Abstract
Sucrose (Suc):Suc 1-fructosyltransferase (1-SST) is the key enzyme in plant fructan biosynthesis, since it catalyzes de novo fructan synthesis from Suc. We have cloned 1-SST from onion (Allium cepa) by screening a cDNA library using acid invertase from tulip (Tulipa gesneriana) as a probe. Expression assays in tobacco (Nicotiana plumbaginifolia) protoplasts showed the formation of 1-kestose from Suc. In addition, an onion acid invertase clone was isolated from the same cDNA library. Protein extracts of tobacco protoplasts transformed with this clone showed extensive Suc-hydrolyzing activity. Conditions that induced fructan accumulation in onion leaves also induced 1-SST mRNA accumulation, whereas the acid invertase mRNA level decreased. Structurally different fructan molecules could be produced from Suc by a combined incubation of protein extract of protoplasts transformed with 1-SST and protein extract of protoplasts transformed with either the onion fructan:fructan 6G-fructosyltransferase or the barley Suc:fructan 6-fructosyltransferase.
Fructans (polyfructosylsucrose) consist of polymers of Fru attached to Suc and serve as an important storage carbohydrate in approximately 15% of flowering plant species (Hendrey and Wallace, 1993). The Fru residues are either linked by a (2–1) β-d-glycosidic bond, as in inulin derived from Cichorium intybus L. (Bonnett et al., 1994), or by a (2–6) β-d-glycosidic bond, as in levans (e.g. Phleum pratense L.; Suzuki and Pollock, 1986). In most grasses branched fructans containing both types of linkages are produced (e.g. Triticum aestivum L.; Carpita et al., 1989). In plants of the Liliales, to which onion (Allium cepa) and tulip (Tulipa gesneriana) belong, a special type of fructan is produced, the inulin neoseries. In this type of fructan the Glc moiety of Suc contains fructosyl residues on both C1 and C6, resulting in a polymer with β (2–1)-linked fructosyl chains on either end of the Suc molecule (Shiomi, 1989).
For synthesis of the linear inulin at least two enzymes are required (Pollock and Cairns, 1991): 1-SST initiates fructan synthesis by catalyzing the transfer of a fructosyl residue from Suc to another Suc molecule, resulting in the formation of the trisaccharide 1-kestose (G1–2F1–2F, also called isokestose), and 1-FFT elongates the fructosyl chain. It has been shown that incubation of a combination of purified 1-SST and 1-FFT from Jerusalem artichoke with Suc results in the production of inulin with a polymer length of up to 20 fructosyl residues (Koops and Jonker, 1996; Lüscher et al., 1996). For production of the inulin neoseries, the type of fructan found in onion, a third enzyme is needed, 6G-FFT (Shiomi, 1989; Wiemken et al., 1995). Recently, we cloned the gene encoding the 6G-FFT from onion (Vijn et al., 1997). This fructosyltransferase catalyzes the transfer of a Fru residue of 1-kestose to C6 of the Glc moiety of Suc, forming neokestose (F2–6G1–2F). Transgenic chicory plants harboring the onion 6G-FFT under the control of the cauliflower mosaic virus 35S RNA promoter produce inulin of the neoseries in addition to linear inulin (Vijn et al., 1997).
In this paper we describe the cloning and functional characterization of onion cDNA clones encoding a 1-SST and an acid invertase. Furthermore, we show that with specific combinations of fructosyltransferases, structurally different fructan molecules can be produced from Suc.
