Abstract
Transcription activator-like effector nucleases (TALENs) are programmable nucleases that join FokI endonuclease with the modular DNA-binding domain of TALEs. Although zinc-finger nucleases enable a variety of genome modifications, their application to genetic engineering of livestock has been slowed by technical limitations of embryo-injection, culture of primary cells, and difficulty in producing reliable reagents with a limited budget. In contrast, we found that TALENs could easily be manufactured and that over half (23/36, 64%) demonstrate high activity in primary cells. Cytoplasmic injections of TALEN mRNAs into livestock zygotes were capable of inducing gene KO in up to 75% of embryos analyzed, a portion of which harbored biallelic modification. We also developed a simple transposon coselection strategy for TALEN-mediated gene modification in primary fibroblasts that enabled both enrichment for modified cells and efficient isolation of modified colonies. Coselection after treatment with a single TALEN-pair enabled isolation of colonies with mono- and biallelic modification in up to 54% and 17% of colonies, respectively. Coselection after treatment with two TALEN-pairs directed against the same chromosome enabled the isolation of colonies harboring large chromosomal deletions and inversions (10% and 4% of colonies, respectively). TALEN-modified Ossabaw swine fetal fibroblasts were effective nuclear donors for cloning, resulting in the creation of miniature swine containing mono- and biallelic mutations of the LDL receptor gene as models of familial hypercholesterolemia. TALENs thus appear to represent a highly facile platform for the modification of livestock genomes for both biomedical and agricultural applications.
Keywords: Tal-effector nuclease, biotechnology, gene-editing
The ability to knockout or precisely alter genes is fundamental for determining gene function and genetic engineering. For livestock, gene-KO strategies enable refinement of traits for xenotransplantation and biomedical products (1) and have been used to produce valuable models of human disease (2). There are further objectives in animal agriculture that would benefit from gene-KO, including functional characterization of high-impact genes identified in association studies, engineering disease resistance, reducing the threat of zoonotic disease transmission, alteration of production traits, and enhancement of animal welfare. Until recently, homologous recombination (HR) in primary fibroblasts followed by somatic-cell nuclear transfer was the exclusive method for the production of KO pigs and cattle. However, generation of KO cell lines by HR is inefficient and the length of time for gestation and reproductive maturation for livestock represent significant barriers to homozygous gene inactivation or the engineering of multiple loci. In addition, HR often requires the use of a linked selection-marker, which can confound functional studies (3) and complicate regulatory approval of engineered food products.
Zinc finger nucleases (ZFNs) offer an alternative platform for germ-line KO. ZFNs are fusion proteins consisting of a modular DNA-binding domain tethered to a FokI endonuclease monomer. When two ZFNs bind their target in an appropriate orientation, FokI monomers can dimerize and introduce a DNA double-strand break (4). Lesions are often repaired by nonhomologous end-joining (NHEJ) that typically results in small insertions or deletions (indels) (5), two-thirds of which cause a frame-shift that disables encoded proteins. Thus, in contrast to HR, where genes are disabled by targeted introduction of selection cassettes, ZFNs disable genes without introduction of exogenous DNA. Generation of germ-line KO has been successful in many model animal systems (6) as well as in pigs (7, 8). Particularly exciting in livestock was the report of ZFN-mediated, biallelic KO of the porcine GGTA1 gene using commercial ZFN reagents (9), wherein biallelic-null cells could be enriched by FACS for the absence of a GGTA1-dependent surface epitope. Double-strand DNA breaks dramatically enhance HR (10), leading to homology-dependent repair of ZFN lesions for precise gene alteration in human cells (11), mice, and rats (12, 13). Although the potential of ZFN genome modification of livestock is great, design and assembly is labor-intensive and limited by available target sites (6).
Transcription activator-like effector nucleases (TALENs), like ZFNs, consist of assembled DNA-binding motifs coupled to the FokI nuclease (6, 14). Active, custom-designed TALENs have been reported to induce indel frequencies between 2% and 55% of targeted chromosomes (15, 16). As with ZFNs, TALEN-mediated double-strand breaks also stimulate HR in human cells at levels similar to those achieved with ZFNs (11, 15). Most importantly, TALENs appear to be superior to ZFNs in terms of simple and straightforward design and assembly strategies (17), such that manufacture of effective TALENs is significantly cheaper and faster than effective ZFNs. Here, we demonstrate efficiency and versatility of TALENs for a variety of genome modifications to livestock genomes. We have achieved mono- and biallelic KO of genes and large chromosomal rearrangements. Finally, toward our goal of producing a large animal model of atherosclerosis, we’ve used TALENs to develop Ossabaw miniature swine (18) containing inactivating alleles of the LDL receptor (LDLR) gene. To our knowledge, this report of genetic engineering and cloning of these valuable biomedical animals is unique, and is also a unique example of TALEN-mediated gene modification of livestock.
Results
Evaluation of TALENs in Livestock Embryos.
