IS at relapse (mg/day) | IS at remission (mg/day) | CD14+ (n = 6) | RO− (n = 4) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 10y4m | 78 | Yes | MC | 13y0m | 3rd | 5 | 40 | 10 | 0 | 0 | Taken | Taken |
| 2 | M | 5y4m | 35 | Yes | MC | 6y0m | 3rd | 16 | 0 | 35 | 0 | CyA90 | Taken | Taken |
| 3 | M | 5y11m | 186 | Yes | MC | 18y6m | 9th | 23 | 10 | 15 | 0 | 0 | Taken | Taken |
| 4 | M | 7y3m | 188 | Yes | MC | 19y10m | 13th | 23 | 60 | 5 | 0 | 0 | Taken | Taken |
| 5 | M | 9y9m | 109 | Yes | ND | 11y7m | 4th | 12 | 0 | 0 | CyA180 | CyA160 | Taken | NT |
| 6 | M | 11y11m | 61 | Yes | ND | 12y5m | 4th | 71 | 0 | 20 | MZV125 | MZV 75 | Taken | NT |
| Mean | 8y4m | 109.5 | 13y5m | 6th | 25 |
PSL, prednisolone IS, immunosuppressant; M, Male; y, years; m, months; MC, minimal change; ND, not done; CyA, cyclosporin A; MZB, mizoribine; NT, not taken
aFollow-up period till June 2011
bIntervals of sampling between relapse and remission
Table 2.
Characteristics of patients with minimal change nephrotic syndrome at sampling who underwent gene expression analysis
| Patient no. | Sex | Age at onset | Total follow-up perioda (months) | Frequent relapser | Biopsy | Sampling age at relapse | Relapse times at sampling (n) | Intervalsb (weeks) | PSL at relapse (mg/day) | PLS at remission (mg/day) | IS at relapse (mg/day) | IS at remission (mg/day) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | M | 8y4m | 121 | Yes | MC | 11y11m | 8 | 1 | 10 | 60 | MZV150 | MZV150 |
| 8 | M | 10y4m | 78 | Yes | MC | 13y8m | 5 | 2 | 0 | 0 | 0 | 0 |
| 9 | M | 2y5m | 154 | Yes | MC | 17y9m | 20 | 3 | 60 | 40 | MZV150 | MZV150 |
| 10 | F | 4y3m | 44 | No | ND | 6y2m | 2 | 26 | 0 | 0 | 0 | 0 |
| 11 | F | 4y10m | 130 | Yes | ND | 15y0m | 8 | 1 | 0 | 30 | 0 | 0 |
| 12 | F | 10y3m | 36 | Yes | ND | 14y0m | 4 | 13 | 0 | 20ADT | CyA50 | CyA50 |
| 13 | F | 4y7m | 37 | Yes | MC | 6y7m | 7 | 8 | 0 | 0 | CyA80 | CyA80 |
| Mean | 6y5m | 85.7 | 12y1m | 7.7 | 7.7 |
F, Female
aFollow-up period till June 2011
bIntervals of sampling between relapse and remission
Cell separation
We used monocytes, which are precursors of the antigen-presenting cells derived from the myeloid cell series, and naive T helper cells (Th0s), which are derived from the lymphoid system, as material for the analyses. PBMCs were isolated from 20-mL samples of anti-coagulated blood that had been obtained by gradient separation using the Lymphprep™ Tube system (Axis-Shield PoC AS, Oslo, Norway). Monocytes and Th0s were separated from PBMCs that had been magnetically labeled with CD14, CD4, and CD45RO microBeads (Miltenyi Biotec) using an autoMACS Pro Separator (Miltenyi Biotec, Bergisch Gladbach, Germany) . To obtain monocytes, the CD14-positive (CD14+) fraction was collected as monocytes, whereas the CD14-negative, CD4-positive, and CD45RO-negative (CD14-CD4+CD45RO-) fractions were collected as Th0s. Flow cytometric analysis with a MACSQuant® Analyzer (Miltenyi Biotec) revealed that the precision of cell separation was 96.2 % for CD14+ cells and 94.46 % for CD4-positive and CD45RA-positive cells as a CD45RO-negative fraction.