MATERIALS AND METHODS
Preparing and Screening of a cDNA Library of Onion
Onion (Allium cepa L., BMCCB) seeds were germinated on soil and grown for 4 weeks at 21°C with a light/dark cycle of 16 h/8 h. Subsequently, leaves were cut off (2 h after the dark-to-light switch), placed in a 5% Suc solution, and illuminated continuously for up to 24 h. Samples were taken after 0, 4, 8, 12, 16, and 24 h. mRNA isolation systems (PolyATtract I and II, Promega) were used to isolate poly(A+) RNA from leaves illuminated for 4 h. A cDNA library was made using a cloning kit (ZAP-cDNA Gigapack III Gold Cloning Kit, Stratagene). The amplified cDNA library was screened with a random 32P-labeled 1340-bp 5′ fragment of the acid invertase cDNA from tulip (Tulipa gesneriana) (accession no. X95651; a kind gift of Dr. A.D. de Boer, ATO-DLO, Wageningen, The Netherlands) according to the Stratagene protocol. Overnight hybridization was performed in buffer containing 5× Denhardt's solution, 6× SSC, and 0.5% SDS at 55°C, and the filters were washed three times in 0.5× SSC and 0.1% SDS at 55°C. Sequencing of the clones was performed with an automatic DNA-sequencing apparatus (48 cm, model 373, Applied Biosystems) using a dye-terminator cycle-sequencing kit (Prism, with Amplitaq DNA polymerase, Applied Biosystems).
Expression of Isolated cDNAs in Tobacco Protoplasts
The complete inserts of the isolated cDNA clones were subcloned into pMon999 (Monsanto, St. Louis, MO), which contains the cauliflower mosaic virus 35S RNA promoter and a nopaline synthase terminator sequence. Isolation and transformation of tobacco (Nicotiana plumbaginifolia) protoplasts was performed according to the method of Goodall et al. (1990). Prior to PEG transformation of 6 × 105 protoplasts with 50 μg of plasmid DNA the protoplasts were washed four times by floating in wash buffer (0.4 m Suc, 5 mm KCl, 125 mm CaCl2•2H2O, and 5 mm Glc, pH 5.8). After overnight incubation at 22°C in the dark, protoplasts were pelleted and resuspended in 50 mm Mes buffer, pH 5.6. Protoplasts were lysed by freezing in liquid nitrogen. Cell debris were removed by centrifugation at 13,000g for 5 min. Protein extracts were desalted by passing the supernatants over Biogel P-10 columns at 350g for 5 min (Simmen et al., 1993). Finally, 135 μL of purified protein extract was obtained per protoplast transfection. Total protein was measured using the method of Bradford (1976), with BSA as the standard.
In Vitro Fructosyltransferase Activity Assay and Detection of Products
Protein extracts from transformed protoplasts were incubated in the presence of 0.02% NaN3 with an appropriate sugar substrate (see Results) in 50 mm Mes buffer, pH 5.6, at 27°C for up to 24 h. The reaction was stopped by heating the samples at 95°C for 4 min. For fructosyltransferase activity assays with a single transfection extract, 45 μL of purified protein extract was used. The total amount of protein corresponding to this volume was 15 μg from transfections with empty vector, onion 6G-FFT, and barley 6-SFT, and 22 μg from transfections with AcN2 and AcT1. For the combined enzyme incubations, 18 μL of each protein extract was used. The end volume of all incubations was 50 μL. Products formed were analyzed by HPLC using a CarboPac PA-100 column (Dionex, Sunnyvale, CA) and a DX-500 gradient chromatography system with a pulsed amperometric detector (Dionex). Five microliters from each incubation was injected, including 0.5 μg of trehalose as an internal standard (final injection volume, 10 μL).
Induction of Fructan Accumulation in Onion Leaves
Leaves of 4-week-old onion plants grown at 21°C with a light/dark cycle of 16 h/8 h were cut off (2 h after the dark-to-light switch), placed in a 5% Suc solution, and then illuminated continuously for up to 24 h. Sugars were isolated by extracting the leaves three times with 80% ethanol at 60°C. The extracts were vacuum desiccated and the sugar pellet was dissolved in water at 0.5 μL/mg fresh weight. Sugar composition was determined by TLC. One microliter of the sugar solution was spotted on TLC foil (Silicagel F1500, Schleicher & Schuell), which was developed three times in acetone:water (87:13, v/v) (Wagner et al., 1983) and then stained with a Fru-specific urea-phosphoric acid spray (Wise et al., 1955).
