Abstract
Trisomy 21 is associated with hematopoietic abnormalities in the fetal liver, a preleukemic condition termed transient myeloproliferative disorder, and increased incidence of acute megakaryoblastic leukemia. Human trisomy 21 pluripotent cells of various origins, human embryionic stem (hES), and induced pluripotent stem (iPS) cells, were differentiated in vitro as a model to recapitulate the effects of trisomy on hematopoiesis. To mitigate clonal variation, we isolated disomic and trisomic subclones from the same parental iPS line, thereby generating subclones isogenic except for chromosome 21. Under differentiation conditions favoring development of fetal liver-like, γ-globin expressing, definitive hematopoiesis, we found that trisomic cells of hES, iPS, or isogenic origins exhibited a two- to fivefold increase in a population of CD43+(Leukosialin)/CD235+(Glycophorin A) hematopoietic cells, accompanied by increased multilineage colony-forming potential in colony-forming assays. These findings establish an intrinsic disturbance of multilineage myeloid hematopoiesis in trisomy 21 at the fetal liver stage.
Induced pluripotent stem (iPS) cells are a promising platform for in vitro modeling of human diseases. Differentiation of iPS cells is especially useful in settings where appropriate patient material is unavailable and animal models fail to recapitulate human phenotypes. For in vitro disease modeling to be meaningful, experimental methods need to be developed to generate iPS-derived differentiated cells of a developmental stage that is biologically relevant to the phenotype or disease under study. In addition, because of inherent differences among independent pluripotent cell lines and observed variability of in vitro differentiation experiments, suitable controls are essential. The importance of control cells is compounded when comparing “disease” ES/iPS cells to wild-type controls with different genetic backgrounds, particularly in situations where no single gene is known that might be used to revert a disease phenotype. An ideal control for in vitro disease modeling are cells that are isogenic except for a defined genetic defect, but such cells are understandably difficult to generate.
We have explored the potential of human pluripotent cells to model hematopoietic disturbances associated with trisomy 21. Trisomy 21 is the most common viable human aneuploidy. In addition to physical and cognitive deficiencies, individuals with trisomy 21 are at increased risk of developing both lymphoid and myeloid leukemias (1). Most striking is the extraordinarily elevated incidence of acute megakaryocytic leukemia (AMKL) in young children with trisomy 21, which is estimated at ∼500-fold (2, 3). Five to 10% of trisomy 21 infants develop a preleukemic condition called transient myeloproliferative disease (TMD), characterized by expansion of immature megakaryoblasts (4). Although most cases of TMD spontaneously resolve, ∼10–20% progress to frank AMKL (5). Both TMD and AMKL are invariably associated with diverse somatic mutations in the transcription factor Gata1, all leading to expression of an amino-truncated polypeptide, termed Gata1s (6, 7).
Consistent with the presumed intrauterine origin of TMD, abnormalities in hematopoiesis within trisomy 21 fetal livers (FL) have been described, including alterations in progenitor populations in the absence of Gata1s mutations (8, 9). Current findings in the field suggest a model whereby baseline hematopoiesis at the FL stage is perturbed in trisomy 21, and further aggravated by acquired Gata1s mutation, leading to clinical TMD either in utero or in the neonatal period. Upon acquisition of additional somatic “hits,” TMD cells may be transformed to generate AMKL (3).
To date, animal models have not elucidated mechanisms underlying the relationship between trisomy 21, abnormal hematopoiesis, and TMD/AMKL. Existing mouse models of human trisomy 21 fail to generate phenotypes reminiscent of TMD (10, 11). Because fetal and adult megakaryocyte progenitors exhibit differences in signaling pathway dependence, for example in response to insulin growth factor (IGF) signaling (12), abnormalities in adult hematopoiesis described in such mouse models may have little relevance to the pathogenesis of TMD/AMKL in trisomy 21 in humans, which likely has its origin in FL-like cells.
In contrast to the above negative observations, FL cells of mice expressing the Gata1s mutation are modestly hyperproliferative. These fetal-type, Gata1s-responsive progenitors are presumed to represent the target cells for TMD in trisomy 21 (13). However, introduction of the Gata1s into trisomy 21 mouse models fails to elicit TMD or AMKL (10, 13).
Given the limitations of existing mouse models for trisomy 21, in vitro hematopoietic differentiation of human pluripotent cells is an appealing approach to examine abnormalities associated with trisomy 21 or TMD/AMKL in humans. For in vitro differentiation to yield biologically relevant results, it is necessary to use a differentiation protocol that is robust, consistent, and capable of generating cells similar to hematopoietic progenitor cells of the FL.
