Abstract
Spatial memory enhances an organism’s navigational ability. Memory typically resides within the brain, but what if an organism has no brain? We show that the brainless slime mold Physarum polycephalum constructs a form of spatial memory by avoiding areas it has previously explored. This mechanism allows the slime mold to solve the U-shaped trap problem—a classic test of autonomous navigational ability commonly used in robotics—requiring the slime mold to reach a chemoattractive goal behind a U-shaped barrier. Drawn into the trap, the organism must rely on other methods than gradient-following to escape and reach the goal. Our data show that spatial memory enhances the organism’s ability to navigate in complex environments. We provide a unique demonstration of a spatial memory system in a nonneuronal organism, supporting the theory that an externalized spatial memory may be the functional precursor to the internal memory of higher organisms.
Keywords: extracellular slime, protist, reactive navigation, amoeboid organism
In simple terms, memory can be defined as the storage and retrieval of information relating to past events (1, 2). The concept of an externalized memory has been applied to the pheromone trails used by many species of ant. By depositing pheromone, individual ants externalize their memory of the route from the nest to a food source. Externalizing their memory not only allows individual ants to efficiently communicate the location of food sources found to their nestmates, but also frees foragers from having to store memories of the route within themselves (3). The network of pheromone trails within the environment is the externalized collective memory of the entire colony (4). The deposition of, and reaction to, chemical markers in the environment was most likely a functional precursor to internal memory, allowing biological systems with primitive information-processing systems to solve tasks requiring spatial memory (5, 6). One such task that would clearly benefit from some form of spatial memory is navigation.
Autonomous artificial systems, such as robots, may use a preconstructed symbolic map to navigate through their environment, or may build their own map from the data they acquire during exploration. This approach is expensive, however, requiring additional on-board memory capacity, supplementary data processing capabilities, or both. “Reactive navigation” is an alternative mechanism that allows effective navigation, requiring the robot to interact only with its immediate environment, and to maintain only a state memory (7–9). However, without some form of memory, goal-oriented autonomous robots using reactive navigation have difficulty navigating toward a goal in complex environments and often become trapped by obstacles (8–10). Providing the robots with a spatial memory relating to the local environment only has proved sufficient for the autonomous units to efficiently solve complex navigational challenges (8).
The slime mold Physarum polycephalum is a unicellular, multinucleate protist that relies on reactive navigation to explore its environment. The vegetative state of P. polycephalum (known as a plasmodium) is composed of many smaller oscillating units. Each unit oscillates at a frequency dependent upon both the local environment and its interactions with neighboring oscillators (11). When the slime mold senses attractants, such as food, via specific binding to receptor molecules presented on the outer membrane surface (12), the oscillation frequency in the area closest to the food increases, causing cytoplasm to flow toward the attractant (13). Additionally, binding of attractant molecules to sections of the surface membrane reduces the tension at that section, leading to a difference in internal hydrostatic pressure, such that cytoplasm flows toward the source of attractants (12). When repellents such as salts and light are detected, oscillation frequency decreases and membrane tension increases (12). The collective behavior of the oscillators, each passing on information to entrain its neighbors, drives the organism’s locomotion.
As it moves, the plasmodium leaves behind a thick mat of nonliving, translucent, extracellular slime (Fig. 1). This extracellular slime is a high molecular weight, polyanionic glycoprotein (14), consisting largely of sulfated galactose polymers (15). As the plasmodium is foraging, we found that it strongly avoids areas that contain extracellular slime. This avoidance behavior is a “choice” because when no previously unexplored territory is available, the slime mold no longer avoids extracellular slime (see further). The slime mold’s behavioral response strongly suggests that it can sense extracellular slime upon contact, and uses its presence as an externalized spatial memory system to recognize and avoid areas it has already explored.