RNA Extraction and Northern Analysis
Total RNA was isolated according to the method of Verwoerd et al. (1989). The RNA gel-blot analysis was performed according to the protocol for Hybond N membrane (version 2.0, Amersham). 5′ Fragments of the fructosyltransferase encoding cDNAs were used as probes: a 910-bp EcoRI/XhoI fragment of pAcN2 (1-SST), a 577-bp EcoRI/KpnI fragment of pAcT1 (acid invertase), a 582-bp EcoRI/PstI fragment of pAc2 (6G-FFT) (Vijn et al., 1997), and a 1500-bp fragment containing the coding region of the 18S rRNA of Arabidopsis (18SrRNA) (Pruitt and Meyerowitz, 1986). The northern blots were hybridized overnight at 65°C in the same hybridization buffer used for screening of the cDNA library and were washed three times in 0.5× SSC and 0.1% SDS at 65°C.
RESULTS
Isolation of Onion cDNA Clones Encoding Fructosyltransferases
Based on the known homology between fructosyltransferases and invertases (Sprenger et al., 1995; Vijn et al., 1997), an onion cDNA library prepared from leaves induced for fructan synthesis was screened with a 32P-labeled insert of an acid invertase cDNA clone from tulip (accession no. X95651). Fourteen positive clones, although containing cDNA inserts with different lengths, encoded the same gene. The clone with the longest cDNA insert, called pAcN2, was sequenced completely. pAcN2 has an insert of 2202 bp and contains a poly(A+) stretch at the 3′ end. It contains one long open reading frame encoding a 623-amino acid polypeptide and a 63-bp 5′-untranslated region. The deduced polypeptide contains five potential glycosylation sites and has a pI of 5.2. The amino acid sequence of AcN2 shows 49% identity to the chicory 1-SST polypeptide, 53% identity to the acid invertase of tulip, and 58% identity to the onion 6G-FFT polypeptide (Fig. 1), suggesting that AcN2 encodes a fructosyltransferase as well.
Figure 1.
Comparison of deduced amino acid sequence of AcN2 and AcT1 from onion with onion 6G-FFT, chicory 1-SST, and acid invertase of tulip. AcSST, Deduced amino acid sequence of AcN2 (accession no. AJ006066); AcInv, deduced amino acid sequence of AcT1 (accession no. AJ007067); Ac6G-FFT, from onion (Vijn et al., 1997; accession no. Y07838); CiSST, 1-SST from chicory (de Halleux and Van Cutsem, 1997; accession no. U81520); TgInv, vacuolar invertase from tulip (accession no. X95651). The N-terminal end of the mature 1-SST protein of chicory is indicated by an arrow. Identical amino acids are shaded. The putative glycosylation sites in AcSST and AcInv are underlined.
Two other positive cDNA clones, also containing different-sized cDNA inserts, encoded a gene that is highly homologous, but not identical to the AcN2-encoded gene. The clone containing the longest cDNA insert, pAcT1, was sequenced completely. pAcT1 has an insert of 2438 bp and contains a poly(A+) stretch at the 3′ end. It has one long open reading frame encoding a 690-amino acid polypeptide and a 59-bp 5′-untranslated region. The deduced polypeptide contained three potential glycosylation sites and the pI was 4.7. The amino acid sequence of AcT1 shows 47% identity to chicory 1-SST, 61% identity to AcN2 and the acid invertase of tulip, and 63% identity to the onion 6G-FFT polypeptide (Fig. 1), suggesting that AcT1 also encodes a fructosyltransferase.
The N-terminal end of mature chicory 1-SST has been determined (Van den Ende et al., 1996) and starts at the 98th codon (de Halleux and Van Cutsem, 1997) (Fig. 1). The high homology of AcN2 and AcT1 with the fructosyltransferases starts downstream of the 98th codon of chicory 1-SST, suggesting that these proteins will also be processed from a larger preproprotein.
