Abstract
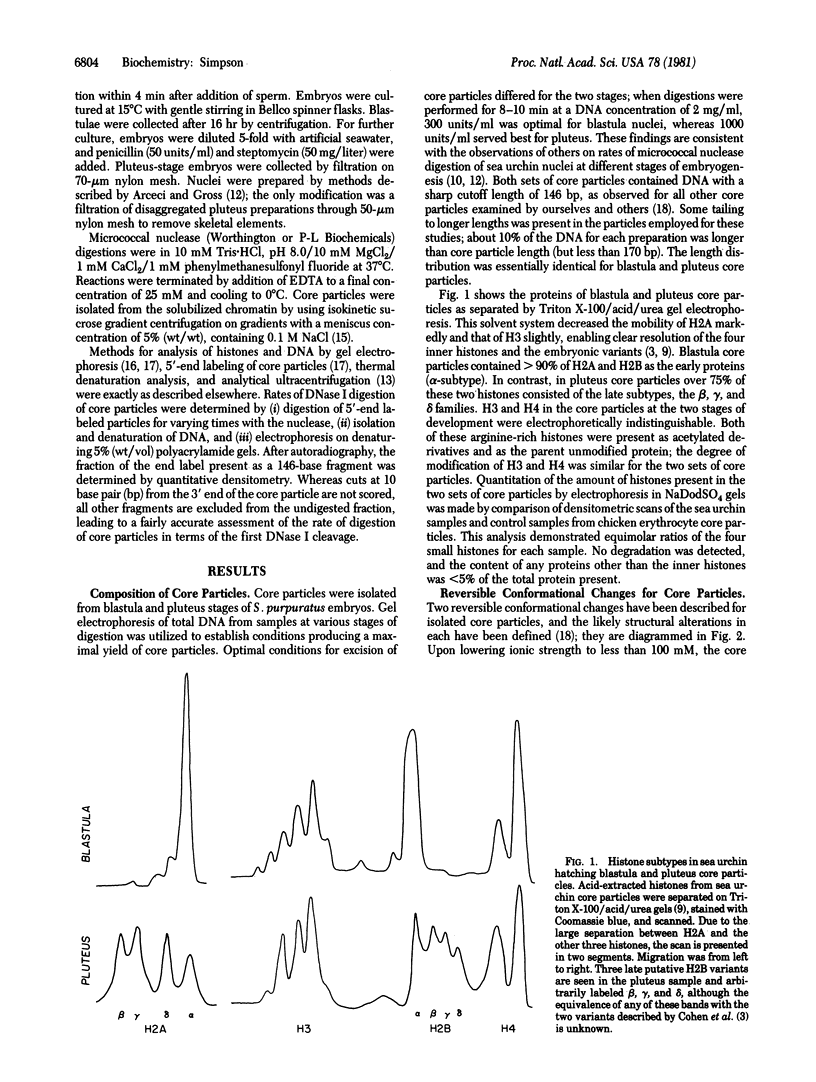
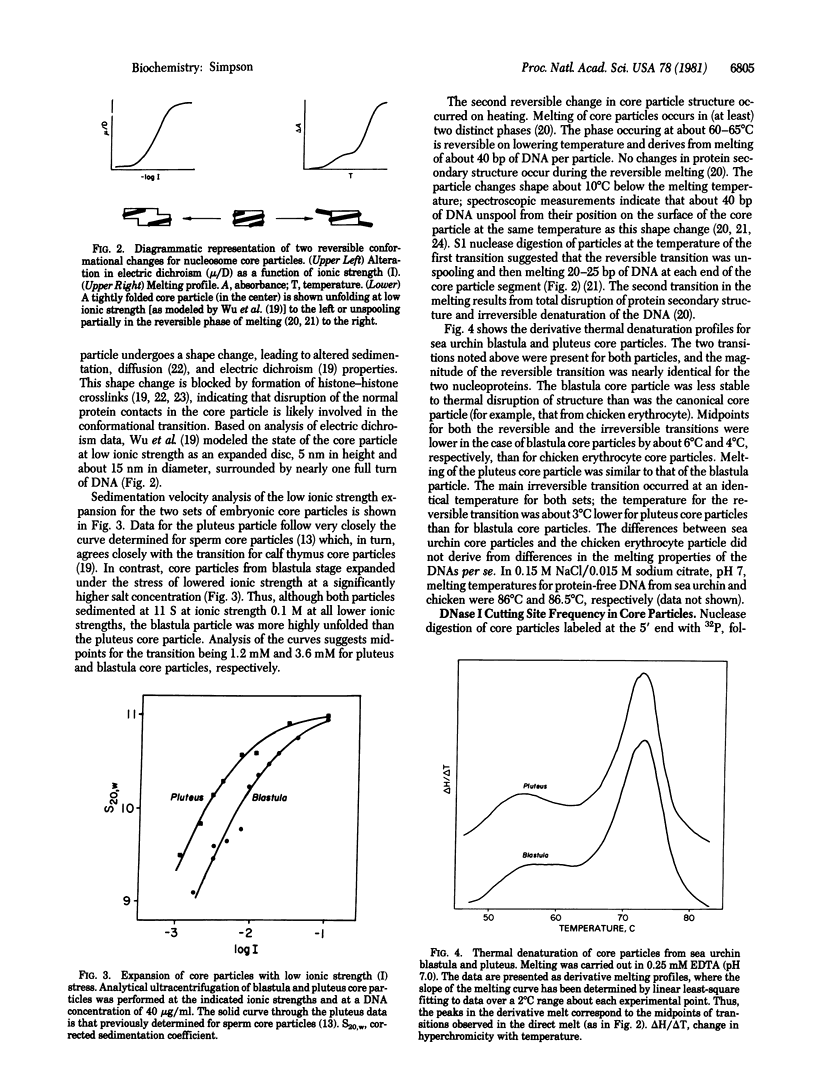
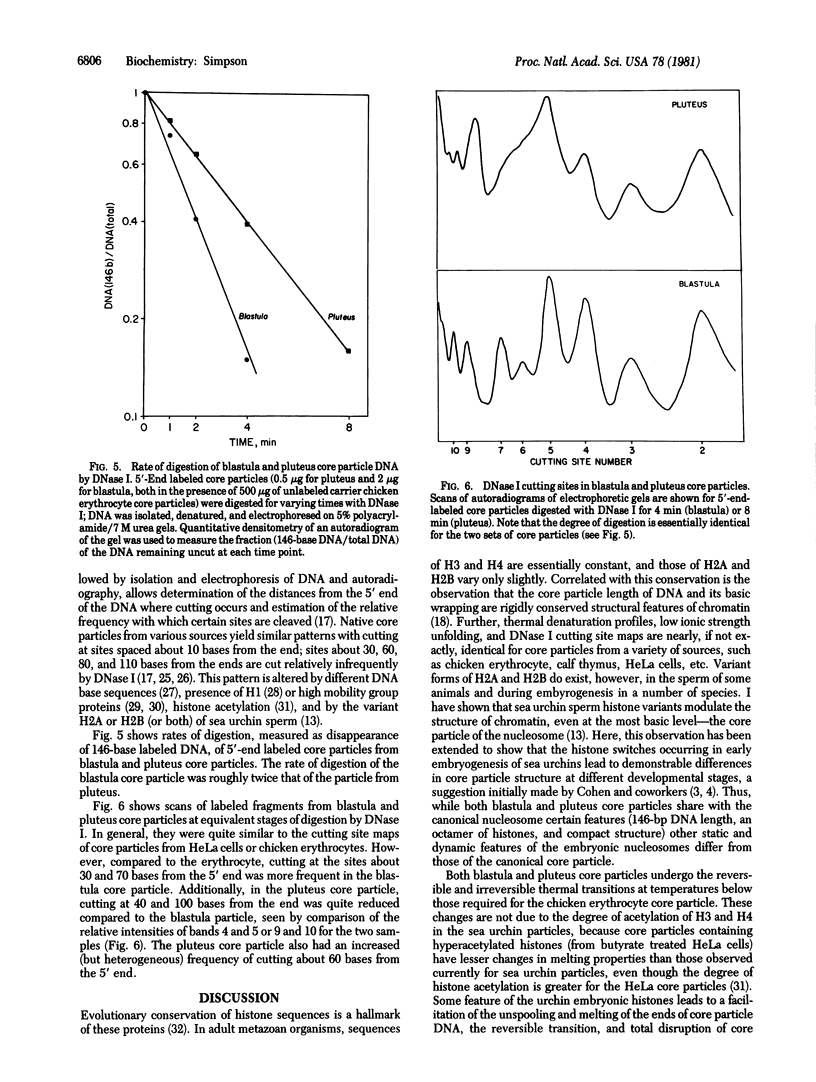
Switches of the types of histones synthesized and incorporated into chromatin occur during sea urchin embryogenesis. In an attempt to define the possible effects of these variant histones on chromatin structure, I have isolated and characterized nucleosome core particles from Strongylocentrotus purpuratus blastula (nearly 100% early histones) and pluteus (75% late histones). Both particles contain 146-base-pair lengths of DNA wrapped around an octamer of H2A, H2B, H3, and H4. Although sharing these similarities with the canonical core particle, the nucleosome structures have certain features that differ from those of typical adult tissues. Both the reversible and the irreversible conformational transitions occurring on heating core particles are destabilized in the embryonic particles vs. "typical" core particles. The blastula core particle unfolds more easily than pluteus (or other) nucleosomes under the stress of low ionic strength. The rate of DNase I digestion of pluteus core particles is about half that of particles from blastula; certain cutting sites differ in their susceptibility between the two embryonic particles and between these two and the canonical core particle. The data demonstrate that the variant histones synthesized during early embryogenesis have demonstrable effects on chromatin structure, even at this basic level.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alfageme C. R., Zweidler A., Mahowald A., Cohen L. H. Histones of Drosophila embryos. Electrophoretic isolation and structural studies. J Biol Chem. 1974 Jun 25;249(12):3729–3736. [PubMed] [Google Scholar]
- Arceci R. J., Gross P. R. Histone variants and chromatin structure during sea urchin development. Dev Biol. 1980 Nov;80(1):186–209. doi: 10.1016/0012-1606(80)90508-4. [DOI] [PubMed] [Google Scholar]