We have differentiated several independent disomic and trisomic human ES (hES) and iPS cell lines to characterize hematopoiesis in the context of trisomy 21. In an effort to minimize clonal variation, we have isolated and characterized disomic and trisomic subclones that are isogenic, with the exception of chromosome 21. These cells validate experimental findings in hES and iPS lines, and constitute a unique tool for further elucidation of trisomy 21 phenotypes that may be elicited in culture. Our findings reveal an intrinsic disturbance of multiple myeloid lineages in FL-like trisomy 21 cells differentiated in vitro from pluripotent human cells.
Results
Experimental Strategy.
Based on the variable extent and quality of in vitro differentiation reported for human pluripotent cell clones and potential differences between ES and iPS cells, we examined lines of diverse origins to minimize these challenges (Table S1). Accordingly, we have analyzed independently derived disomic and trisomic hES (CSES2 and CSES13) and iPS (DS2-iPS1, DS2-iPS10, DS1-iPS4, MRC5-IPS7) cell lines (14, 15), as well as disomic and trisomic iPS subclones that are isogenic except for the presence of an additional chromosome 21 (detailed below).
Isolation and Characterization of Isogenic Di- and Trisomic iPS Cells.
In general, we and others have observed that hES and iPS cells trisomic for chromosome 21 are karyotypically stable (14, 16, 17). However, during routine quality-control analysis of serially passaged trisomic DS1-iPS4 iPS cells, we observed the emergence of a mixed population consisting of cells disomic and trisomic for chromosome 21. DNA microsatellite analysis excluded inadvertent contamination of the original trisomic line. Indeed, during passage, trisomic cells lost one copy of chromosome 21 and gave rise to disomic derivatives (Fig. 1A). From these mixed DS1-iPS4 cultures, single cells were isolated and plated by limiting dilution for expansion of cell clones. FISH confirmed that isolated subclones were entirely disomic or trisomic for chromosome 21 (Fig. 1A and Fig. S1). No other chromosomal abnormalities associated with the disomic or trisomic subclones were observed in karyotype analysis (Fig. 1B).
Fig. 1.
Isolation and characterization of isogenic disomic and trisomic clones. (A) Overview depicting subcloning of disomic and trisomic isogenic iPS cells from a mixed culture of cells. (B) Subcloned isogenic disomic and trisomic clones express Oct4, Tra1-60, and exhibit stable karyotypes lacking chromosomal abnormalities other than trisomy 21. (Magnification: 20×.)
Disomic and trisomic isogenic subclones expressed markers indicative of pluripotency, as revealed by immunofluorescence with antibodies directed to Oct4, Nanog, Tra1-60, and Tra1-81 (Fig. 1B and Fig. S1). Functional pluripotency was assessed by teratoma formation. All analyzed subclones developed differentiated teratomas consisting of all three germ layers (Fig. S1). Semiquantitative PCR was performed on all isolated subclones to confirm silencing of transduced reprogramming factors, and no differences were observed between disomic and trisomic subclones (Fig. S2). Four disomic and four trisomic DS1-iPS4 isogenic subclones were chosen for subsequent study. FISH was used to confirm genotypes before differentiation experiments.
FL-Type Hematopoietic Differentiation of Human Pluripotent Cells.
To examine the effect of trisomy 21 on hematopoietic development, disomic and trisomic hES, iPS, and isogenic iPS cells were differentiated into hematopoietic cells via embryoid bodies (EB) (Fig. 2A) (18, 19). The differentiation protocol we used yields hemangioblast cells poised to develop into hematopoietic or endothelial progenitors, and subsequently more mature hematopoietic cells. The use of chemically defined serum-free media in this feeder-free culture system results in robust, reproducible differentiation, while minimizing variability associated with feeder cell-dependent protocols.
Fig. 2.
Fetal-like definitive hematopoietic differentiation via EBs. (A) Schematic detailing the differentiation of human pluripotent cells into hematopoietic lineages via EBs. (B) Globin expression in cells isolated from day 10 EBs and CFU-E colonies relative to GAPDH. CD34+ bone marrow and CD34+ FL represent positive control samples from differentiated CD34+ bone marrow and FL cells, respectively. N.D., not detected.