Determination of Enzymatic Activity in Tobacco Protoplasts
The amino acid sequence comparisons suggest that both AcN2 and AcT1 encode fructosyltransferases, but do not indicate specific enzymatic activities. Therefore, the cDNA inserts of pAcN2 and pAcT1 were placed under control of the cauliflower mosaic virus 35S RNA promoter and transiently expressed in tobacco protoplasts. Protein extracts of transformed protoplasts were incubated with either 20 or 100 mm Suc for 24 h at 27°C, and the sugar products were analyzed by HPLC. Several independent protoplast transformations and enzyme activity assays were performed, with comparable results.
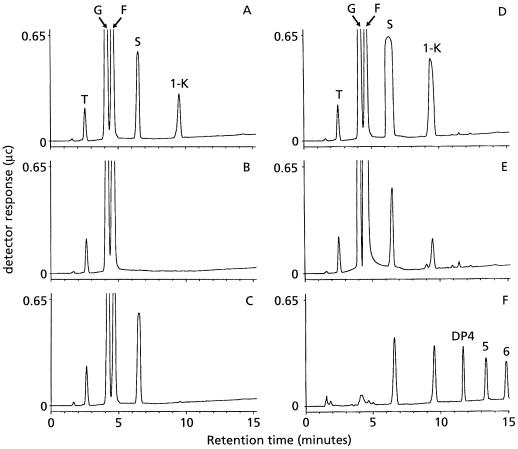
The product formed by AcN2-transformed protoplasts after incubation with 20 mm Suc was 1-kestose (Fig. 2A). Higher levels of 1-kestose were produced when the protein extract was incubated with 100 mm Suc (Fig. 2D). These results strongly suggest that AcN2 encodes a 1-SST. Incubation of AcT1-transformed protoplasts with 20 mm Suc resulted in a complete Suc hydrolysis showing invertase activity (Fig. 2B). It is well known that at elevated Suc levels invertase has some fructosyltransferase activity (Straathof et al., 1986; Obenland et al., 1993), and this was observed when AcT1 protein extract was incubated with 100 mm Suc (Fig. 2E). These results strongly suggest that AcT1 encodes an invertase. According to the high sequence homology to the acid invertase of tulip (Fig. 1) and the low pI (4.7) of the deduced AcT1 polypeptide, AcT1 most likely codes for an acid invertase.
Figure 2.
Analysis of sugar products generated by protein extracts of transformed protoplasts after incubation with different Suc concentrations. Sugar products were analyzed by HPLC after a 24-h incubation with 20 mm (A, B, and C) or 100 mm (D and E) Suc at 27°C. A and D, Protein extract from protoplasts transformed with AcN2 under control of the 35S promoter; B and E, protein extract from protoplasts transformed AcT1 under control of the 35S promoter; C, protein extract from protoplasts transformed with empty vector; and F, sugar extract from chicory root. Peaks in the HPLC chromatograms: T, trehalose (internal standard); G, Glc; F, Fru; S, Suc (G1–2F); 1-K, 1-kestose (G1–2F1–2F); DP4–6, inulin fructan with a degree of polymerization of 4 to 6.