We chose to characterize TALEN efficiency for precise alteration of genomes in livestock embryos by using in vitro-prepared bovine and porcine embryos. Three pairs of TALENs were generated using the +231 TALEN scaffold (17, 19) (Fig. 1). In vitro-transcribed mRNA encoding each TALEN-pair was injected into the cytoplasm of bovine embryos (≥50 embryos per condition) at ∼19 h postfertilization and were cultured in vitro until blastocyst stage (Table S1). Whole-genome amplification (WGA) and genotypic analysis revealed indels in one blastocyst injected with ACAN11 and five injected with ACAN12 TALENs (Fig. 1B and Table S1); however, development was significantly impaired in conditions with the greatest indel frequency (max indel frequency ∼10%) (Table S1). Two groups have reported enhancement in TALEN activity using different N- and C-terminal truncations of the native TALE protein (15, 20). Thus, we created a truncated TALEN scaffold (GoldyTALEN, GT) compatible with the Golden Gate cloning procedure reported in Cermak et al. (17) (Fig. 1A) for use in subsequent experiments. An additional TALEN-pair, GDF83.1, was developed using the GT scaffold. Zygotes were injected with TALEN mRNA plus 2 ng/μL of EGFP mRNA as an indicator of successful injection. Indel analysis was performed on individual blastocysts by direct sequencing of PCR amplicons spanning the TALEN recognition site (Fig. S1). Mutation frequency of GT-GDF83.1 TALENs significantly exceeded previous injections. Six of 14 blastocysts (43%) injected with a low mRNA dosage (2 ng/μL) displayed indels without a significant reduction in development rate (Table S1). Three of four blastocysts in the high-dosage group (10 ng/μL) displayed indels, with biallelic modification occurring in two of three mutant blastocysts (Fig. 1B).
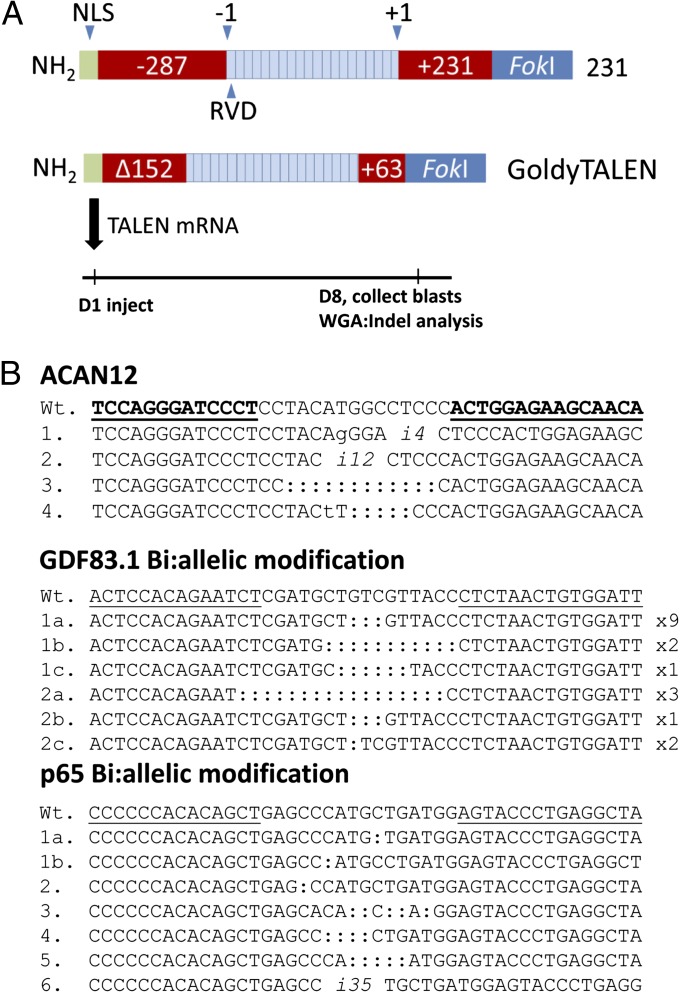
Fig. 1.
TALEN activity in bovine embryos. (A) TALENs for embryo injection generated in either the +231 or GT scaffold. Each scaffold shares a common SV40 nuclear localization signal (NLS) and C-terminal fusion of the FokI homodimer domain. Numbering is relative to the DNA-binding domain; the amino acid before the first repeat variable di-residue repeat (RVD) is labeled “−1” and the amino acid following the last RVD repeat is labeled “+1.” Bovine or swine, in vitro-produced zygotes were injected with TALEN mRNA on day 1 (D1) and cultured in vitro to blastocyst formation. Individual blastocysts (blasts) were collected on day 8, subjected to WGA, and analyzed for indels. (B) TALEN-mediated indels in bovine (ACAN12 and GDF83.1 injected) and porcine (p65-11.1 injected) embryos. Wild-type sequence is shown above with TALEN binding sites underlined. Both deletion and insertion (denoted with an “i” and number of base pairs) events were identified. Mismatch bases are indicated by lowercase text. Only biallelic modifications are shown for GDF83.1 and p65-11.1 embryos. Embryos with a homozygous indel (same indel on each allele) are shown on a single line. Indel alleles of compound biallelic embryos (two or more unique indel alleles) are displayed on multiple lines (e.g., 1a, 1b). Some blastocysts were partially mosaic. Sequence analysis of embryos 1 and 2 injected with bovine GDF83.1 TALENs revealed three unique indel alleles each; the number of reads for each allele is noted to the right of the sequence. In addition to indel alleles, 3 and 1 wild-type sequence reads were observed for GDF83.1 embryos 1 and 2, respectively.
A single set of injections was also conducted in porcine zygotes using TALENs targeted to the porcine RELA gene (p65) for which a tolerance allele for African Swine Fever has been proposed (21). Zygotes were injected with a mixture, including 20 ng/μL of TALEN mRNA along with 5 ng/μL of EGFP mRNA as an indicator of successful injection. In contrast to bovine injections, where all EGFP injected embryos fluoresced, only 35% (71 of 214) were EGFP+. WGA and PCR amplification was successful from 56 of the EGFP+ embryos, and 16 of these (29%) revealed indels by Surveyor assay or sequence analysis. One-third of the mutants (6 of 16) were either homozygous or heterozygous biallelic mutants (Fig. 1B).
TALEN Function in Livestock Fibroblasts.