A critical aspect of in vitro disease modeling relates to characterization of the developmental state of cells generated for analysis. TMD and AMKL in trisomy 21 individuals represent disorders of fetal hematopoiesis. Prior experience with human pluripotent cells has revealed a strong bias toward embryonic or FL-type hematopoiesis, and a deficiency of adult-type (i.e., bone marrow) hematopoiesis (20, 21). In human development, embryonic, fetal, and adult-type hematopoiesis are characterized by expression of stage-specific β-like globins. Embryonic-, fetal-, and adult-stage development is reflected by expression of ε-, γ-, and β-globins, respectively. γ-Globin is expressed during midgestation once hematopoiesis shifts from the embryonic yolk sac to the FL (20). To assess the nature of hematopoiesis achieved in our differentiation protocol, we isolated glycophorin A (CD235)+ cells from day 10 EBs, and performed quantitative RT-PCR for β-like globin transcripts. As shown in Fig. 2B, the predominantly expressed β-like globin was γ-globin. Furthermore, we isolated RNA from CFU-E colonies (see below, Fig. 4) formed by cells from day 10 EBs, and analyzed β-like globin expression. Although some ε-globin was detected in these cells, γ-globin was much more abundant, accounting for roughly 82% of total globin transcripts in CFU-E cells. Appropriate positive controls for quantification included erythroid precursors differentiated from CD34+ cells isolated from human bone marrow or FL (22). Expression of γ-globin in the in vitro differentiation protocol we used indicates that the cells generated are characteristic of FL-type definitive hematopoiesis. Therefore, the protocol gives rise to cells well suited for interrogation of the effects of trisomy 21 on FL hematopoiesis.
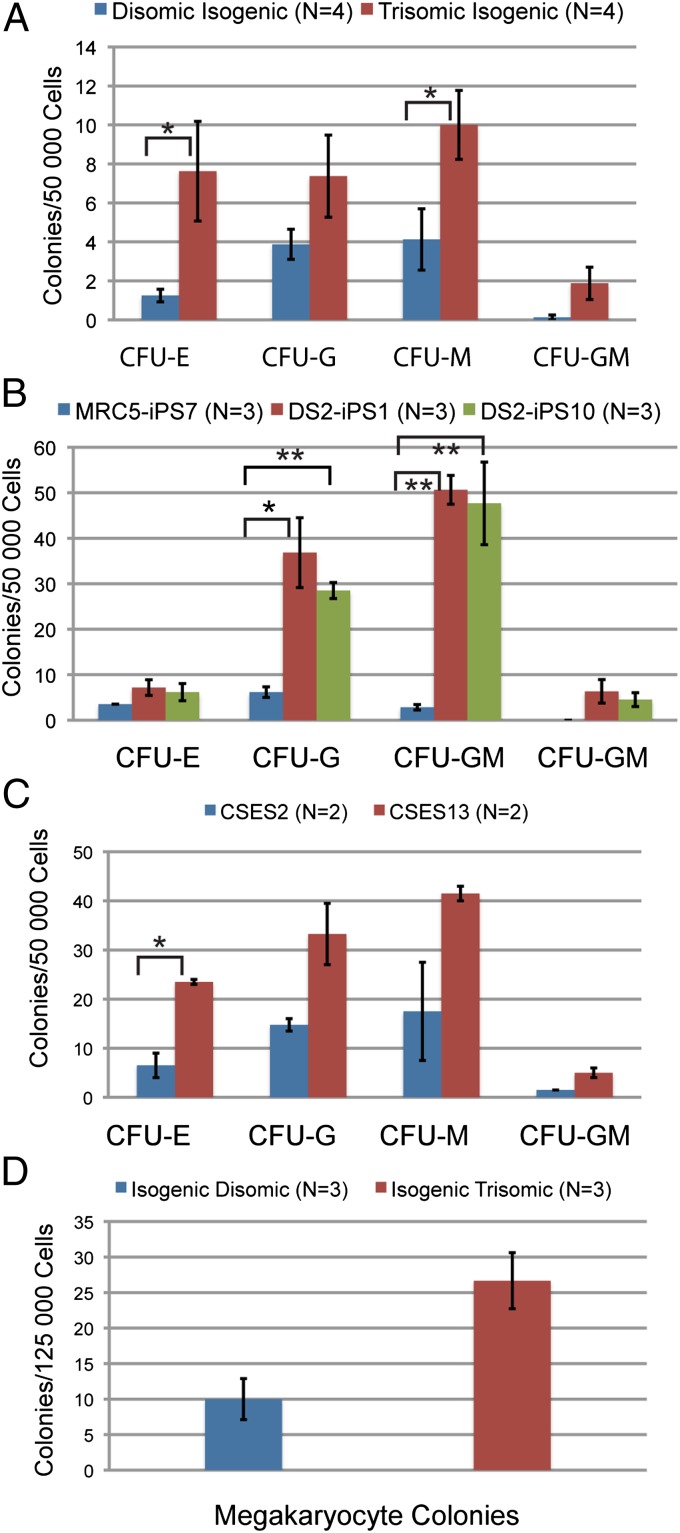
Fig. 4.
Colony-forming potential of differentiated pluripotent cells. Colonies formed by isogenic (A), ES (B), and independent iPS (C) in methylcellulose colony-forming assay. (D) Megakaryocyte colonies formed by isogenic cells. Error bars represent SEM, and P values were determined by the Student t test; *P < 0.05, **P < 0.005.
Altered Hematopoietic Populations During Differentiation of Trisomic Pluripotent Cells.