- Childs G., Maxson R., Kedes L. H. Histone gene expression during sea urchin embryogenesis: isolation and characterization of early and late messenger RNAs of Strongylocentrotus purpuratus by gene-specific hybridization and template activity. Dev Biol. 1979 Nov;73(1):153–173. doi: 10.1016/0012-1606(79)90144-1. [DOI] [PubMed] [Google Scholar]
- Cohen L. H., Newrock K. M., Zweidler A. Stage-specific switches in histone synthesis during embryogenesis of the sea urchin. Science. 1975 Dec 5;190(4218):994–997. doi: 10.1126/science.1237932. [DOI] [PubMed] [Google Scholar]
- Gordon V. C., Knobler C. M., Olins D. E., Schumaker V. N. Conformational changes of the chromatin subunit. Proc Natl Acad Sci U S A. 1978 Feb;75(2):660–663. doi: 10.1073/pnas.75.2.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon V. C., Schumaker V. N., Olins D. E., Knobler C. M., Horwitz J. The temperature and pH dependence of conformational transitions of the chromatin subunit. Nucleic Acids Res. 1979 Aug 24;6(12):3845–3858. doi: 10.1093/nar/6.12.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross K. W., Jacobs-Lorena M., Baglioni C., Gross P. R. Cell-free translation of maternal messenger RNA from sea urchin eggs. Proc Natl Acad Sci U S A. 1973 Sep;70(9):2614–2618. doi: 10.1073/pnas.70.9.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Keichline L. D., Wassarman P. M. Developmental study of the structure of sea urchin embryo and sperm chromatin using micrococcal nuclease. Biochim Biophys Acta. 1977 Mar 2;475(1):139–151. doi: 10.1016/0005-2787(77)90348-3. [DOI] [PubMed] [Google Scholar]
- Keichline L. D., Wassarman P. M. Structure of chromatin in sea urchin embryos, sperm, and adult somatic cells. Biochemistry. 1979 Jan 9;18(1):214–219. doi: 10.1021/bi00568a033. [DOI] [PubMed] [Google Scholar]
- Leffak I. M., Grainger R., Weintraub H. Conservative assembly and segregation of nucleosomal histones. Cell. 1977 Nov;12(3):837–845. doi: 10.1016/0092-8674(77)90282-3. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Mardian J. K., Paton A. E., Bunick G. J., Olins D. E. Nucleosome cores have two specific binding sites for nonhistone chromosomal proteins HMG 14 and HMG 17. Science. 1980 Sep 26;209(4464):1534–1536. doi: 10.1126/science.7433974. [DOI] [PubMed] [Google Scholar]