Although our in vitro data demonstrates that direct embryo modification is likely to be a viable approach to livestock genome modification, the need to engineer alleles precisely led us to explore TALEN function in cultured livestock cells to meet two objectives: (i) to serve as a quality control measure for assembled TALEN-pairs before injection and (ii) to develop methods for TALEN-mediated genetic modification of livestock by cloning. To determine the best TALEN architecture for livestock fibroblasts, binding domains of six TALEN-pairs were placed in the context of the +231 and GT scaffolds (Fig. 1A). Each TALEN-pair was transfected into primary livestock fibroblasts, and the efficiency of genome modification was measured at day 3 by the Surveyor assay. The most active TALEN-pairs, DMDE7.1 and ACAN12, displayed cleavage efficiencies of 38% and 25% (Fig. 2A). The TALEN scaffold had a significant effect on nuclease activity in fibroblasts. In total, four of six loci targeted with the GT scaffold cleaved at 3.5% or greater but only the DMDE7.1 TALEN-pair cleaved above 1% with the +231 scaffold (Fig. 2). As noted in previous studies (15, 22), a 72-h incubation at 30 °C after transfection had a positive effect on target cleavage in livestock fibroblasts. We applied these findings to the design and testing of additional TALEN-pairs. In total, 23 of 36 (64%) TALEN-pairs were detectably active (> 1.0% NHEJ) at 15 genes scattered across the pig and cow genomes (autosomes and sex chromosomes) (Table S2). Three-quarters of the active pairs cleaved with high efficiency (19–40% NHEJ), with an average of 25%.
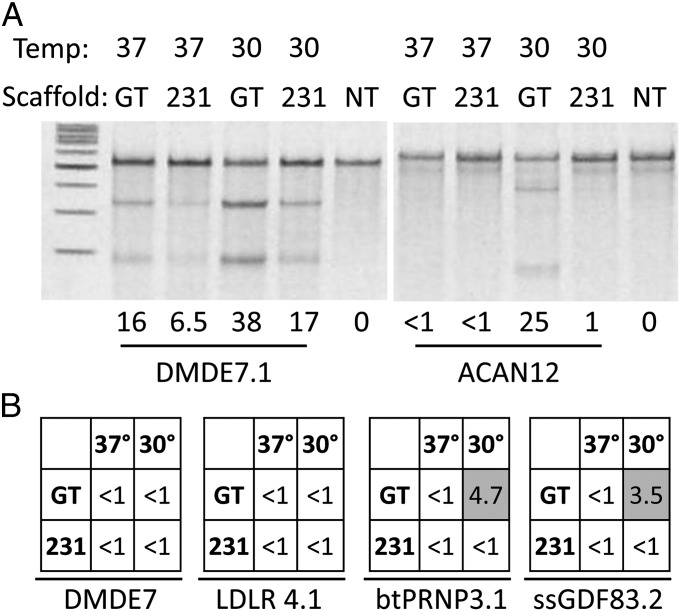
Fig. 2.
Comparison of TALEN scaffolds for gene-editing in livestock fibroblasts. (A) The Surveyor assay was conducted on transfected fibroblasts transfected with either DMDE7.1 or ACAN12 TALEN-pairs. Scaffold and temperature treatment is indicated above the gel and percent NHEJ is indicated below. NT, not treated. (B) Activity of four additional TALEN-pairs, identified at the bottom of each matrix, with either the +231 or GT scaffold (left column in each matrix) at either 30 or 37 °C.
Extended Culture and Indel Enrichment by Transposon Cotransfection.
An ideal resource for cloning would be cell populations in which the majority of cells harbor indels that remain stable over extended periods in culture. Although an average genomic modification level of 25% (i.e., ∼44% of cells with at least one modified allele; calculations described in Table 1) would be reasonable for direct somatic-cell nuclear transfer of cell populations, a method to enrich for modified cells or isolation of specific modified clones would be useful. To this end, we evaluated the stability of cells containing indels after an extended period in culture and tested an indel enrichment strategy based on transposon coselection (23). To apply this approach to TALEN-gene modification, a Sleeping Beauty transposon carrying a selection marker plus SB100X transposase was added to transfections with TALEN-encoding plasmids at a ratio of 1:5. Transient TALEN expression and transposon integration can only occur in cells that have been successfully transfected, thus providing a mechanism for enrichment for transfected cells. In addition, selection is an extremely reliable method for clonal isolation in primary fibroblasts, a cell type averse to low culture density and dilutional cloning (24).
Table 1.
Genotype distribution in fibroblast clones
| TALEN-pair | % NHEJ day 3 (Surveyor) | Predicted % mod clones | Predicted % biallelic mod | Observed mod clones (%) | Observed biallelic mod (%) | |
| LDLRE2.1 | Pig ♂ | 19 | 34.5 | 10.5 | 30/81 (37) | 5/26 (19) |
| LDLRE2.1 | Pig ♀ | 21.5 | 38.3 | 12 | 23/76 (30) | 8/23 (35)* |
| LDLRE2.1 | Pig ♂ | 14.4 | 26.7 | 7.7 | 12/94 (13) | 2/12 (≥17)† |
| LDLRE2.1-2x‡ | Pig | 19.7 | 35.5 | 10.9 | 8/24 (33) | 2/8 (≥25) † |
| LDLRE4.2 | Pig ♂ | 20 | 36 | 11.1 | 4/48 (8.3) | 1/4(25) † |
| LDLRE4.2 | Pig ♀ | 19 | 34.4 | 10 | 8/47 (17) | 0/8† |
| DMDE6 | Pig | 25 | 43.8 | 15.6 | 17/35 (49) | - |
| DMDE7.1 | Pig | 27 | 47 | 15.6 | 12/29 (41) | 3/10 (30) |
| DMDE7.1-2x‡ | Pig | 22 | 39.2 | 12.4 | 22/41 (54) | 7/22 (≥32)*† |
| GHRHR2.3 | Pig | 29 | 50 | 17 | 26/43 (60) | 15/26 (≥58)*§ |
| ACAN12 | Cow | 29 | 50 | 17 | 27/35 (77) | 2/6 (NA) ¶ |
| btGDF83.1 | Cow | 17 | 31 | 9.3 | 7/24 (29) | 0/7 |
If chromosome modification is an independent event, the frequency of biallelic modification can be predicted by the following equation: biallelic = (day 3% NHEJ)2. By the same assumption, monoallelic modifications (mod) can be predicted by this equation: monoallelic modification = (day 3% NHEJ × 2) × (1 − day 3% NHEJ). Expected % mod clones (both mono- and biallelic) was calculated by summing mono- and biallelic modification proportions derived from the equations above.