Using this in vitro differentiation protocol, we examined the consequences of trisomy 21 on fetal hematopoiesis. Isogenic disomic and trisomic clones were induced to form EBs, and cells were isolated from day 8 EBs, a stage at which a CD31/KDR+ population containing hematopoietic progenitor cells appears (18). Disomic and trisomic isogenic clones generated comparable frequencies of CD31+ cells, 11.2 ± 1.4% and 11.7 ± 0.6% of total cells, respectively (Fig. 3A). In hematopoietic-specific progenitor populations defined as CD31/KDR+ in day 8 EBs, there was no significant difference between disomic (2.4 ± 0.6%) and trisomic (1.9 ± 0.3%) isogenic clones. Similarly, in cells isolated from day 10 EBs, there was no difference in CD31+ populations between disomic (20.6 ± 2.2) and trisomic (21.8 ± 1.9) isogenic subclones.
Fig. 3.
Hematopoietic populations of in vitro differentiated disomic and trisomic pluripotent cells. (A) CD31/KDR+ cells derived from day 8 EBs. (B and C) CD43 and CD235+ populations isolated from day 10 and day 11 EBs, respectively. Error bars represent SEM, and P values were determined by the Student t test; **P < 0.005.
CD43 (leukosialin), an early hematopoietic progenitor-specific marker, is expressed during in vitro differentiation of hES cells (23). CD43-expressing hematopoietic cells derived from differentiated hES/iPS cells express the erythroid marker CD235 (glycophorin A) (24). With the differentiation protocol that we used, CD43 and CD235 expression is first detected in a subpopulation of CD31+ cells from day 10 EBs (Fig. 3B). In differentiated isogenic iPS cells, there was a small but not statistically significant increase in CD43/CD235+ cells in trisomic (2.9 ± 0.4%) compared with disomic (2.0 ± 0.4%) controls. However, in day 11 EBs the CD235/CD43+ population was significantly larger for isogenic trisomic cells (12.7 ± 1.1% vs. 5.2 ± 0.5% in isogenic disomic cells, P ≤ 0.005) (Fig. 3C). The frequency of CD43+CD235− cells was also increased in trisomic isogenic clones (2.3 ± 0.2%) compared with disomic control (1.3 ± 0.3%, P ≤ 0.005) cells (Fig. 3C). A small population of CD235+CD43− cells, which presumably represent more mature erythroid lineage cells, was observed, but there was no significant difference between isogenic trisomic (1.9 ± 0.2%) and disomic (1.8 ± 0.2%) clones. To ensure that observed phenotypes were not because of re-expression of reprogramming factors used to generate the iPS cells, semiquantitative RT-PCR was performed, and no expression of endogenous or transduced Oct4, KLF4, or Sox2 was observed in differentiated cells isolated from day 11 EBs (Fig. S2). Low levels of endogenous Myc expression were detected in disomic and trisomic subclones (Fig. S2).
Hematopoietic differentiation was also analyzed in trisomic ES and iPS cells, as well as corresponding controls (Table S1). These cell lines exhibited similar differentiation trends as the isogenic disomic and trisomic cells, but with different kinetics. In cells isolated from day 10 EBs, a time at which no significant difference was observed in the isogenic clones, trisomic ES cells exhibited an increase in CD43/CD235+ cells (7.6 ± 0.8%) compared with disomic ES cells (1.0 ± 0.4%, P ≤ 0.02). In addition, the population of CD43+CD235− cells in trisomic clones was also expanded (4.6 ± 0.3% compared with 0.9 ± 0.1%, P ≤ 0.01) (Fig. 3B). This increase was maintained in cells isolated from day 11 EBs, as trisomic ES cells generated 13.0 ± 1.0% CD235/CD43+ cells compared with 4.9 ± 1.4% in disomic ES cells. Similarly, in day 10 EBs, the trisomic iPS cell lines DS2-iPS1 and DS2-iPS10 generated 8.3 ± 3.0% and 4.7 ± 1.3% CD43/CD235+ cells respectively, whereas disomic iPS cells (MRC5-IPS7) generated 2.0 ± 0.7% CD235/CD43+ cells (Fig. 3C).
Increased Colony-Forming Potential of Trisomic iPS Cells.
To enumerate hematopoietic progenitor populations, we conducted methylcellulose colony-forming assays using unsorted cells from day 10 EBs. Isogenic trisomic cells displayed increased multilineage colony-forming potential as CFU-E, CFU-G, CFU-M, and CFU-GM outputs were all greater than isogenic disomic controls (Fig. 4A). Megakaryocyte colony-forming potential (CFU-Meg) was also greater in isogenic trisomic clones compared with isogenic disomic clones (26.7± 3.9 vs. 10.0 ± 2.9, P ≤ 0.05) (Fig. 4D).