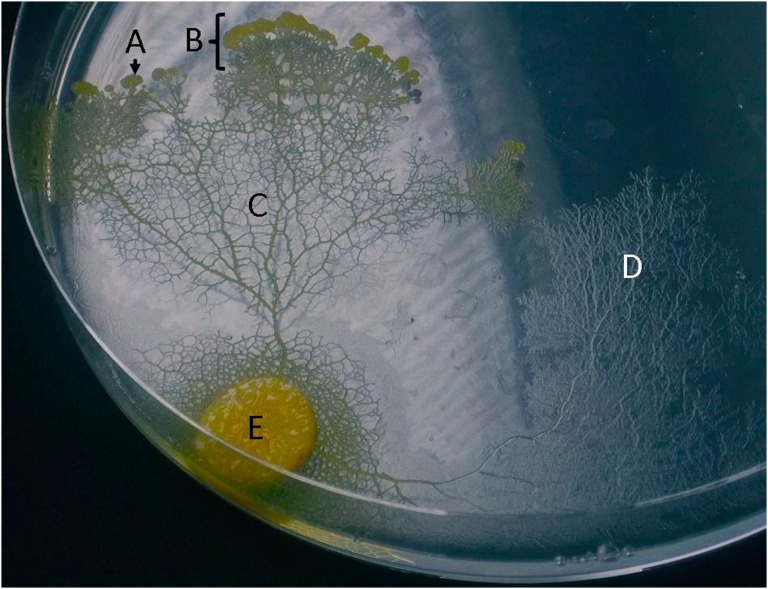
Fig. 1.
Photograph of P. polycephalum plasmodium showing (A) extending pseudopod, (B) search front, (C) tubule network, and (D) extracellular slime deposited where the cell has previously explored. The food disk containing the inoculation of plasmodial culture is depicted at (E).
To examine the importance of memory in spatial navigation in complex environments, we challenged the slime mold with a navigational task common in mobile robot navigation: the U-shaped trap problem. In this problem, the robot requires an external spatial memory to avoid getting trapped in a U-shaped obstacle (8–10). We tested the importance of an external spatial memory in the slime mold by challenging the organism with a U-shaped trap problem when it could use the presence of extracellular slime to navigate and when it could not.
Results
We first determined that the slime mold avoids areas it has visited before by providing P. polycephalum plasmodia with a choice between agar containing extracellular slime and blank agar in an agar Y-maze (Fig. 2). At the terminus of each arm we placed an identical food source. Hence, the only factor in the plasmodia’s decision of which arm to traverse was the presence or absence of extracellular slime. When one arm contained extracellular slime, 39 of 40 plasmodia chose the blank agar arm (P < 0.001, binomial test). We therefore concluded that while the plasmodium is foraging, it strongly avoids areas that contain extracellular slime. When both arms of the Y-maze contained extracellular slime, plasmodia did not show a preference for one arm over the other (P = 0.93, n = 23, split decisions: n = 1, binomial test), indicating that the avoidance response is overridden in the absence of choice. This finding indicates that the avoidance behavior is a choice because when no previously unexplored territory is available, the slime mold no longer avoids extracellular slime. Plasmodia did not show a bias toward either the left or right arm when both arms contained blank agar (P = 0.636, n = 40, binomial test). The slime mold’s behavioral response strongly suggests that it can sense extracellular slime upon contact, and uses its presence as an externalized spatial memory system to recognize and avoid areas it has already explored.
Fig. 2.
Y-mazes testing the extracellular slime avoidance response. A 3-cm2 piece of plasmodial search front was placed onto a 1-cm2 surface of 1% agar. Plasmodia were given a choice of 1% agar arms (Arm A and Arm B) leading to identical 10% wt/vol powdered oat-agar food sources.
We then tested the slime mold’s navigational ability using the U-shaped trap problem. The set-up (Fig. 3) consisted of a Petri dish with an acetate U-shaped trap placed between the plasmodium and a well in the agar containing food: an attractive 2% (wt/vol) glucose solution, the “goal” (16–18). The diffusing glucose solution produced an attraction gradient through the agar, drawing the plasmodium toward the glucose source and into the U-shaped trap. The U-shaped trap, essentially an acetate sheet laid on the agar surface, did not interfere with the diffusion of glucose through the agar, but the dry surface of the acetate prevented the plasmodium from moving over it.