*A 95% Confidence interval exceeds expected biallelic-null hypothesis that cleavage and repair are of each chromosome is an independent event.
†Biallelic KO were identified by sequencing of PCR products. Only overlapping or homozygous deletions can be identified using this technique.
‡Fibroblasts were transfected and recovered twice within 2 wk with the same TALEN-pair.
§Five of 15 biallelic colonies were confirmed as double frame-shift alleles.
¶Only colonies with distinguishable gross deletions in the PCR amplicon were analyzed.
We first evaluated cotransfection with Mirus LT1, a commonly used cationic lipid transfection reagent. Whereas gene modification was below detection 14 d posttransfection without selection, genome modification levels in transposon-selected populations were 31%, 13%, and 20% for DMD7.1, DMD6, and LDLR2.1 TALEN-pairs, respectively (Fig. 3). Thus, despite low-transfection efficiency with cationic lipids (<5% for this experiment), efficient modification can be achieved using transposon coselection. We next applied transposon coselection to cells transfected by nucleofection where >90% transfection efficiency is routine. Without selection, the proportion of cells modified after 14 d in culture was 50--90% lower than levels measured at day 3, suggesting an attrition of modified cells. In contrast, transposon coselection was in all cases effective for maintenance of modified cells transfected by nucleofection but, with the exception of ACAN12, did not significantly increase the frequency of modified cells (Fig. 3C). Transposon coselection thus appears to be an effective enrichment method when transfection efficiency is low and an effective maintenance method when transfection efficiency is high.
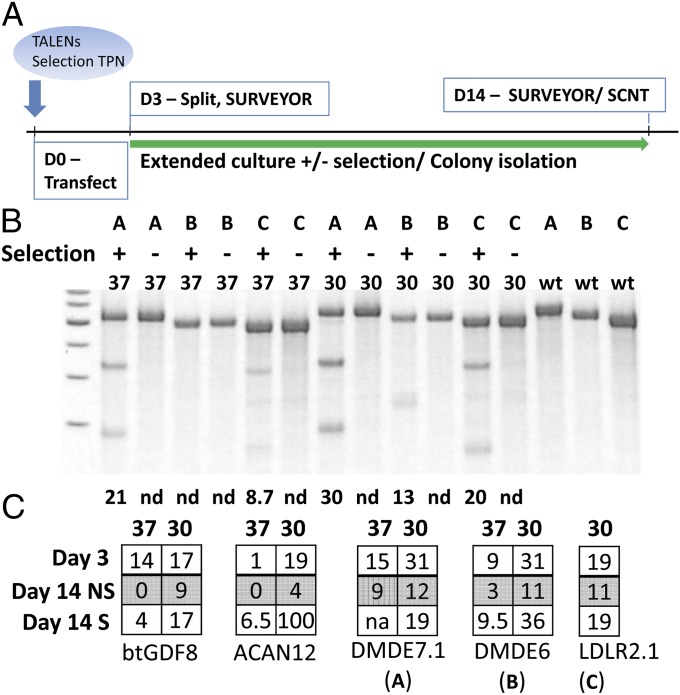
Fig. 3.
Transposon coselection for indel enrichment. (A) The experimental timeline. Day zero (D0), cells were transfected with a mixture of plasmids, including an expression cassette for each TALEN, a transposon encoding a selection marker, and transposase expression cassette. Transfected cells were cultured for 3 d at either 30 or 37 °C before splitting, collection of a sample for Surveyor assay, and replating for extended culture with and without selection for transposon integration. Cells cultured for 14+ d were collected for Surveyor assay. (B) Fibroblasts were transfected using Mirus LT1 reagent and Surveyor assay was performed on day 14 populations. Temperature treatment, selection and TALEN identification (identified by single letters (A, B, and C) as indicated in C are shown above the gel. (C) Fibroblasts were transfected by nucleofection and the percent NHEJ was measured at day 3, and in day 14 nonselected (NS) and selected (S) populations. Temperature treatment is indicated above each matrix. ND, not detected; WT, wild-type amplicon, Surveyor-treated.
Isolation of Mono- and Biallelic KO Clones.
We have previously shown that transgenic primary fibroblast colonies can reliably be isolated and expanded when plated with wild-type fibroblasts and subjected to drug selection using transposon coselection (23). To evaluate this approach, we isolated puromycin-resistant colonies from cells treated with six TALEN-pairs and evaluated their genotypes by the Surveyor assay or direct sequencing of PCR products spanning the target site (Table 1). The proportion of indel-positive clones was similar to our predictions calculated from the percent NHEJ measured in the parent populations 3 d after transfection (Table 1). Biallelic modified clones were identified for five of six TALEN-pairs, occurring in up to 35% of indel-positive cells (Table 1). Notably, among modified clones, the frequency of biallelic modification exceeded predictions based on day 3 modification levels and the assumption that each chromosome cleavage/repair would be an independent event (observed 17–35% vs. predicted 10–16%). Among clones with biallelic modifications, a significant portion (15 of 23, 65%), were homozygous for the same indel, suggesting that sister chromatid exchange may be common (Fig. S2).