These results were validated as assays performed with independent iPS cell lines (MRC5-IPS7-disomic, DS2-iPS1, DS2-iPS10-trisomic) also demonstrated increased colony-forming potential in these trisomic clones for CFU-GM, CFU-G, and CFU-M lineage (Fig. 4B). Similarly, we observed increased multilineage colony-forming potential in trisomic ES cells compared with disomic ES controls (Fig. 4C). In general, the independent iPS and ES cell lines (either disomic or trisomic) exhibited greater colony-forming potential than the isogenic iPS cells (Compare Fig. 4 B and C with A). This observation emphasizes the importance of isogenic cells as controls, as we and other groups have observed that different “wild-type” human iPS and ES lines exhibit extensive variability in colony forming potential (24).
Gene-Expression Profiling of Differentiated Pluripotent Cells.
Gene-expression profiling with the isogenic clones was performed on RNA isolated from CD235+ cells generated by day 11 EBs. Expression analysis was performed on a panel of genes on chromosome 21, insulin-like growth factor (IGF) signaling genes, and hematopoietic progenitor markers (Fig. S3). Although it has been speculated that the increased copy number of the cancer-associated hematopoietic transcription factors on chromosome 21 ETS, ERG, and RUNX1 might contribute to TMD or AMKL (25), we observed no consistent change in their transcript levels between differentiated isogenic disomic and trisomic cells. It has been demonstrated that IGF signaling is increased in human DS-AMKL (12); hence, we also analyzed the expression of IGF signaling genes, but observed no significant differences between disomic and trisomic cells.
Discussion
Use of Isogenic Cells to Control in Vitro Differentiation.
Experimental variability of in vitro differentiation of human pluripotent cells complicates simple comparison of clones of diverse origins. Several factors contribute to these observations, including inherent genetic differences between individuals, different methods for derivation of hES cells, the origin of cells used for reprogramming to an iPS state, and the specific reprogramming protocol used. We and others have observed significant clonal variability upon in vitro differentiation of distinct pluripotent cell lines (Fig. 3) (26). In the absence of a “rescue” assay, in which introduction of a single gene product corrects a disease phenotype, generation of isogenic lines should mitigate the effects of genetic diversity or independent derivation of cell lines, as illustrated by creation of isogenic iPS Parkinson disease cells through gene targeting (27). Isogenic cells serve as critical controls for in vitro differentiation experiments, and provide a framework for comparing results with independent hES and iPS lines.
FL-Type Hematopoiesis from Pluripotent Cells.
Several protocols for in vitro hematopoietic differentiation of human pluripotent cells have been reported (18, 20, 23, 28–30). The milieu of cytokines added to the cultures may account in part for the different lineages produced in the various protocols. Protocols vary considerably in the capacity to recapitulate primitive or definitive hematopoiesis. Several groups have reported generation of differentiated cells expressing ε- and γ-globin; in contrast, only very low levels of β-globin expression have been observed (20, 29). Thus, existing protocols tend to reflect embryonic and fetal, but not adult, hematopoiesis.
As revealed by the predominant expression of γ-globin (Fig. 2B) in CD235+ cells, our differentiation protocol yields cells most comparable to definitive cells within the human FL. This observation is significant, because evidence suggests that the target cell population of trisomy 21/TMD resides in the FL. In addition to reports of perturbed hematopoiesis in human trisomy 21 FL, analysis of Gata1s knock-in mice identified a FL hematopoietic progenitor cell population that is hyperproliferative in response to Gata1s expression (8, 9, 13). Adult Gata1s-expressing animals fail to show prominent hematological abnormalities, indicating that fetal and adult hematopoietic progenitors respond differently to Gata1s expression (13). Further insights into the pathogenesis of TMD will necessitate study of Gata1s within the context of human FL progenitors. Therefore, to be biologically relevant, in vitro disease modeling should develop hematopoietic cells similar to those of the FL, such as those generated by our differentiation protocol.
In an accompanying manuscript, Chou et al. used a similar experimental approach to analyze hematopoiesis in trisomy 21 pluripotent cells (31). The authors report enhanced erythropoiesis, reduced myelopoiesis, and normal megakaryocyte production under differentiation conditions that generate largely ε-globin expressing, embryonic erythroid cells. Although the precise experimental parameters that lead to embryonic versus fetal-like hematopoiesis in the in vitro differentiation system are unknown, we believe that the principal differences are readily accounted for by the different stages of hematopoietic development achieved in the two laboratories. It is well established that in vitro hematopoietic differentiation of human ES or iPS cells fails to generate fully adult-type hematopoietic cells, but rather gives rise to embryonic- or fetal-stage cells in various published protocols. Rather than a drawback of the in vitro systems, we concur with Chou et al. that the observed differences in our findings present a unique opportunity to examine the consequences of trisomy 21 at two distinct stages of hematopoiesis (31). Therefore, trisomy 21 exerts distinct effects on embryonic and fetal hematopoiesis. Insight into such a biological difference cannot be obtained in human embryos because of inaccessibility of the embryonic stage for analysis. Furthermore, the contrast in the findings of our work and that of Chou et al., highlight the importance of defining the developmental stage of differentiated cells, an aspect that is generally overlooked in the majority of in vitro disease modeling studies.