- McCarty K. S., Jr, Vollmer R. T., McCarty K. S. Improved computer program data for the resolution and fractionation of macromolecules by isokinetic sucrose density gradient sedimentation. Anal Biochem. 1974 Sep;61(1):165–183. doi: 10.1016/0003-2697(74)90343-1. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Rau D. C., Charney E., Felsenfeld G. Orientation of the nucleosome within the higher order structure of chromatin. Cell. 1980 Nov;22(1 Pt 1):87–96. doi: 10.1016/0092-8674(80)90157-9. [DOI] [PubMed] [Google Scholar]
- Newrock K. M., Alfageme C. R., Nardi R. V., Cohen L. H. Histone changes during chromatin remodeling in embryogenesis. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):421–431. doi: 10.1101/sqb.1978.042.01.045. [DOI] [PubMed] [Google Scholar]
- Newrock K. M., Cohen L. H., Hendricks M. B., Donnelly R. J., Weinberg E. S. Stage-specific mRNAs coding for subtypes of H2A and H2B histones in the sea urchin embryo. Cell. 1978 Jun;14(2):327–336. doi: 10.1016/0092-8674(78)90118-6. [DOI] [PubMed] [Google Scholar]
- Noll M. DNA folding in the nucleosome. J Mol Biol. 1977 Oct 15;116(1):49–71. doi: 10.1016/0022-2836(77)90118-8. [DOI] [PubMed] [Google Scholar]
- Ruderman J. V., Gross P. R. Histones and histone synthesis in sea urchin development. Dev Biol. 1974 Feb;36(2):286–298. doi: 10.1016/0012-1606(74)90052-9. [DOI] [PubMed] [Google Scholar]
- Sandeen G., Wood W. I., Felsenfeld G. The interaction of high mobility proteins HMG14 and 17 with nucleosomes. Nucleic Acids Res. 1980 Sep 11;8(17):3757–3778. doi: 10.1093/nar/8.17.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale R. L., Aronson A. I. Chromatin-associated proteins of the developing sea urchin embryo. I. Kinetics of synthesis and characterization of non-histone proteins. J Mol Biol. 1973 Apr 25;75(4):633–645. doi: 10.1016/0022-2836(73)90297-0. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Bergman L. W. Structure of sea urchin sperm chromatin core particle. J Biol Chem. 1980 Nov 25;255(22):10702–10709. [PubMed] [Google Scholar]