mRNA Levels of 1-SST and Invertase during Induced Fructan Accumulation in Onion Leaves
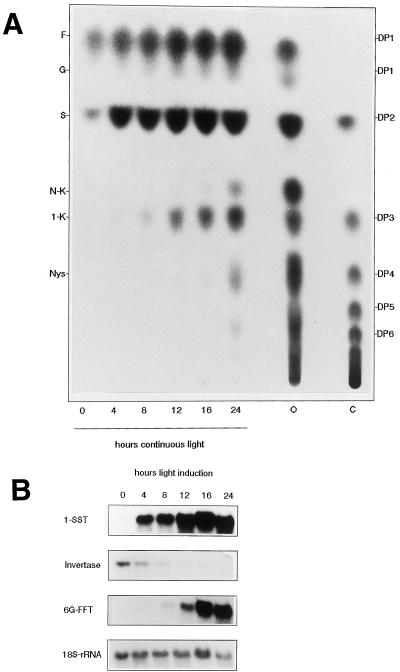
Normally, onion leaves do not accumulate fructan. However, by supplying the leaves with excessive Suc, fructan synthesis can be induced (Vijn et al., 1997). Excessive Suc can be supplied by removing the sink tissue and feeding the leaves with Suc, by placing them under continuous illumination, or both. mRNA accumulation of 1-SST (AcN2) and invertase (AcT1) during fructan accumulation in onion leaves was determined by northern blotting. Fructan accumulation was induced in 4-week-old onion leaves from which the sinks had been removed and that were placed in 5% Suc solution and illuminated continuously for up to 24 h. After 4 h of continuous illumination, 1-SST mRNA was clearly detected and increased up to 12 h, after which time it stabilized (Fig. 3B). This expression pattern corresponded with 1-kestose production, which was detected after 4 h and increased up to 12 h, after which time it stabilized (Fig. 3A). Invertase mRNA was present in the leaves at the moment they were cut and placed in the Suc solution (Fig. 3B). During fructan accumulation the mRNA level decreased (Fig. 3B). The expression of onion 6G-FFT, which catalyzes the formation of neokestose from Suc and 1-kestose, corresponded with neokestose formation. 6G-FFT mRNA was detected at 12 h and increased after this time (Fig. 3B). Neokestose production was detected at 16 h and increased up to 24 h (Fig. 3A).
Figure 3.
A, Fructan accumulation in onion leaves. TLC of sugars extracted from onion leaves after several time intervals of continuous illumination and feeding of 5% Suc. F, Fru; G, Glc; S, Suc; N-K, neokestose; 1-K, 1-kestose; Nys, nystose; DP1-DP6, inulin fructan with a degree of polymerization of 1 to 6; O, sugar products of onion bulb; C, inulin from chicory. B, mRNA levels of 1-SST, invertase, and 6G-FFT during fructan accumulation in onion leaves. Northern blots contain approximately 20 μg of total RNA from onion leaves treated similarly as the ones used in A. cDNA clones encoding onion 1-SST, invertase, and 6G-FFT were used as probes. An Arabidopsis 18S rRNA probe was used as a loading control.
In Vitro Synthesis of Structurally Different Fructan Molecules
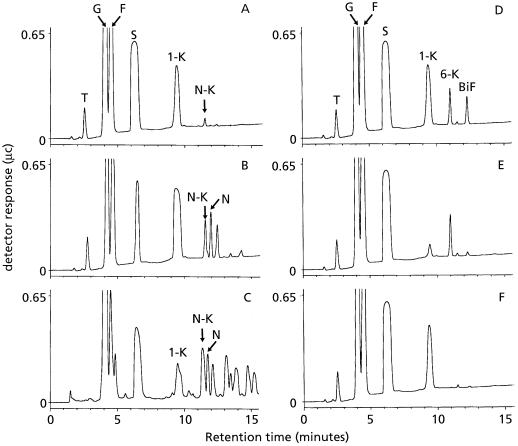
The cloning of 1-SST enables synthesis of fructan molecules from Suc. Here we show that 1-SST in combination with other fructosyltransferases for which the specific enzymatic activity is known can synthesize structurally different fructan molecules from Suc. Combined incubation of protein extracts of protoplasts transformed with onion 1-SST and onion 6G-FFT with 100 mm Suc resulted in the formation of 1-kestose and neokestose (Fig. 4A). Incubation of only protein extract of onion 6G-FFT transformed protoplasts with 100 mm Suc did not give any additional products (data not shown), but when this protein extract was incubated with 20 mm Suc and 20 mm 1-kestose as the substrates, neokestose, nystose, and some products with a higher degree of polymerization were formed (Fig. 4B). This shows that in vitro, 1-SST and 6G-FFT together are capable of producing fructan of the inulin neoseries from Suc. The combined incubation of protein extracts with only Suc has a higher Suc-to-1-kestose ratio than the single incubation of 6G-FFT protein extract with 20 mm Suc and 20 mm 1-kestose. A low Suc concentration is preferable for 6G-FFT activity, which explains the presence of products with a higher degree of polymerization in Figure 4B.