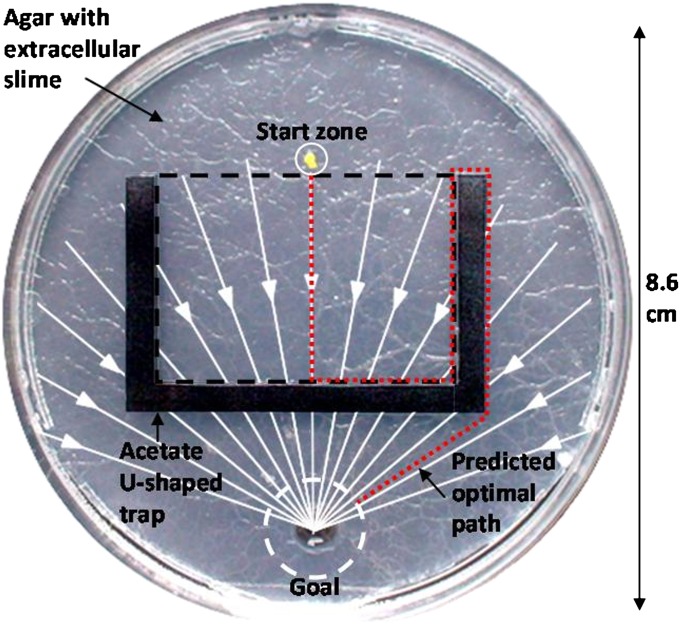
Fig. 3.
Set-up for the U-shaped trap navigational task. The agar surface was either blank or coated in extracellular slime. An attractive 2% (wt/vol) solution of glucose was placed in a well in the agar (the goal). A 1-mm2 piece of plasmodial search front was placed in the start zone and drawn into the bounds (dashed square) of the acetate U-shaped trap by the gradient of diffused glucose solution (white lines/arrows). The red dashed line shows the predicted optimal path; the minimum trajectory of the plasmodium responding to chemotaxis while using its externalized spatial memory system to escape the trap.
We challenged our slime mold to reach the glucose goal on substrates of either blank agar or agar containing fresh extracellular slime. The coating of extracellular slime masked the plasmodium’s own trail, thus interfering with its ability to use the presence of slime as an externalized spatial memory. We therefore expected the slime mold’s ability to navigate in complex environments to be diminished. In particular, we expected plasmodia to take longer to reach the goal on the substrate of extracellular slime, and to spend longer within the bounds of the U-shaped trap, compared with plasmodia on blank agar. We further expected the plasmodia on the extracellular slime-coated substrate to spend more time moving over areas they had explored previously, as their own slime trail was masked by the substrate coating, and to subsequently take an overall longer route from the start zone to the goal.
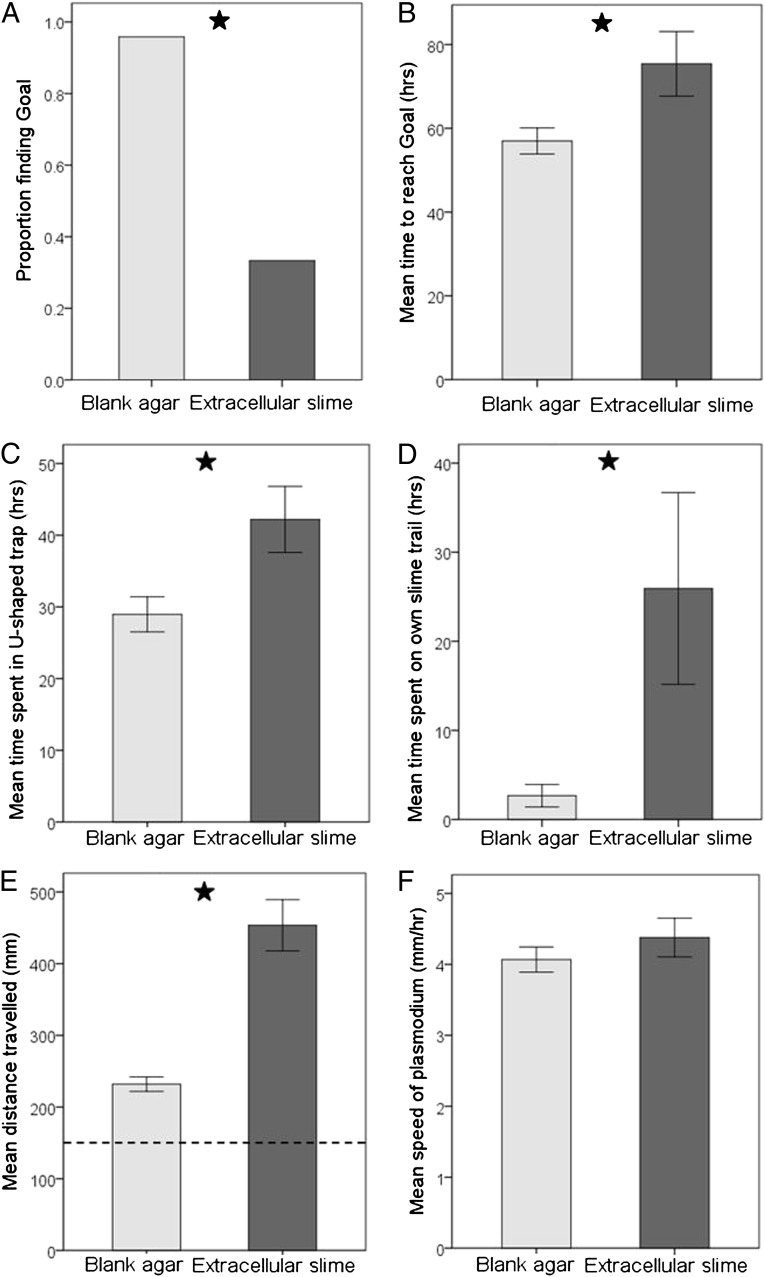
Of the 24 plasmodia run on a substrate of blank agar, 96% reached the goal within the experimental time limit of 120 h (Fig. 4A), but only 33% of 24 plasmodia reached the goal when the agar was coated with extracellular slime [χ2(1,48) = 20.493, P < 0.001, two-sample binomial test]. Of the plasmodia that successfully reached the goal within the allotted time (Fig. 4B), those on blank agar found the goal significantly faster than those on agar coated with extracellular slime [t(29) = −2.667, P = 0.012, independent samples t test, n = 23 for blank agar, n = 8 for extracellular slime]. The U-shaped trap held plasmodia for a significantly longer period when the agar was coated with extracellular