Abnormal FL-Type Hematopoiesis from Trisomy 21 Cells.
TMD and AMKL in association with trisomy 21 reflect step-wise oncogenesis. Defining the contributions of each genetic event—trisomy 21, Gata1s mutation, and additional somatic hits—should provide critical biological insights and suggest novel, targeted therapeutic approaches. Limitations of murine models for human trisomy 21 have necessitated the use of alternative approaches to uncover the mechanisms underlying TMD/AMKL development in relationship to trisomy 21. Using human pluripotent cells, and a differentiation protocol that generates hematopoietic progenitor cells resembling definitive fetal-type cells of the FL, we have observed that trisomy 21 is accompanied by an expansion of hematopoietic progenitor cells, similar to that reported in the human FL (8, 9). Specifically, we have identified a significant increase in CD43/CD235+ cells in trisomic clones. Increased colony-forming potential in erythroid, myeloid, and megakaryocytic lineages indicates that trisomy 21 may result in enhanced progenitor populations with common myeloid progenitor-like activity, or perhaps hematopoietic stem cell (HSC)-like activity. The findings of an accompanying manuscript by Roy et al. demonstrate that trisomy 21 human FL cells exhibit an increase in phenotypic HSCs, and a concomitant decrease in B-cell potential (32). In vitro differentiated human ES/iPS cells do not consistently generate lymphocytes; thus, we are unable at present to functionally discriminate between HSC and common myeloid progenitor-like cells. Therefore, our results and those of Roy et al. are in agreement that trisomy 21 is associated with increased production of cells representing multiple, nonlymphoid hematopoietic lineages (32).
We have observed increases in hematopoietic progenitor populations in trisomic ES, iPS and isogenic iPS cells. These consistent observations provide persuasive evidence that the described phenotypes can be attributed directly to the presence of an additional copy of chromosome 21, and not another genetic effect. Furthermore, our results validate isogenic cells as an appropriate model to study the effects of trisomy 21 in isolation. These isogenic cells will be particularly important for further molecular analysis in identifying genetic targets responsible for the effects of trisomy 21 on hematopoiesis. Furthermore, advances in disease modeling of other tissues, for example in study of development of early-onset Alzheimer’s Disease in trisomy 21 (16), illustrates how these cells may be used to explore other trisomy 21-associated phenotypes relevant to the central nervous system or cardiovascular development. Taken together with the findings of Roy et al. and Chou et al., our findings establish abnormalities in hematopoiesis because of trisomy 21, and provide tools for further elucidating how an extra copy of chromosome 21 leads to an altered transcription network and phenotypic consequences.
Experimental Procedures
HES/iPS Cell Culture and Subcloning of Isogenic iPS Cells.
hES and iPS lines were maintained on irradiated CF-1 feeder cells (Global Stem), in standard hES media (15). To isolate isogenic subclones, cultures of mixed disomic/trisomic cells (as determined by FISH, performed by Cell Line Genetics) were plated on matrigel-coated plates (Invitrogen). After 5 d of culture in mTeSR1 media (Stem Cell Technologies), cells were treated with Accutase (Millipore) for 10 min at 37 °C until single-cell suspensions were obtained. Cells were then plated out in limiting dilution onto 24-well matrigel-coated plates. Resultant isolated colonies from single cells were expanded for further characterization.
Characterization of Isogenic iPS Cells.
Karyotype, FISH and DNA microsatellite analysis of isogenic clones was performed by Cell Line Genetics (www.clgenetics.com). Subsequently, FISH was performed routinely in our laboratory with a chromosome 21 specific probe (Vysis LSI 21 probe; Vysis).
Teratoma Formation.
For teratoma formation, 2 × 106 cells were isolated from matrigel-coated plates and resuspended in 25 μL undiluted matrigel and 25 μL collagen solution (3 mg/mL; Stem Cell Technologies) before intramuscular injection into Rag2−/−γC−/−mice. After 6–9 wk, tumors were isolated, fixed in Bouin’s solution, and sent for sectioning, H&E staining, and analysis (Rodent Histopathology Core, Harvard Medical School).
Hematopoietic Differentiation.
Pluripotent cells were differentiated into hematopoietic cells as previously described and are detailed in the Table S2 (18, 19). Cytokines were purchased from R&D Systems, with the exception of FGF (StemGent) and EPO (EPOGEN).