Figure 4.
Analysis of sugar products generated by either single or combined incubation of protein extracts from transformed protoplasts with different substrates. Sugar products were analyzed by HPLC after 24 h of incubation at 27°C. A, Combined incubation of protein extracts from protoplasts transformed with onion 1-SST and 6G-FFT with 100 mm Suc; B, single incubation of protein extract from protoplasts transformed with onion 6G-FFT with 20 mm Suc and 20 mm 1-kestose; C, sugar products from onion bulb; D, combined incubation of protein extracts from protoplasts transformed with onion 1-SST and barley 6-SFT with 100 mm Suc; E, single incubation of protein extract from protoplasts transformed with barley 6-SFT with 100 mm Suc; and F, single incubation of protein extract from protoplasts transformed with onion 1-SST with 100 mm Suc. Peaks in the HPLC chromatograms: T, trehalose (internal standard); G, Glc; F, Fru; S, Suc (G1–2F); 1-K, 1-kestose (G1–2F1–2F); N-K, neokestose (F2–6G1–2F); N, nystose (G1–2F1–2F1–2F); 6-K, 6-kestose (G1–2F6–2F); BiF, bifurcose (G1–2F1[6–2F]1–2F).
The combination of protein extracts of protoplasts transformed with onion 1-SST and with barley 6-SFT resulted in the formation of 1-kestose, 6-kestose, and bifurcose from Suc (Fig. 4D). The single incubation of protein extract of 6-SFT-transformed protoplasts with Suc resulted in the production of 6-kestose, a low amount of 1-kestose production, and a very low amount of bifurcose (Fig. 4E). These results show that the cloning of 1-SST enables synthesis of structurally different fructan molecules from Suc when combined with other fructosyltransferases
DISCUSSION
In this paper we describe the isolation of the onion cDNA clone pAcN2, which encodes the key enzyme in fructan synthesis: 1-SST. This enzyme initiates de novo fructan synthesis in plants by catalyzing the transfer of a fructosyl residue from Suc to another Suc molecule, resulting in the production of 1-kestose and Glc. We also isolated the cDNA clone pAcT1, which encodes an acid invertase.
The cDNA clones were isolated by screening an onion cDNA library with a cDNA encoding a tulip acid invertase. The deduced amino acid sequence of AcN2 shows 49% identity to chicory 1-SST, 53% identity to tulip acid invertase, and 58% identity to 6G-FFT of onion. The deduced amino acid sequence of AcT1 shows 61% identity to tulip acid invertase and to onion 1-SST polypeptide and 63% identity to onion 6G-FFT. These high sequence homologies only show that the cDNAs most likely encode fructosyltransferases, but do not discriminate between specific enzymatic activities, so the cDNAs were transiently expressed in tobacco protoplasts. Incubation of protein extract of AcN2-transformed protoplasts with Suc showed the formation of 1-kestose. When protein extract of AcT1-transformed protoplasts was incubated with 20 mm Suc, a complete Suc hydrolysis was observed, whereas with 100 mm Suc, in addition to hydrolysis of the Suc, some 1-kestose was also produced. This is in agreement with the enzymatic activity of purified barley acid invertase, which also has 1-SST activity at high Suc concentrations (Obenland et al., 1993). Since in these enzymatic assays the same amount of total protein in the AcN2 and AcT1 protoplast extracts was used, these results strongly suggest that AcN2 encodes a 1-SST, whereas AcT1 encodes an invertase. Whether the cloned 1-SST can also catalyze fructosyl transfer to water is not known. The expression system used to characterize the enzymatic activities of the cloned genes does contain endogenous invertases that compete for the same substrate. To characterize the enzymatic activities of the cloned 1-SST and invertase in more detail, it will be necessary either to purify the proteins or to express them in a system lacking β-fructosidase activity.