Chromosomal Deletions and Inversions with TALENs.
We next investigated whether the simultaneous delivery of two TALEN-pairs targeting the same chromosome could induce either large chromosomal deletions or inversions. We chose TALEN-pairs DMDE6 and DMDE7.1 because a high percentage of Duchene’s Muscular Dystrophy (DMD) is caused by gross deletions (25), providing the opportunity to mimic the human condition in a porcine model. Day 3 gene-modification levels were high for each TALEN-pair (24% for DMDE6 and 23% DMDE7.1), albeit slightly lower than when either TALEN-pair was transfected individually (Fig. 4B). To determine if the sequence between the two TALEN-pairs had been deleted, we designed PCR primers to span this region. If the 6.5-kb sequence had been removed, we expected to see a PCR product of ∼500 bp. A fragment approximating 500 bp was observed in replicates in which both TALEN-pairs were introduced, but was absent when either TALEN-pair was introduced alone (Fig. 4C). We next assayed the cell population for inversion events by PCR amplification across presumptive new 5′- and 3′-junctions. Products were observed at the expected sizes for both the 5′- and 3′-junctions of the predicted inversion, but only when both TALEN-pairs were introduced (Fig. 4D). Both deletion and inversion events were recovered at relatively high frequencies (10.3% and 4.1%, respectively; n > 1,000) (Table S3) in colonies generated using the transposon coselection strategy. Deletion and inversion events occurred with remarkable fidelity, with 41 of 43 (95%) of putative inversion-positive colonies confirmed by PCR amplification of both the 5′- and 3′-junctions. Sequencing of PCR products confirmed both deletion and inversion events with addition or deletion of very few nucleotides at their junctions (Fig. S3).
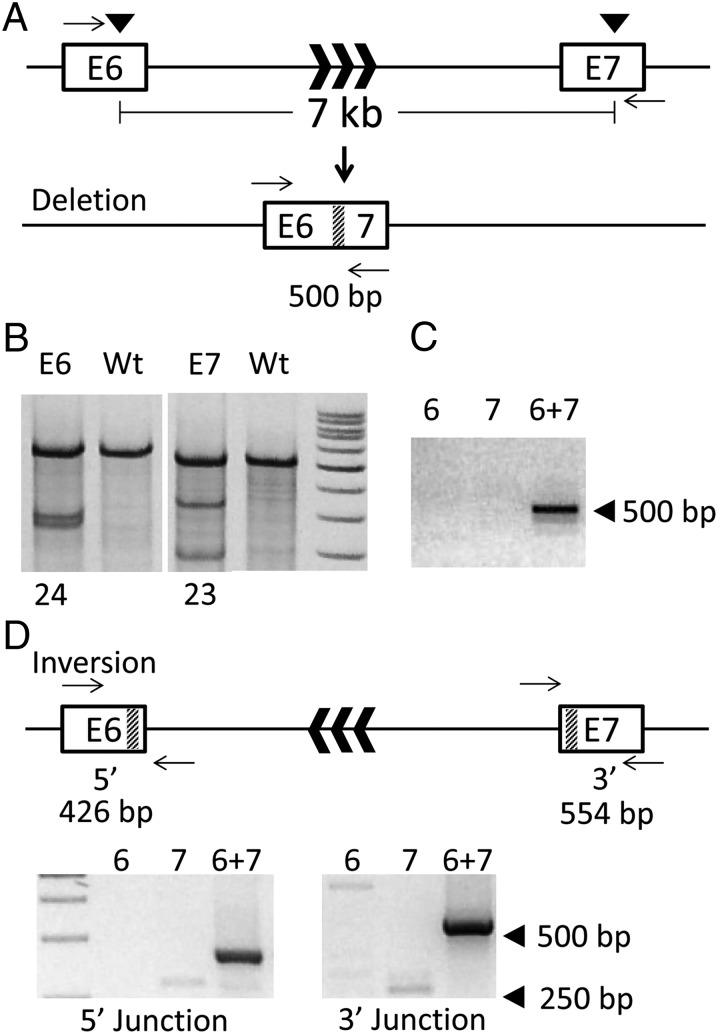
Fig. 4.
TALEN-induced deletions and inversions. (A) DMD locus. Transcriptional orientation is denoted by black chevrons in intron-6. (B) Surveyor assay of cells transfected simultaneously with TALENs targeted to exon 6 and 7 reveal NHEJ indels at both sites. (C) PCR with primers (black arrows) flanking the presumptive deletion yield a ∼500-bp product when both exon-6 and exon-7 TALENs were introduced simultaneously, but not when transfected singly. (D) The predicted outcome of an inversion event of the sequence between the TALEN target sites. Primers flanking the presumptive flanking sites at the 5′- and 3′-ends of the inversion locus are shown (black arrows) along with predicted product size. PCR products were observed at both 5′- and 3′-junctions only when both exon-6 and exon-7 TALENs were introduced simultaneously.
Production of TALEN-Modified Swine by Cloning.