Isolation of Single Cells from Embryoid Bodies and FACS Analysis.
To isolate single cells, EBs were treated with 0.2% collagenase IV in 20% (vol/vol) FCS for 1 h at 37 °C. Then, EBs were treated with 0.05% Trypsin for 5 min at 37 °C before being passed through a 20-G needle six times. Cells were washed in 10% (vol/vol) FCS/IMDM and filtered before staining for FACS analysis. The following directly conjugated antibodies were used for FACS staining: anti–CD235-PE, anti–CD235-APC, anti–CD43-FITC, anti–CD34-APC, (BD Pharmingen), anti–CD31-Alexa488 (Invitrogen). Live cells were selected by staining with 7-AAD or DAPI. Cells were stained for 30 min at 4 °C in the dark, and washed twice with PBS/2% (vol/vol) FCS before FACS analysis.
Colony-Forming Assays.
Colony-forming assays were performed using Methocult H4434 Classic media (Stem Cell Technologies); 50,000 unsorted cells from day 10 EBs were used for assays as per the manufacturer’s directions. For each cell line, two plates were used for the assay, and results were averaged when colonies were scored 10–14 d after plating. Results are presented as the average of assays ± the SEM. For megakaryocyte colonies, the Megacult kit (Stem Cell Technologies) was used, with 125,000 unsorted cells from day 10 EBs used for each assay. After 10 d, cultures were fixed and stained as directed by the manufacturer. Megacult assays were also performed in duplicate for each cell line; results were averaged, and error bars represent SEM.
Quantitative RT-PCR.
RNA was isolated from various sorted cell populations using TRIzol (Invitrogen) extraction followed by purification with an RNeasy kit (Qiagen). cDNA was amplified using iScript cDNA synthesis kit as directed (Bio-Rad) and quantitative PCR reactions were performed using IQ Sybr Green Supermix (Bio-Rad). Primers for ε-globin, β-globin, γ-globin, and GAPDH have previously been described (33). All reactions were performed in triplicate on the same plate, and relative expression was calculated relative to GAPDH expression. For positive controls, RNA was extracted from differentiated erythroid precursors from CD34+ cells isolated from FL or bone marrow (22). For analysis of reprogramming factor expression, semiquantitative RT-PCR was performed as previously described (15).
Expression Profiling.
Multiplex quantitative PCR was performed as previously described, using RNA isolated from day 11 EBs formed by isogenic disomic and trisomic cells (34). Primers used for analysis are listed in Table S3.
Supplementary Material
Acknowledgments
This work was funded by National Institutes of Health Grant U01 HL 10001 as part of the Progenitor Cell Biology Consortium of the National Heart, Lung and Blood Institute; a seed grant of the Harvard Stem Cell Institute and the Howard Hughes Medical Institute; a Lady Tata Memorial Trust fellowship (to G.M.); and Leukemia and Lymphoma Research and Kay Kendal Leukemia Fund fellowships (to T.F.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215468109/-/DCSupplemental.
References
- 1.Bruwier A, Chantrain CF. Hematological disorders and leukemia in children with Down syndrome. Eur J Pediatr. 2011;171:1301–1307. doi: 10.1007/s00431-011-1624-1. [DOI] [PubMed] [Google Scholar]
- 2.Klusmann JH, et al. Treatment and prognostic impact of transient leukemia in neonates with Down syndrome. Blood. 2008;111:2991–2998. doi: 10.1182/blood-2007-10-118810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roy A, Roberts I, Norton A, Vyas P. Acute megakaryoblastic leukaemia (AMKL) and transient myeloproliferative disorder (TMD) in Down syndrome: A multi-step model of myeloid leukaemogenesis. Br J Haematol. 2009;147:3–12. doi: 10.1111/j.1365-2141.2009.07789.x. [DOI] [PubMed] [Google Scholar]
- 4.Pine SR, et al. Incidence and clinical implications of GATA1 mutations in newborns with Down syndrome. Blood. 2007;110:2128–2131. doi: 10.1182/blood-2007-01-069542. [DOI] [PubMed] [Google Scholar]
- 5.Massey GV, et al. Children’s Oncology Group (COG) A prospective study of the natural history of transient leukemia (TL) in neonates with Down syndrome (DS): Children’s Oncology Group (COG) study POG-9481. Blood. 2006;107:4606–4613. doi: 10.1182/blood-2005-06-2448. [DOI] [PubMed] [Google Scholar]
- 6.Wechsler J, et al. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32:148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- 7.Rainis L, et al. Mutations in exon 2 of GATA1 are early events in megakaryocytic malignancies associated with trisomy 21. Blood. 2003;102:981–986. doi: 10.1182/blood-2002-11-3599. [DOI] [PubMed] [Google Scholar]
- 8.Tunstall-Pedoe O, et al. Abnormalities in the myeloid progenitor compartment in Down syndrome fetal liver precede acquisition of GATA1 mutations. Blood. 2008;112:4507–4511. doi: 10.1182/blood-2008-04-152967. [DOI] [PubMed] [Google Scholar]