CD14+ monocytes were obtained from six patients, while CD14-CD4+CD45RO- Th0 cells were also obtained from four of these patients (Table 1) for the MIAMI analysis both at relapse and following complete remission. Genomic DNA (gDNA) was extracted from these cells as previously described [16]. Consistent amounts of extracted gDNA (300 ng from monocytes and 250 ng from Th0s) from each selected cell type were pooled and used for subsequent DNA methylation analysis using MIAMI in order to exclude the influences of epigenetic differences among the cell types, to identify specific changes due to clinical courses of MCNS, and to reduce the noise caused by individual differences. Total RNA was extracted from Th0s purified from seven other patients with MCNS (Table 2) using a ToTALLY RNA™ kit (Ambion, Austin, TX). Complementary DNA (cDNA) was synthesized from total RNA of each Th0s sample using the High Capacity RNA-to-cDNA Master Mix (Applied Biosystems, Foster City, CA) for the qRT-PCR.
MIAMI analysis
The MIAMI method, which provides high-throughput global analysis of DNA methylation, was performed as described previously using 1.8 and 1.0 μg of pooled gDNA isolated from the monocytes of six patients and from the Th0s of four patients, respectively [17, 18]. Briefly, this technique utilizes isoschizomers (HpaII and MspI) that recognize the same recognition site (CCGG). Pooled gDNA was digested with HpaII, a methylation-sensitive restriction enzyme that cleaves only unmethylated DNA, and then adapter-ligated and amplified by PCR with primers designed against the adapter sequences. The samples were then further digested with MspI, a methylation-insensitive enzyme that digests CCGG sites irrespective of their methylation status, and amplified again with the same set of primers (HpaII–MspI treatment). The second treatment with MspI yields amplicons from unmethylated DNA fragments only. Hence, only HpaII cleavable unmethylated DNA fragments are amplified, and these can then be quantified based on their respective fluorescence intensity by microarray analysis. The amplified products were then labeled with Cy3 (remission samples) or Cy5 (relapse samples) and co-hybridized to a microarray spotted with 38,172 sixty-mer oligonucleotides covering the vicinity of the transcription start sites (TSSs) of 14,978 genes.
Following hybridization, the microarray was scanned, and the obtained fluorescence intensities were quantified and normalized. The same pooled gDNA samples were treated first with MspI instead of HpaII (MspI–MspI treatment) and analyzed on a duplicate array to correct for false-positives caused by single nucleotide polymorphisms or incomplete digestion.
Bisulfite-pyrosequencing analysis
In the bisulfite-pyrosequencing method, unmethylated cytosine residues are converted into uracil, whereas methylated cytosines remain unchanged. Analysis of the methylation status in this manner exploits the quantitative nature of pyrosequencing by reporting the ratio of cytosine to thymine at each analyzed CpG site, which reflects the proportion of methylated DNA. This analysis was performed using pooled samples in accordance with established protocols. Briefly, gDNA extracts from patients were digested with EcoRI (Takara Bio, Otsu, Japan) and subjected to bisulfite treatment using the EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA). The analyzed CpG sites were in the closest HpaII recognition sites at the 5′- and 3′- ends of the probes designed in the TSSs vicinity of GATA2 (A_17_P02574948), PBX4 (A_17_P10909964), and NYX (A_17_P11717994) used in the MIAMI analysis. Amplification and sequencing primers for pyrosequencing were designed with PyroMark Assay Design software ver. 2.0 (Qiagen, Venlo, the Netherlands) (Electronic Supplementary Material Table 1).
The targeted DNA segments were amplified using a hot start protocol with a touchdown PCR system (Veriti™ Thermal Cycler; Applied Biosystems), and the strand serving as the pyrosequencing template was biotinylated. Following denaturation, the biotinylated single-stranded PCR amplicons were isolated and allowed to hybridize with a sequencing primer. Pyrosequencing (PyroMark Gold Q96 Reagents; Qiagen) was then performed using the PyroMark Q24 system (Qiagen) according to the manufacturer’s protocol. The sequencing assay was validated using an internal control (a non-CpG cytosine within the target methylation sequence region).