slime [t(46) = −2.539, P = 0.016, independent samples t test, n = 24 for both treatments] (Fig. 4C). The average amount of time spent traveling over areas they had previously explored was almost 10-times greater in the extracellular slime-coated treatment than in the blank agar treatment [t(46) = −2.147, P = 0.042, independent samples t test, n = 24 for both treatments] (Fig. 4D). Plasmodia traveled a significantly shorter distance in the blank agar treatment, and were much closer to the predicted optimal path length [t(46) = −5.964, P < 0.001, independent samples t test, n = 24 for both treatments] (Fig. 4E) than in the extracellular slime treatment (see Movies S1 and S2 to view plasmodia negotiating U-shaped traps on blank and extracellular slime-coated agar). Speed differences resulting from the presence of extracellular slime were not responsible for the results, as the mean speed of the plasmodia did not differ between treatments [t(46) = −0.956, P = 0.345, independent samples t test, n = 24 for both treatments] (Fig. 4F).
Fig. 4.
Results for the U-shaped trap navigational task on the treatments of blank agar and agar coated with extracellular slime. (A) Proportion of replicates reaching the goal within the time limit of 120 h. (B) Mean time to reach the goal. (C) Mean time spent within the bounds of the U-shaped trap. (D) Mean time plasmodia spent traveling over their own slime trail. (E) Mean distance traveled by plasmodia (dashed horizontal line indicates the length of the predicted optimal path). (F) Mean speed of plasmodia. Error bars are ± SEM. Stars indicate significant difference between treatments (α = 0.05).
To test if the coating of extracellular slime interfered with the perception of glucose food cues by the plasmodia, we used the same set-up but without the trap (n = 13 for blank agar, n = 16 for extracellular slime). We found no evidence for reduced glucose cue perception because of the presence of the extracellular slime coat [time taken to reach the target: t(27) = −0.904, P = 0.374, independent samples t test; total distance traveled: t(27) = −0.732, P = 0.471, independent samples t test; speed: t(27) = 0.141, P = 0.889, independent samples t test]. Only one replicate in each treatment failed to reach the target in the time limit of 120 h.