During induced fructan accumulation in onion leaves, 1-SST (AcN2) mRNA accumulation corresponded with Suc accumulation and 1-kestose production, whereas the invertase (AcT1) mRNA level decreased upon Suc accumulation. The accumulation of 6G-FFT mRNA corresponded to neokestose production. Most likely, mRNA synthesis of 1-SST is induced by a high Suc concentration, whereas for the induction of 6G-FFT mRNA synthesis an additional signal is necessary. Furthermore, these results strongly suggest that in vivo the enzymes encoded by AcN2 and AcT1 have the same enzymatic activity as that determined in vitro.
Combined incubation of protein extracts of protoplasts transformed with the onion 1-SST and 6G-FFT with 100 mm Suc resulted in the formation of 1-kestose and neokestose. The combined incubation of protein extracts of protoplasts transformed with onion 1-SST and barley 6-SFT resulted in the formation of 1-kestose, 6-kestose, and bifurcose. In chicory it has been shown that introduction of the onion 6G-FFT or barley 6-SFT results in the production of inulin of the neoseries and branched fructans, respectively, in addition to linear inulin (Sprenger et al., 1997; Vijn et al., 1997). These findings showed that the type of fructan made by a plant can be changed by introduction of a fructosyltransferase with a different enzymatic activity than the endogenous fructosyltransferases. Here we show that it is possible to synthesize in vitro structurally defined fructan molecules from Suc by combining fructosyltransferases with specific enzymatic activities. By introducing specific sets of fructosyltransferases, it will now be possible to accumulate structurally defined fructan molecules in plants that normally do not accumulate fructan.
ACKNOWLEDGMENTS
We thank Tony van Kampen (Agricultural University, Wageningen, The Netherlands) for providing excellent sequence data of the isolated cDNAs, Douwe de Boer (ATO-DLO, Wageningen, The Netherlands) for his kind gift of the acid invertase cDNA clone of tulip, and M. Hirayama (Meiji Seika Kaisha, Ltd., Saitama, Japan) for the gift of 1-kestose.
Abbreviations:
- 1-SST
Suc:Suc 1-fructosyltransferase
- 6G-FFT
fructan:fructan 6G-fructosyltransferase
- 6-SFT
Suc:fructan 6-fructosyltransferase
Footnotes
LITERATURE CITED
- Bonnett GD, Sims IM, John JAS, Simpson RJ. Purification and characterization of fructans with β-2,1- and β-2,6-glycosidic linkages suitable for enzyme studies. New Phytol. 1994;127:261–269. doi: 10.1111/j.1469-8137.1994.tb04277.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Kanabus J, Housley TL. Linkage structure of fructans and fructan oligomers from Triticum aestivum and Festuca arundinaceae leaves. J Plant Physiol. 1989;134:162–168. [Google Scholar]
- de Halleux S, Van Cutsem P. Cloning and sequencing of the 1-SST cDNA from chicory root. Plant Physiol. 1997;113:1003. [Google Scholar]
- Goodall GJ, Wiebauer K, Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- Hendry GAF, Wallace RK. The origin, distribution, and evolutionary significance of fructans. In: Suzuki M, Chatterton NJ, editors. Science and Technology of Fructans. Boca Raton, FL: CRC Press; 1993. pp. 119–139. [Google Scholar]
- Koops AJ, Jonker HH. Purification and characterization of the enzymes of fructan biosynthesis in tubers of Helianthus tuberosus Colombia. II. Purification of sucrose:sucrose 1-fructosyltransferase and reconstitution of fructan synthesis in vitro with purified sucrose:sucrose 1-fructosyltransferase and fructan:fructan 1-fructosyltransferase. Plant Physiol. 1996;110:1167–1175. doi: 10.1104/pp.110.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher M, Erdin C, Sprenger N, Hochstrasser U, Boller T, Wiemken A. Inulin synthesis by a combination of purified fructosyltransferases from tubers of Helianthus tuberosus. FEBS Lett. 1996;385:39–42. doi: 10.1016/0014-5793(96)00343-2. [DOI] [PubMed] [Google Scholar]