Quantitative real-time PCR
The qRT-PCR analysis was performed using the TaqMan PCR method with a 7900HT Fast Real-Time PCR System (Applied Biosystems) with cDNA from Th0s separated from the other patients with MCNS at relapse and subsequent remission. The three genes previously analyzed using MIAMI, namely, GATA2 (Hs00231119_m1), PBX4 (Hs00257935_m1), and NYX (Hs00360869_m1), were assayed. Each gene was assayed four times for each sample. Aliquots of cDNA equivalent to 300 ng of total RNA were used in the RT-PCR reactions. GAPDH (Hs99999905_m1) was used as an endogenous control for normalizing the RNA concentrations. Differences in the CT values between the tested genes and endogenous control (ΔCT) were calculated and used for subsequent statistical analyses.
Statistical analysis
Statistical analyses of the distance distributions from the MIAMI regression line for each of the probes used in the assays was performed using a non-parametric Mann–Whitney U test (Fig. 1). Comparisons between the expression levels for each group were analyzed using the Wilcoxon matched-pairs signed-rank test (Fig. 4a, b). All statistical analyses were performed using GraphPad PRISM5 software (GraphPad Software, La Jolla, CA) with the significance level set at P < 0.05.
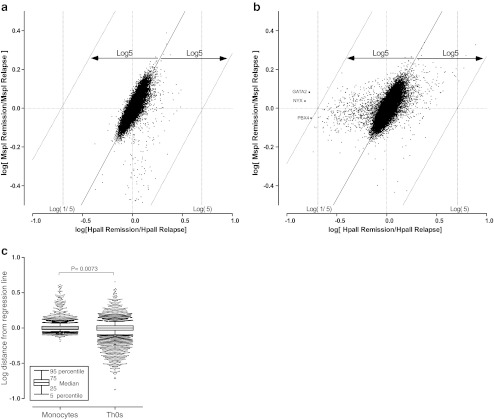
Fig. 1.
a, b Scatter plots of the signals obtained for each probe in monocytes (a) and naive T helper cells (b). Log [(HpaII intensity) Remission/(HpaII intensity) Relapse] values are plotted on the x-axis and log[(MspI intensity) Remission/(MspI intensity) Relapse] values are plotted on the y-axis. The threshold values are determined at log5 of the horizontal distance from the center of the mass and at log5 of the horizontal distance to the regression line of the plots in accordance with the original MIAMI method of Hatada et al. [17, 18]. Points located on the right side and beyond the distance of these lines are judged to be more highly methylated; those located to the left are judged to be significantly less methylated in the relapse samples compared with the remission samples. The three gene probes were found to be less methylated in relapse samples than in remission samples in naive T helper cells (b), whereas no significant signal was detected in monocytes (a). c Distance distributions of all probes are from the regression line. Each dot indicates the log distance of the indicated probes plotted out to the 90th percentile of signals from the regression line. These distributions were significantly different (P = 0.0073) between monocytes and naive T helper cells (Th0s)
Fig. 4.
Expression of GATA2 and PBX4 genes in Th0s from patients with minimal change nephrotic syndrome at relapse and in remission. Nyctalopin (NYX) was not detected in Th0s. a GATA2 expression was low and did not change significantly (P = 0.0781) between relapse and remission. b PBX4 expression did not differ significantly (P = 0. 2188) between relapse and remission
Results
Patients for DNA methylation analysis
We first investigated whether any genome-wide changes in DNA methylation occurred between relapse and remission in the patients with MCNS. The mean total follow-up period until June 2011 was 109.5 months (range 35–188 months). All patients were steroid responsive and frequent relapsers within their total clinical course. Renal biopsy was performed during the follow-up period in four patients who had been administered cyclosporine because of their frequent relapses in order to histologically evaluate the side effects of cyclosporine. The histological findings were consistent with the diagnosis of MCNS. The ages at sampling for relapse and the total number of relapses until samples were collected are listed in Tables 1 and 2. The mean age at relapse sampling was 13 years and 5 months (range 6 years–19 years and 10 months), and the mean number of relapses at the time of sampling was six (range 3–13 relapses). The mean sampling interval from the relapse to the subsequent remission was 25 (range 5–71) weeks. The therapeutic conditions were similar at the time of relapse and remission sampling in all subjects except for patients 2 and 6, who received a corticosteroid (patients 2 and 6) and immunosuppressant (patient 2) at remission (Tables 1, 2).