Discussion
We predicted that the loss of externalized spatial memory would hinder the slime mold’s ability to escape the U-shaped trap. Indeed, when the agar substrate was covered in extracellular slime, the plasmodia took much longer to reach the goal compared with plasmodia that could use the presence of extracellular slime to determine where they had been before. We also showed that in simple environments—those without U-shaped traps—the use of externalized spatial memory is not necessary for effective navigation. In complex environments, however, the use of an externalized spatial memory system greatly enhances navigational ability.
Nakagaki et al. have shown previously that P. polycephalum is capable of solving labyrinthine mazes (19, 20) and other shortest-path problems (21–23). When solving a maze or connecting several food sources using the most efficient network, the slime mold first explores its entire environment, with cytoplasm simultaneously covering all exploration space, before retracting cytoplasm from areas that do not contain food. The result is the construction of a single tubule when connecting two food sources only (19, 20), or an efficient tubule network between food-source nodes (22, 23). Although such problems appear more complex than the U-shaped trap used in the present study, the organism essentially first constructs a map of its environment before constructing a solution. In essence, maze solving and network construction are analogous to programming an autonomous robot with a symbolic map of its environment. In contrast, our work has shown that in the absence of global information about its environment, the slime mold uses the presence of extracellular slime to navigate through a complex environment simply by avoiding areas it has visited before.
Spatial navigation has been studied extensively in central place foragers, animals that continually return with their food to a fixed location. The need to return to the nest or den has led to selection for many sophisticated navigation mechanisms. The desert ant Cataglyphis, while traveling out from its nest to forage, continuously calculates distance traveled by counting its steps and keeps track of angles turned, integrating this information into a constantly updated vector that will lead the ant home via the shortest path (24, 25). Many insects, including species of ants, bees, and wasps, use landmarks to memorize their route to and from the nest (26, 27). It is thus not surprising that insects are a popular model system to study navigation, providing insights into how simple mechanisms can produce robust and seemingly complex behavior (28).
Results obtained from insect studies challenge the prevailing paradigm that navigation requires learning or otherwise sophisticated high-level spatial modules (28). We go a step further by showing that even an organism without a (central) nervous system can effectively navigate complex environments. When foraging for immobile resources, an organism’s search efficiency is expected to increase with its ability to avoid areas that have previously been explored (29, 30). Slime molds achieve this with a simple behavioral mechanism: preferentially avoiding areas that contain extracellular slime from recent exploration.
Our study shows, in a unicellular organism, how an externalized spatial memory increases navigational ability in a way similar to that often implemented in autonomous mobile robots (10, 31, 32). Although many advances in the development of autonomous agents are based on biological phenomena (7, 33–35), our study was inspired by the way reactive robots use feedback from their immediate environment to avoid becoming trapped. Our study is uinque in providing empirical evidence of a spatial memory system in a nonneuronal, reactive organism, lending strong support to the theory that feedback from chemicals deposited in the environment was the first step toward the evolution of memory in organisms with more sophisticated neurological capabilities than our slime mold.
Materials and Methods
Biological Material.
P. polycephalum plasmodia were maintained in the dark at 22 °C on large 1% agar plates embedded with 10% (wt/vol) rolled oat flakes (Coles Smart Buy Rolled Oats). Original cultures were obtained from Southern Biological Supplies, and laboratory stocks were subcultured onto new agar-oat plates daily.
Experimental Procedure.
Avoidance response.
We gave P. polycephalum plasmodia a choice between agar containing extracellular slime and blank agar in a Y-maze (Fig. 2). Each arm was 4 cm in length. At the terminus of each arm we placed a food source of 10% (wt/vol) powdered oat-agar mix. Y-mazes were constructed from 1% agar. The base of the Y-maze consisted of a 1-cm2 agar block containing 3 cm2 of plasmodial search front. To obtain extracellular slime we allowed a culture of slime mold to migrate (8–12 h) across a 1% agar surface, leaving behind it a trail of extracellular slime. The culture used to generate the extracellular slime was the same laboratory stock as the plasmodium to be tested.