Having demonstrated the ability to efficiently generate knockouts, gross deletions, and inversions in vitro, we next sought to examine the utility of TALEN-modified swine cells for animal production. Transposon coselected Ossabaw swine colonies with mono- and biallelic modification of the class A domain 1 of the LDLR gene were pooled disproportionately (pool A, four genotypes; pool B, three genotypes; pool C, five genotypes) and cloned by chromatin transfer (26). Pregnancy was established in seven of nine transfers (one of two for pool A, two of three for pool B, and four of four for pool C). Six pregnancies were maintained to term, resulting in 18 live-born piglets, including one stillborn from pool A, nine healthy and two stillborn piglets from pool B, and eight healthy, one stillborn, and one euthanized (because of a cloning defect) piglets from pool C (Fig. 5A). At 60+ days 17 piglets remained in good health. In total, four of eight colony genotypes from pools B and C were represented in cloned offspring. Ten of the 11 piglets from pool B carried biallelic modifications of LDLR (B1) (Fig. 5) derived from a colony harboring both an insertion-induced frame-shift (289_290ins34) in one allele and a 3-bp deletion (285_287delATG) in the other allele, the former prematurely truncating the LDLR ORF, the latter removing a universally conserved aspartic acid residue (D47del) implicated in familial hypercholesterolemia (27, 28). A second colony genotype from pool B (B2; 211_292del128) was observed in a stillborn piglet. Two of five colony genotypes were represented in piglets from pool C, including 289_290del10 and 289_290insA, each resulting in frame-shift mutations expected to prematurely truncate the LDLR ORF.
Fig. 5.
Genotypes of LDLR-knockout Ossabaw swine. The genotypes of 22 cloned piglets were evaluated by cloning and sequencing of amplicons spanning the LDLR2.1 target site. Four distinct genotypes were identified and labeled B1 and B2 for pigs derived from the B pool and C1 and C2 for pigs derived from the C pool. The wild-type sequence is shown above and the TALEN recognition sites are indicated in bold. Inserted bases are indicated by underlined text or denoted with an “i” and number of base pairs.
Discussion
Our examination of TALEN activity in livestock embryos produced some intriguing results. First, only one-third of pig zygotes cytoplasmically injected with the TALEN/EGFP mRNA fluoresced at a detectable level. Although this low rate was unexpected, others have reported similar findings by either EGFP mRNA injections of porcine oocytes (29, 30) or cytoplasmic injection of plasmids into bovine zygotes (31). In contrast, all EGFP-injected bovine embryos fluoresced. Pig and cattle injections took place at different locales; thus, we are unable to conclude whether this difference is a result of technical or species-specific differences. Regardless, we speculate that selecting for EGFP+ embryos will be a valuable first-level method for enrichment of TALEN-modified embryos. Second, although we did not directly compare TALEN efficiency based on scaffold alone, the GT-TALENs had greater activity than the +231 scaffold in livestock zygotes. In contrast to +231 injected zygotes, biallelic modification was common (25% of modified embryos) in zygotes injected with GT–TALEN-pairs in both pigs and cattle. Finally, there was a pronounced dosage response using the GT-btGDF83.1 TALENs in terms of activity (43% vs. 75% mutant blastocysts) and development (24% vs. 8% blast formation rate) in low- and high-dosage groups, respectively. However, the fivefold difference between low and high dosage indicates TALENs function over a broad dosage range, where a balance between activity and development could be sought. Thus, we predict that cytoplasmic injection of GT-TALEN mRNA will be an efficient platform for genetic modification of livestock.
Although direct modification of zygotic genomes may have some advantages, somatic cell modification followed by cloning also provides a significant advantage by permitting the isolation of cells containing precise modifications before the expense of animal production. We therefore evaluated the use of TALENs for gene modification in primary fibroblasts, the standard cell-type used for cloning. In agreement with others, we found the truncated TALEN scaffold was superior for gene modification in primary fibroblasts (15, 20). The efficiency of TALEN modification could also be improved by culture at 30 °C for 72 h following transfection (22). Combined use of these enhancement strategies resulted in highly successful TALEN production; 64% of synthesized pairs were active in fibroblasts with a NHEJ frequency from 1.8 to 40%. Assuming a TALEN-pair introduces NHEJ at an average efficiency of 20%, about five heterozygous KOs could be recovered after screening only 24 colonies.
Our transposon coselection strategy (23) in cell populations was extremely successful for enrichment of indel-positive cells when transfection was inefficient, and for maintenance of indel-positive cells when transfection was efficient. When cationic lipid transfection was used, enrichment for modified cells was likely a result of the elimination of nontransfected cells, >95% of total cells. However, coselection was also capable of preventing an observed 50–90% loss of TALEN-modified cells after 14 d postnucleofection. The reason for modified-cell attrition is not clear, although it may be that without selection, cells with a low level of transgene expression have a growth advantage over cells with higher levels of expression. Given that coselection was able to enrich and maintain modified cells, it seems unlikely that nuclease off-target activity or toxicity (20, 32) is the cause for attrition. Instead, selection for antibiotic resistance is likely biased toward high levels of ectopic gene expression, thereby enriching for cells in which TALEN expression was also high, and cleavage more likely to have occurred. Potentially, transposition mobilizes the same factors that are involved with repair after double-stranded DNA cleavage by nucleases. Because transposition occurs in only a low percentage of transfected cells, as well as cells in animal tissues (32–34), coselection might enrich for a subpopulation of cells that were amenable to NHEJ.
By whatever mechanism, transposon coselection was very effective for the clonal isolation of both mono- and biallelic TALEN-modified cells. In fact, an analysis of coselected clones revealed the frequency of biallelic modifications to exceed predictions, assuming each TALEN-induced cleavage/repair was an independent event. This finding has also been observed for cells treated with ZFN’s (11, 35, 36). Furthermore, approximately two-thirds of the biallelic modified clones were homozygous for the same indel, suggesting that gene conversion of TALEN-mediated genetic changes from the sister chromatid is common, a bias previously observed by others (37). The efficiency of producing clones with biallelic KO observed here represents a significant improvement over traditional approaches, for which line breeding or sequential targeting and recloning are required to generate homozygous KO animals (38). An additional advantage of nuclease-mediated biallelic KO is that linked selection markers are not theoretically required. For example, when the expression of the targeted gene results in a distinguishable epitope on the cell surface (9), cells harboring a biallelic KO can be isolated by FACS sorting. Alternatively, cotransfection of cells with a nonintegrating reporter system enabled the isolation of cells displaying evidence of nuclease activity at 3 d posttransfection (39), although inefficient isolation of modified primary cell clones (two of six, both monoallelic indels) suggests low viability among sorted cells. In contrast, transposon coselection allowed efficient isolation of stable cells containing mono- and biallelic indels without FACS or a physically linked selection-marker. These targeted modifications can be easily segregated away from the selection transposons in a single generation of breeding during line propagation (23).