MIAMI analysis
The scatter plots of the signals from each probe in monocytes and Th0s are shown in Fig. 1a and b. The values for log [(HpaII intensity) remission/(HpaII intensity) relapse] are plotted on the x-axis, which represents the relative methylation changes upon relapse compared with those at remission. The values for the log [(MspI intensity) remission/(MspI intensity) relapse] are plotted on the y-axis and represent the control of the enzyme effects at sample digestion. The threshold values were determined according to the original MIAMI method described by Hatada et al [17, 18]. Three probes were found to be less methylated in the relapse Th0s samples compared with those taken at remission, whereas none of the probes had a significant detectable signal in monocytes (Fig. 1a and b). The distance distributions of all probes from the regression line were significantly different (P = 0.0073) between monocytes and Th0s (Fig. 1c). These results indicate that the DNA methylation status undergoes changes between relapse and remission in Th0s from MCNS patients and that the regulation of DNA methylation differs between monocytes and Th0s.
The three genes detected in Th0s were GATA binding protein 2 (GATA2) at chromosome (Ch) 3 q21.3, pre-B-cell leukemia homeobox 4 (PBX4) at Ch 19 p12, and nyctalopin (NYX) at Ch X p11.4. The locations of the probes used to detect these three genes are shown in Fig. 2. The remission-to-relapse signal intensity ratios measured by MIAMI for the three probes following HpaII treatment are shown in Table 3 and indicate the unmethylated intensity ratio between relapse and remission.
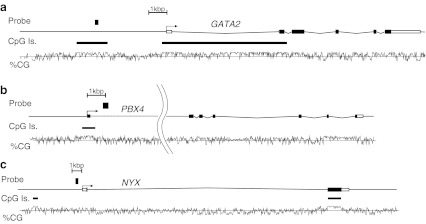
Fig. 2.
Gene maps of GATA2 (a), PBX4 (b), and NYX (c), including the position of the probes used in the MIAMI analysis. The number of CpG (covalent addition of a methyl group to a cytosine residue followed by a guanine) islands and the CG percentages throughout the genomic regions of these genes are also indicated. GATA binding protein 2 (GATA2) maps to chromosome (Ch) 3 q21.3, pre-B-cell leukemia homeobox 4 (PBX4) maps to Ch19 p12, and nyctalopin (NYX) maps to Ch X p11.4. Filled squares indicate the position of the probes with significant changes to the methylation status in Th0s. Probe A_17_P02574948 for GATA2 recognizes the region 3.6 kb upstream of the transcription start site of this gene, which is within a CpG island. The probe A_17_P10909964 sequence in PBX4 is located within the first intron of this gene, and probe A_17_P11717994 for NYX maps to the region just upstream of the transcription start site
Table 3.