- Obenland DM, Simmen U, Boller T, Wiemken A. Purification and characterization of three soluble invertases from barley (Hordeum vulgare L.) leaves. Plant Physiol. 1993;89:549–556. doi: 10.1104/pp.101.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock CJ, Cairns AJ. Fructan metabolism in grasses and cereals. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:77–101. [Google Scholar]
- Pruitt RE, Meyerowitz EM. Characterization of the genome of A. thaliana. J Mol Biol. 1986;187:169–183. doi: 10.1016/0022-2836(86)90226-3. [DOI] [PubMed] [Google Scholar]
- Shiomi N. Properties of fructosyltransferases involved in the synthesis of fructan in Liliaceous plants. J Plant Physiol. 1989;134:151–155. [Google Scholar]
- Simmen U, Obenland D, Boller T, Wiemken A. Fructan synthesis in excised barley leaves. Identification of two sucrose:sucrose fructosyltransferases induced by light and their separation from constitutive invertases. Plant Physiol. 1993;101:459–468. doi: 10.1104/pp.101.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger N, Bortlik K, Brandt A, Boller T, Wiemken A. Purification, cloning, and functional expression of sucrose:fructan 6-fructosyltransferase, a key enzyme of fructan synthesis in barley. Proc Natl Acad Sci USA. 1995;92:11652–11656. doi: 10.1073/pnas.92.25.11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger N, Schellenbaum L, van Dun K, Boller T, Wiemken A. Fructan synthesis in transgenic tobacco and chicory plants expressing barley sucrose:fructan 6-fructosyltransferase. FEBS Lett. 1997;400:355–358. doi: 10.1016/s0014-5793(96)01418-4. [DOI] [PubMed] [Google Scholar]
- Straathof AJJ, Kieboom APG, van Bekkum H. Invertase catalyzed fructosyl transfer in concentrated solutions of sucrose. Carbohydr Res. 1986;146:154–159. doi: 10.1016/0008-6215(86)85033-9. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Pollock CJ. Extraction and characterization of the enzymes of fructan biosynthesis in timothy grass (Phleum pratense L.) Can J Bot. 1986;64:1884–1887. [Google Scholar]
- Van den ende W, Van Wonterghem D, Dewil E, Verheart P, De Loof A, Van Laere A (1996) Purification and characterization of 1-SST, the key enzyme initiating fructan biosynthesis in young chicory roots (Cichorium intybus). Physiol Plant 98: 455–466
- Verwoerd TL, Dekker BMM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijn I, van Dijken A, Sprenger N, van Dun K, Weisbeek P, Wiemken A, Smeekens S. Fructan of the inulin neoseries is synthesized in transgenic chicory plants (Cichorium intybus L.) harbouring onion (Allium cepa L.) fructan:fructan 6G-fructosyltransferase. Plant J. 1997;11:387–398. doi: 10.1046/j.1365-313x.1997.11030387.x. [DOI] [PubMed] [Google Scholar]
- Wagner W, Keller F, Wiemken A. Fructan metabolism in cereals: induction in leaves and compartmentation in protoplasts and vacuoles. Z Pflanzenphysiol. 1983;112:359–372. [Google Scholar]
- Wiemken A, Sprenger N, Boller T (1995) Fructan: an extension of sucrose by sucrose. In HG Pontis, GL Salerno, EJ Echeverria, eds, Sucrose Metabolism, Biochemistry, Physiology, and Molecular Biology. American Society of Plant Physiologists, Rockville, MD, pp 178–189
- Wise CS, Dimler RJ, Davis HA, Rist CE. Determination of easily hydrolyzable fructose units in dextran preparations. Anal Biochem. 1955;27:33–36. [Google Scholar]