- Simpson R. T., Künzler P. Cromatin and core particles formed from the inner histones and synthetic polydeoxyribonucleotides of defined sequence. Nucleic Acids Res. 1979 Apr;6(4):1387–1415. doi: 10.1093/nar/6.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Mechanism of a reversible, thermally induced conformational change in chromatin core particles. J Biol Chem. 1979 Oct 25;254(20):10123–10127. [PubMed] [Google Scholar]
- Simpson R. T., Shindo H. Conformation of DNA in chromatin core particles containing poly(dAdT)-poly(dAdT) studied by 31 P NMR spectroscopy. Nucleic Acids Res. 1979 Sep 25;7(2):481–492. doi: 10.1093/nar/7.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Structure of chromatin containing extensively acetylated H3 and H4. Cell. 1978 Apr;13(4):691–699. doi: 10.1016/0092-8674(78)90219-2. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P. Mapping DNAase l-susceptible sites in nucleosomes labeled at the 5' ends. Cell. 1976 Oct;9(2):347–353. doi: 10.1016/0092-8674(76)90124-0. [DOI] [PubMed] [Google Scholar]
- Skoultchi A., Gross P. R. Maternal histone messenger RNA: detection by molecular hybridization. Proc Natl Acad Sci U S A. 1973 Oct;70(10):2840–2844. doi: 10.1073/pnas.70.10.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Weischet W. O., Tatchell K., Van Holde K. E., Klump H. Thermal denaturation of nucleosomal core particles. Nucleic Acids Res. 1978 Jan;5(1):139–160. doi: 10.1093/nar/5.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Preparation and physical characterization of a homogeneous population of monomeric nucleosomes from HeLa cells. Nucleic Acids Res. 1976 Sep;3(9):2255–2266. doi: 10.1093/nar/3.9.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]