Signal intensity ratio in HpaII-treated samples for the indicated gene probes
| Cell type | Gene | Probe | Aa | Assessedb |
|---|---|---|---|---|
| Naive T cells | GATA2 | A_17_P02574948 | 0.167722527 | −1 |
| PBX4 | A_17_P10909964 | 0.174326395 | −1 | |
| NYX | A_17_P11717994 | 0.151913072 | −1 | |
| Monocytes | GATA2 | A_17_P02574948 | 2.523381325 | 0 |
aSignal intensity ratio calculated by [(HpaII intensity) Remission/(HpaII intensity) Relapse] indicating an unmethylated intensity ratio for remission vs. relapse
b−1, Signal intensity ratio at this probe is located further left than the threshold lines indicating significantly lower methylation in the relapse samples than in the remission samples; 0, signal intensity ratio at this probe is located within the threshold lines
Bisulfite-pyrosequencing analysis
To confirm the results of the MIAMI analysis, we directly analyzed the methylation ratio using bisulfite-pyrosequencing for the CpGs that were in the closest CCGG HpaII recognition sites on either side of the recognition sequences of all 3 probes tested in MIAMI analysis (Fig. 3a). At all sites in this analysis, the methylation ratios in Th0s were found to be lower at relapse than in remission (Fig. 3b). The methylation ratios for the GATA2 probe A_17_P02574948 in monocytes were also tested and found to be higher at relapse than in remission on both sides of the probe (Fig. 3c). The methylation status at every CpG site determined by bisulfite-pyrosequencing analysis accorded well with the earlier MIAMI data (Table 3).
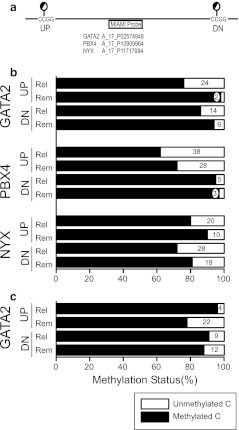
Fig. 3.
DNA methylation ratio analysis by bisulfite-pyrosequencing. To validate the MIAMI data, methylation ratios for CpGs in the closest CCGG HpaII recognition sites on both the 5′ (UP) and 3′ (DN) side of the probe sequences were determined using bisulfite-pyrosequencing (a). These ratios were lower in minimal change nephrotic syndrome (MCNS) relapse (Rel) samples than in remission (Rem) samples at all sites for the three probes in Th0s (b). The methylation ratios for the GATA2 probe A_17_P02574948 in monocytes (c) were higher in Rel than in Rem at both sides of the probe. The methylation pattern at every CpG site determined using bisulfite-pyrosequencing accorded well with the MIAMI results (Table 3), which indicate an unmethylated intensity ratio between relapse and remission
qRT-PCR analysis
According to the MIAMI analysis results, expression of the 3 genes under examination was determined in Th0s from the other patients with MCNS (Table 2) at relapse and subsequent remission using qRT-PCR (Fig. 4). GATA2 expression was low and did not change significantly between relapse and remission (P = 0.0781; Fig. 4a). PBX4 expression was also detectable but did not change significantly (P = 0. 2188; Fig. 4b). NYX was not amplified with the TaqMan probe.
Discussion
A number of different approaches have been adopted in attempts to elucidate the mechanisms underlying alterations in the glomerular capillary permeability in MCNS. These have included studies of the capillary loop membrane, including the podocytes [19–23], searches for secretory factors which may change the permeability of the membrane [4, 24, 25], and investigations of immunocytes as a source of these factors [26, 27]. In the study reported here, we focused on immunocytes and their epigenetic regulation. We specifically isolated and studied Th0s and monocytes rather than using PBMCs, which usually include a wide range of cell types at different differentiation stages, as the latter cells would likely have a distinct epigenotype that could change even through the normal differentiation of Th0s to T helper cells, subset 2 (Th2) [28, 29]. We targeted naive or precursor cells in our search for possible predisposing causes of MCNS to exclude other influences on disease activity, such as Th2 activation, which has been reported in MCNS [30–33].
Differences in epigenetic patterns have been reported to occur even in genetically identical twins, which may be due to the influence of environmental factors [34]. In our experiments, it was possible to eliminate the effects of environmental or aging factors, which can impact on epigenotype variation among individual subjects, by comparing the DNA methylation differences in samples from the same individual obtained within short intervals (Tables 1, 2). However, it was impossible to exclude the effects of treatment with corticosteroids and cyclosporine A, which one and four patients received, respectively, although we could not find clear evidence of the impact of those medicines on the regulation of DNA methylation in immunocytes.