- 9.Chou ST, et al. Trisomy 21 enhances human fetal erythro-megakaryocytic development. Blood. 2008;112:4503–4506. doi: 10.1182/blood-2008-05-157859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alford KA, et al. Perturbed hematopoiesis in the Tc1 mouse model of Down syndrome. Blood. 2010;115:2928–2937. doi: 10.1182/blood-2009-06-227629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirsammer G, et al. Highly penetrant myeloproliferative disease in the Ts65Dn mouse model of Down syndrome. Blood. 2008;111:767–775. doi: 10.1182/blood-2007-04-085670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klusmann JH, et al. Developmental stage-specific interplay of GATA1 and IGF signaling in fetal megakaryopoiesis and leukemogenesis. Genes Dev. 2010;24:1659–1672. doi: 10.1101/gad.1903410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, et al. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat Genet. 2005;37:613–619. doi: 10.1038/ng1566. [DOI] [PubMed] [Google Scholar]
- 14.Biancotti JC, et al. Human embryonic stem cells as models for aneuploid chromosomal syndromes. Stem Cells. 2010;28:1530–1540. doi: 10.1002/stem.483. [DOI] [PubMed] [Google Scholar]
- 15.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y, Kirwan P, Smith J, MacLean G, Orkin SH, Livesey FJ. A human stem cell model of early Alzheimer's disease pathology in Down syndrome. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003771. 124ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayshar Y, et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 18.Kennedy M, D’Souza SL, Lynch-Kattman M, Schwantz S, Keller G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood. 2007;109:2679–2687. doi: 10.1182/blood-2006-09-047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grigoriadis AE, et al. Directed differentiation of hematopoietic precursors and functional osteoclasts from human ES and iPS cells. Blood. 2010;115:2769–2776. doi: 10.1182/blood-2009-07-234690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dias J, et al. Generation of red blood cells from human induced pluripotent stem cells. Stem Cells Dev. 2011;20:1639–1647. doi: 10.1089/scd.2011.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang CJ, et al. Production of embryonic and fetal-like red blood cells from human induced pluripotent stem cells. PLoS ONE. 2011;6:e25761. doi: 10.1371/journal.pone.0025761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sankaran VG, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 23.Vodyanik MA, Thomson JA, Slukvin II. Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. Blood. 2006;108:2095–2105. doi: 10.1182/blood-2006-02-003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi KD, et al. Hematopoietic and endothelial differentiation of human induced pluripotent stem cells. Stem Cells. 2009;27:559–567. doi: 10.1634/stemcells.2008-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan I, Malinge S, Crispino J. Myeloid leukemia in Down syndrome. Crit Rev Oncog. 2011;16:25–36. doi: 10.1615/critrevoncog.v16.i1-2.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woods NB, et al. Brief report: Efficient generation of hematopoietic precursors and progenitors from human pluripotent stem cell lines. Stem Cells. 2011;29:1158–1164. doi: 10.1002/stem.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soldner F, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill KL, Kaufman DS. Hematopoietic differentiation of human embryonic stem cells by cocultivation with stromal layers. Curr Protoc Stem Cell Biol. 2008 doi: 10.1002/9780470151808.sc01f06s6. Chapter 1:Unit 1F.6. [DOI] [PubMed] [Google Scholar]
- 29.Ma F, et al. Generation of functional erythrocytes from human embryonic stem cell-derived definitive hematopoiesis. Proc Natl Acad Sci USA. 2008;105:13087–13092. doi: 10.1073/pnas.0802220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno-Gimeno I, Ledran MH, Lako M. Hematopoietic differentiation from human ESCs as a model for developmental studies and future clinical translations. Invited review following the FEBS Anniversary Prize received on 5 July 2009 at the 34th FEBS Congress in Prague. FEBS J. 2010;277:5014–5025. doi: 10.1111/j.1742-4658.2010.07926.x. [DOI] [PubMed] [Google Scholar]
- 31.Chou, et al. Trisomy 21 associated defects in human primitive hematopoiesis revealed through induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:9101–9106. doi: 10.1073/pnas.1211175109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy A, et al. Perturbation of fetal liver hematopoietic stem and progenitor cell development by trisomy 21. Proc Natl Acad Sci USA. 2012;109:17579–17584. doi: 10.1073/pnas.1211405109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sankaran VG, et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460:1093–1097. doi: 10.1038/nature08243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo G, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.