Structural variation in the form of chromosomal deletions, inversions, and copy number accounts for a significant portion of human genetic variation (40). In this study, we found that large deletions and inversions could be generated in fibroblasts by a single cotransfection of two TALEN-pairs that targeted the same chromosome. The efficiency of deletions and inversion in pig fibroblasts was similar to that reported by others using ZFN to generate chromosomal deletions in immortalized human cells (41, 42). However, whereas others found cointroduction of two ZFN pairs often resulted in unintended chromosomal rearrangements in addition to the desired rearrangement (42), we did not observe such events, perhaps because we were targeting the hemizygous DMD locus in male cells. The majority of useful rearrangements will likely occur on autosomes; therefore, founder lines will have to be carefully screened to avoid confounding rearrangements. Chromosomal rearrangements in livestock may also have applications beyond modeling of human disease. Deletions could be useful for functional query of gene clusters or used for deletion mapping of elusive genetic differences identified in association studies. Targeted inversions could also theoretically allow fixation of neighboring trait-determining alleles in a manner analogous to balancer chromosomes commonly used for genetic studies in lower eukaryotes. This process could serve as a means to fix a desired trait or traits in livestock for agricultural purposes or to use nondisjunction to control the spread of genetically modified genomes.
The combination of TALENs plus the transposon components had no apparent impact on the utility of cells for cloning. We achieved a pregnancy rate of 78% with mono- and biallelic modified Ossabaw fibroblasts, a rate similar to our previous results with transposon transgenic Landrace cells (23). This result is especially encouraging considering that the cloning of Ossabaw swine, which are superior models of metabolic syndrome, had not previously been reported. Of the 18 live-born clones, 8 contained monoallelic mutations and 10 contained biallelic modifications of the LDLR gene. These results demonstrate that TALENs can be used to generate animals with either monoallelic or biallelic gene modification by cloning.
TALEN-mediated genome engineering clearly has the capacity to revolutionize genetics and genome engineering in livestock species by introducing a variety of genomic changes, including KO, biallelic KO, large chromosomal deletions/inversions, and potentially, precise allelic introgression. TALENs can be easily designed and assembled using molecular biology techniques available in most laboratories. We anticipate that their ease of use and versatility will rapidly expand the field of livestock genome engineering for a variety of purposes.
Materials and Methods
See SI Materials and Methods for more detail and Tables S4 and S5 for a complete list of TALENs and PCR primers used in this study. All TALENs were designed using the TALE-NT software and assembled using methods described in Cermak et al. (17), with some modifications. Four new backbone plasmids were generated to replace pTAL for the final step in TALEN assembly. The first pair of vectors, pC-TAL+231 and pC-GoldyTALEN, direct expression of the +231 (19) and GoldyTALEN scaffolds from the mini Caggs promoter (43). The second pair of vectors, RCIscript-TAL+231 and RCIscript-GoldyTALEN, are useful for in vitro transcription of TALEN mRNA based on the pT3Ts vector previously described (44). The GoldyTALEN vectors have been made available through Addgene and are fully compatible with the D.F.V. Laboratory Golden Gate TALEN Kit also available through Addgene (#1000000016). Intermediary arrays were created and joined into these vectors, as previously described (17).
TALEN mRNA was synthesized from SacI linearized RCIscript vectors using the mMessage Machine T3 Kit (Ambion), as previously described (44), and injected into the cytoplasm at specified concentrations. Fibroblasts were cultured and transfected using the Basic Fibroblast Nucleofection Kit (Amaxa Biosystems/Lonza) or Mirus LT1 reagent (Mirus), as previously described (44). TALEN activity was analyzed by Surveyor Assay (Transgenomic) and measurements were performed as described in Guschin et al. (45). Indels were detected in embryos or individual colonies by Surveyor assay or direct sequencing. PCR assays described in SI Materials and Methods were developed to detect large deletions and inversions.
Animal Husbandry/Cloning.
Pigs were cloned by CT under contract with Minitube of America under Recombinetics, Inc.s’ Institutional Animal Care and Use Committee Protocol 1103A97232 at the University of Minnesota.
Supplementary Material
Acknowledgments
The authors thank Dr. Mark Westhusin, Gayle Williamson, Jane Pryor, and Ali Wilkerson from Texas A&M University for bovine zygote preparation and injection. This study was funded in part by a Small Business Technology Transfer program Grant 1R41HL108440-01 (to Recombinetics, Inc.); grants to C.B.A.W.; Biotechnology and Biological Sciences Research Council Institute Strategic Programme Grant BBS/E/D/07731442; and support from Genus plc (C.B.A.W.).
Footnotes
Conflict of interest statement: S.C.F. is an executive and shareholder in Recombinetics, Inc., a company focused on the commercialization of transcription activator-like effector nucleases for livestock biomedical and agricultural applications. D.F.C. is an employee and shareholder of Recombinetics, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211446109/-/DCSupplemental.