GATA2 is a member of the GATA family of zinc-finger transcription factors (TFs), which play an essential role in the hematopoietic and endocrine systems (http://www.ncbi.nlm.nih.gov/pubmed?term=gata2). PBX4 is a homeodomain protein similar to a TF that is involved in translocations in pre-B-cell leukemia (http://www.ncbi.nlm.nih.gov/pubmed?term=PBX4). NYX is a member of the small leucine-rich proteoglycan family of proteins in which defects can cause congenital stationary night blindness type 1, a rare inherited retinal disorder (http://www.ncbi.nlm.nih.gov/pubmed?term=nyx). Our finding that the DNA methylation ratios were lower in MCNS relapse samples within the promoter regions of these genes suggests that their expression might be higher at relapse than in remission. However, the expression of GATA2 and PBX4 were not higher at relapse in Th0s. One possible explanation for this result is that these two genes are in a primed state for transcription but are not necessarily activated in naive T cells. Changes in DNA methylation may precede changes in gene expression and influence the differentiation of Th0s into effector Th cells and/or influence gene expression after differentiation, which in turn affects the immunological and clinical states of MCNS.
Our findings that DNA methylation levels differ between the tested cell types—one from the myeloid series of cells and the other from the lymphoid system—indicates that it is necessary to separate the different types of immunocytes to investigate the epigenetic regulation of associated diseases, even though they are included in the general PBMC population. Although further analysis is required to determine candidate genes for MCNS, we conclude from our results data that the regulation of DNA methylation in Th0s, but not in monocytes, differs significantly between relapse and remission in affected patients and that epigenetic regulation in Th0s underlies the pathogenesis of MCNS, whose disturbances have been implicated in the development of the disease.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOCX 19 kb)
Acknowledgments
The authors thank Ms. Tomoko Endo, Ms. Chinori Iijima, Ms. Sachiko Hayashi, and Ms. Kiyoe Ishii for their technical assistance. Part of this study was presented at the 2010 Annual Meeting of American Society of Nephrology in Denver, Colorado, November 16–21, 2010. This study was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Research Project Number: 19591238), Kawano Masanori Memorial Foundation for Promotion of Pediatrics, and The Joint Research Program of the Institute for Molecular and Cellular Regulation, Gunma University (Research Project Number: 11027).
Conflict of interest
None.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
References
- 1.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. ChemBioChem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 2.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 3.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 4.Heard E, Clerc P, Avner P. X-chromosome inactivation in mammals. Annu Rev Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- 5.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 6.Shalhoub RJ. Pathogenesis of lipoid nephrosis: a disorder of T-cell function. Lancet. 1974;2:556–559. doi: 10.1016/S0140-6736(74)91880-7. [DOI] [PubMed] [Google Scholar]
- 7.Heslan JM, Branellec A, Laurent J, Lagrue G. The vascular permeability factor is a T lymphocyte product. Nephron. 1986;42:187–188. doi: 10.1159/000183664. [DOI] [PubMed] [Google Scholar]
- 8.Maruyama K, Tomizawa S, Shimabukuro N, Fukuda T, Johshita T, Kuroume T. Studies of vascular permeability factor and inhibitory effect of supernatants derived from T lymphocytes culture in minimal change nephrotic syndrome on rat kidney capillaries. Nephron. 1989;51:73–76. doi: 10.1159/000185246. [DOI] [PubMed] [Google Scholar]
- 9.Koyama A, Fujisaki M, Kobayashi M, Igarashi M, Narita M. A glomerular permeability factor produced by human T cell hybridomas. Kidney Int. 1991;40:453–460. doi: 10.1038/ki.1991.232. [DOI] [PubMed] [Google Scholar]
- 10.International Study of Kidney Disease in Children Nephrotic syndrome in children: prediction of histopathology from clinical and laboratory characteristics at time of diagnosis. Kidney Int. 1978;13:159–165. doi: 10.1038/ki.1978.23. [DOI] [PubMed] [Google Scholar]
- 11.Tarshish P, Tobin JN, Bernstein J, Edelmann CM., Jr Prognostic significance of the early course of minimal change nephrotic syndrome: report of the International Study of Kidney Disease in Children. J Am Soc Nephrol. 1997;8:769–776. doi: 10.1681/ASN.V85769. [DOI] [PubMed] [Google Scholar]
- 12.Audard V, Pawlak A, Candelier M, Lang P, Sahali D. Upregulation of Nuclear Factor-Related Kappa B Suggests a Disorder of Transcriptional Regulation in Minimal Change Nephrotic Syndrome. PLoS One. 2012;7:e30523. doi: 10.1371/journal.pone.0030523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elie V, Fakhoury M, Deschenes G, Jacqz-Aigrain E. Physiopathology of idiopathic nephrotic syndrome: lessons from glucocorticoids and epigenetic perspectives. Pediatr Nephrol. 2011 doi: 10.1007/s00467-011-1947-1. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Dai Y, Peng W, Lu J, Zhang Y, Wang L. Genome-wide analysis of histone H3 lysine 4 trimethylation in peripheral blood mononuclear cells of minimal change nephrotic syndrome patients. Am J Nephrol. 2009;30:505–513. doi: 10.1159/000243811. [DOI] [PubMed] [Google Scholar]
- 15.International Study of Kidney Disease in Children The primary nephrotic syndrome in children. Identification of patients with minimal change nephrotic syndrome from initial response to prednisone. J Pediatr. 1981;98:561–564. doi: 10.1016/S0022-3476(81)80760-3. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi Y, Arakawa H, Suzuki M, Takizawa T, Tokuyama T, Morikawa A. Polymorphisms of interleukin-4-related genes in Japanese children with minimal change nephrotic syndrome. Am J Kidney Dis. 2003;42:271–276. doi: 10.1016/S0272-6386(03)00652-8. [DOI] [PubMed] [Google Scholar]
- 17.Hatada I, Fukasawa M, Kimura M, Morita S, Yamada K, Yoshikawa T, Yamanaka S, Endo C, Sakurada A, Sato M, Kondo T, Horii A, Ushijima T, Sasaki H. Genome-wide profiling of promoter methylation in human. Oncogene. 2006;25:3059–3064. doi: 10.1038/sj.onc.1209331. [DOI] [PubMed] [Google Scholar]
- 18.Hatada I, Morita S, Kimura M, Horii T, Yamashita R, Nakai K. Genome-wide demethylation during neural differentiation of P19 embryonal carcinoma cells. J Hum Genet. 2008;53:185–191. doi: 10.1007/s10038-007-0228-0. [DOI] [PubMed] [Google Scholar]
- 19.Clement LC, Avila-Casado C, Mace C, Soria E, Bakker WW, Kersten S, Chugh SS. Podocyte-secreted angiopoietin-like-4 mediates proteinuria in glucocorticoid-sensitive nephrotic syndrome. Nat Med. 2011;17:117–122. doi: 10.1038/nm.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai KW, Wei CL, Tan LK, Tan PH, Chiang GS, Lee CG, Jordan SC, Yap HK. Overexpression of interleukin-13 induces minimal-change-like nephropathy in rats. J Am Soc Nephrol. 2007;18:1476–1485. doi: 10.1681/ASN.2006070710. [DOI] [PubMed] [Google Scholar]