References
- 1.Aigner B, et al. Transgenic pigs as models for translational biomedical research. J Mol Med (Berl) 2010;88:653–664. doi: 10.1007/s00109-010-0610-9. [DOI] [PubMed] [Google Scholar]
- 2.Rogers CS, et al. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science. 2008;321:1837–1841. doi: 10.1126/science.1163600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abuin A, Bradley A. Recycling selectable markers in mouse embryonic stem cells. Mol Cell Biol. 1996;16:1851–1856. doi: 10.1128/mcb.16.4.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 6.Carlson DF, Fahrenkrug SC, Hackett PB. Targeting DNA With Fingers and TALENs. Mol Ther Nucleic Acids. 2012;1:e3. doi: 10.1038/mtna.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whyte JJ, et al. Gene targeting with zinc finger nucleases to produce cloned eGFP knockout pigs. Mol Reprod Dev. 2011;78:2. doi: 10.1002/mrd.21271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang D, et al. Generation of PPARγ mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning. Cell Res. 2011;21:979–982. doi: 10.1038/cr.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hauschild J, et al. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc Natl Acad Sci USA. 2011;108:12013–12017. doi: 10.1073/pnas.1106422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 12.Cui X, et al. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol. 2011;29:64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- 13.Chen F, et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogdanove AJ, Voytas DF. TAL effectors: Customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 15.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 16.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyson MC, Alloosh M, Vuchetich JP, Mokelke EA, Sturek M. Components of metabolic syndrome and coronary artery disease in female Ossabaw swine fed excess atherogenic diet. Comp Med. 2006;56:35–45. [PubMed] [Google Scholar]
- 19.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mussolino C, et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palgrave CJ, et al. Species-specific variation in RELA underlies differences in NF-κB activity: A potential role in African swine fever pathogenesis. J Virol. 2011;85:6008–6014. doi: 10.1128/JVI.00331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doyon Y, et al. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat Methods. 2010;7:459–460. doi: 10.1038/nmeth.1456. [DOI] [PubMed] [Google Scholar]
- 23.Carlson DF, et al. Strategies for selection marker-free swine transgenesis using the Sleeping Beauty transposon system. Transgenic Res. 2011;20:1125–1137. doi: 10.1007/s11248-010-9481-7. [DOI] [PubMed] [Google Scholar]
- 24.Falanga V, Kirsner RS. Low oxygen stimulates proliferation of fibroblasts seeded as single cells. J Cell Physiol. 1993;154:506–510. doi: 10.1002/jcp.1041540308. [DOI] [PubMed] [Google Scholar]
- 25.Blake DJ, Weir A, Newey SE, Davies KE. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol Rev. 2002;82:291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- 26.Collas P, Robl JM, Sullivan E, Kasinathan P. (2007) Methods for cloning nonhuman mammals using reprogrammed donor chromatin or donor cells US Patent 7,253,334.
- 27.Leigh SE, Foster AH, Whittall RA, Hubbart CS, Humphries SE. Update and analysis of the University College London low density lipoprotein receptor familial hypercholesterolemia database. Ann Hum Genet. 2008;72:485–498. doi: 10.1111/j.1469-1809.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- 28.Leitersdorf E, et al. Deletion in the first cysteine-rich repeat of low density lipoprotein receptor impairs its transport but not lipoprotein binding in fibroblasts from a subject with familial hypercholesterolemia. Proc Natl Acad Sci USA. 1988;85:7912–7916. doi: 10.1073/pnas.85.21.7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishimura Y, Kano K, Naito K. Porcine CPEB1 is involved in Cyclin B translation and meiotic resumption in porcine oocytes. Anim Sci J. 2010;81:444–452. doi: 10.1111/j.1740-0929.2010.00755.x. [DOI] [PubMed] [Google Scholar]
- 30.Ohashi S, et al. Expression of exogenous proteins in porcine maturing oocytes after mRNA injection: Kinetic analysis and oocyte selection using EGFP mRNA. J Reprod Dev. 2001;47:351–357. [Google Scholar]
- 31.Iqbal K, et al. Cytoplasmic injection of circular plasmids allows targeted expression in mammalian embryos. Biotechniques. 2009;47:959–968. doi: 10.2144/000113270. [DOI] [PubMed] [Google Scholar]
- 32.Bell JB, et al. Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat Protoc. 2007;2:3153–3165. doi: 10.1038/nprot.2007.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hackett PB, Ekker SC, Largaespada DA, McIvor RS. Sleeping Beauty transposon-mediated gene therapy for prolonged expression. Adv Genet. 2005;54:189–232. doi: 10.1016/S0065-2660(05)54009-4. [DOI] [PubMed] [Google Scholar]
- 34.Hackett PB, Largaespada DA, Cooper LJ. A transposon and transposase system for human application. Mol Ther. 2010;18:674–683. doi: 10.1038/mt.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santiago Y, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu PQ, et al. Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol Bioeng. 2010;106:97–105. doi: 10.1002/bit.22654. [DOI] [PubMed] [Google Scholar]
- 38.Kuroiwa Y, et al. Sequential targeting of the genes encoding immunoglobulin-mu and prion protein in cattle. Nat Genet. 2004;36:775–780. doi: 10.1038/ng1373. [DOI] [PubMed] [Google Scholar]
- 39.Kim H, et al. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat Methods. 2011;8:941–943. doi: 10.1038/nmeth.1733. [DOI] [PubMed] [Google Scholar]
- 40.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 41.Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20:81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee HJ, Kweon J, Kim E, Kim S, Kim JS. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 2012;22:539–548. doi: 10.1101/gr.129635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark KJ, et al. Enzymatic engineering of the porcine genome with transposons and recombinases. BMC Biotechnol. 2007;7:42. doi: 10.1186/1472-6750-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carlson DF, et al. Efficient mammalian germline transgenesis by cis-enhanced Sleeping Beauty transposition. Transgenic Res. 2011;20:29–45. doi: 10.1007/s11248-010-9386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guschin DY, et al. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.