- 21.Van Den Berg JG, Aten J, Annink C, Ravesloot JH, Weber E, Weening JJ. Interleukin-4 and -13 promote basolateral secretion of H(+) and cathepsin L by glomerular epithelial cells. Am J Physiol Renal Physiol. 2002;282:F26–F33. doi: 10.1152/ajprenal.0102.2001. [DOI] [PubMed] [Google Scholar]
- 22.Kawachi H, Suzuki K, Miyauchi N, Hashimoto T, Otaki Y, Shimizu F. Slit diaphragm dysfunction in proteinuric states: identification of novel therapeutic targets for nephrotic syndrome. Clin Exp Nephrol. 2009;13:275–280. doi: 10.1007/s10157-009-0162-x. [DOI] [PubMed] [Google Scholar]
- 23.Harita Y, Kurihara H, Kosako H, Tezuka T, Sekine T, Igarashi T, Ohsawa I, Ohta S, Hattori S. Phosphorylation of nephrin triggers Ca2+ signaling by recruitment and activation of phospholipase C-{gamma}1. J Biol Chem. 2009;284:8951–8962. doi: 10.1074/jbc.M806851200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chugh S, Yuan H, Topham PS, Haydar SA, Mittal V, Taylor GA, Kalluri R, Salant DJ. Aminopeptidase A: a nephritogenic target antigen of nephrotoxic serum. Kidney Int. 2001;59:601–613. doi: 10.1046/j.1523-1755.2001.059002601.x. [DOI] [PubMed] [Google Scholar]
- 25.Garin EH, Diaz LN, Mu W, Wasserfall C, Araya C, Segal M, Johnson RJ. Urinary CD80 excretion increases in idiopathic minimal-change disease. J Am Soc Nephrol. 2009;20:260–266. doi: 10.1681/ASN.2007080836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sellier-Leclerc AL, Duval A, Riveron S, Macher MA, Deschenes G, Loirat C, Verpont MC, Peuchmaur M, Ronco P, Monteiro RC, Haddad E. A humanized mouse model of idiopathic nephrotic syndrome suggests a pathogenic role for immature cells. J Am Soc Nephrol. 2007;18:2732–2739. doi: 10.1681/ASN.2006121346. [DOI] [PubMed] [Google Scholar]
- 27.Ikeuchi Y, Kobayashi Y, Arakawa H, Suzuki M, Tamura K, Morikawa A. Polymorphisms in interleukin-4-related genes in patients with minimal change nephrotic syndrome. Pediatr Nephrol. 2009;24:489–495. doi: 10.1007/s00467-008-1003-y. [DOI] [PubMed] [Google Scholar]
- 28.Santangelo S, Cousins DJ, Winkelmann NE, Staynov DZ. DNA methylation changes at human Th2 cytokine genes coincide with DNase I hypersensitive site formation during CD4(+) T cell differentiation. J Immunol. 2002;169:1893–1903. doi: 10.4049/jimmunol.169.4.1893. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita M, Ukai-Tadenuma M, Kimura M, Omori M, Inami M, Taniguchi M, Nakayama T. Identification of a conserved GATA3 response element upstream proximal from the interleukin-13 gene locus. J Biol Chem. 2002;277:42399–42408. doi: 10.1074/jbc.M205876200. [DOI] [PubMed] [Google Scholar]
- 30.Yap HK, Cheung W, Murugasu B, Sim SK, Seah CC, Jordan SC. Th1 and Th2 cytokine mRNA profiles in childhood nephrotic syndrome: evidence for increased IL-13 mRNA expression in relapse. J Am Soc Nephrol. 1999;10:529–537. doi: 10.1681/ASN.V103529. [DOI] [PubMed] [Google Scholar]
- 31.Cho BS, Yoon SR, Jang JY, Pyun KH, Lee CE. Up-regulation of interleukin-4 and CD23/FcepsilonRII in minimal change nephrotic syndrome. Pediatr Nephrol. 1999;13:199–204. doi: 10.1007/s004670050592. [DOI] [PubMed] [Google Scholar]
- 32.Sahali D, Pawlak A, Valanciute A, Grimbert P, Lang P, Remy P, Bensman A, Guellaen G. A novel approach to investigation of the pathogenesis of active minimal-change nephrotic syndrome using subtracted cDNA library screening. J Am Soc Nephrol. 2002;13:1238–1247. doi: 10.1681/ASN.V1351238. [DOI] [PubMed] [Google Scholar]
- 33.Komatsuda A, Wakui H, Iwamoto K, Togashi M, Masai R, Maki N, Sawada K. GATA-3 is upregulated in peripheral blood mononuclear cells from patients with minimal change nephrotic syndrome. Clin Nephrol. 2009;71:608–616. doi: 10.5414/cnp71608. [DOI] [PubMed] [Google Scholar]
- 34.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 19 kb)