As soon as the slime mold touched a food source at one of the arms, we considered this arm chosen. When the plasmodium reached both food sources at the same time, we called the decision split and excluded the replicate from analysis. Which arm contained the extracellular slime was alternated between replicates.
Externalized spatial memory navigation task.
To test the slime mold’s navigational ability, we used a U-shaped trap (Fig. 3). We filled Petri dishes (30 cm in diameter) with 1% agar and set half of these aside for use as a blank agar treatment. We inoculated the remaining half with a small amount (3–4 mm2) of P. polycephalum. These plasmodia were allowed to explore the dish, depositing extracellular slime for 48 h. After 48 h we cut an 8.6-cm diameter circle of agar coated in extracellular slime (no plasmodial biomass) and placed into an 8.6-cm diameter Petri dish. We did the same using the blank agar to create our control dishes. We then placed an acetate U-shaped trap, measuring 5.2 cm at the base, with vertical arms measuring 3.5 cm, in the center of the Petri dish and added a 1-mm2 piece of plasmodial search front in the start zone indicated in Fig. 3. We filled a 0.5-cm-diameter well in the agar with 50 μL of 2% (wt/vol ) glucose solution just before adding the plasmodium. The well filled with glucose was the slime mold’s goal.
We used the following metrics to compare the slime mold’s ability to avoid becoming trapped in the U-shaped trap in the absence and presence of extracellular slime: (i) Time taken to reach the goal from when the plasmodium first formed a search front. (ii) Time spent inside the bounds of the U-shaped trap from when the leading plasmodial pseudopod first entered the trap boundary, and ending when it exited the boundary. Where the plasmodium re-entered the trap, total time spent inside the trap was summed. (iii) Amount of time the plasmodium spent on agar containing its own slime trail from when the leading pseudopod first moved over the trail until it left the trail. Where the plasmodium moved over its slime trail multiple times, all time spent on the slime trail was summed. (iv) Total distance traveled, measured by tracing the path of the leading edge of the cell in ImageJ (36) v1.45I. (v) The average speed of the plasmodium, calculated using the total distance traveled and the time taken to reach the goal. Where the plasmodium did not reach the goal after 120 h, we calculated the mean speed by dividing the total distance traveled by the full experimental time of 120 h. We used t test for independent samples to compare our metrics between plasmodia moving on blank agar and those moving on agar containing extracellular slime. We used a two-sample binomial test to compare the proportion of plasmodia that reached the goal between the two treatments. Statistical analyses were performed using IBM SPSS Statistics v19.
Photographs were taken every 5 min with a Sony HDR-HC5 Handycam for 120 h. The camera was mounted 30 cm above the experiments inside a custom built darkbox containing 200 mL of water. This set-up was designed to reduce ambient light and increase humidity in the microenvironment surrounding the slime mold while allowing flash photography from above.
Supplementary Material
Acknowledgments
We thank members of the Behaviour and Genetics of Social Insects Laboratory. This work was supported by the Human Frontier Science Program (M.B.), the Australian Research Council (M.B. and T.L.), the Natural Sciences and Engineering Council of Canada (T.L.), Centre National de la Recherche Scientifique (A.D.), the Fyssen Foundation (A.D.), and the University of Sydney Centre for Mathematical Biology (C.R.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215037109/-/DCSupplemental.
References
- 1.Kilian AE, Muller BS. 2002. Life-like learning in technical artefacts: Biochemical vs. neuronal mechanisms. Proceedings of the Ninth International Conference on Neural Information Processing: Computational Intelligence for the E-Age, eds Wang L, Rajapakse JC, Fukushima K, Lee SY, Yao X (IEEE), pp 296–300, 10.1109/ICONIP.2002.1202181.
- 2.Sweatt JD. Mechanisms of Memory. Sydney: Academic; 2009. [Google Scholar]
- 3.Jackson DE, Martin SJ, Holcombe M, Ratnieks FLW. Longevity and detection of persistent foraging trails in Pharaoh's ants, Monomorium pharaonis (L.) Anim Behav. 2006;71:351–359. [Google Scholar]
- 4.Couzin ID. Collective cognition in animal groups. Trends Cogn Sci. 2009;13:36–43. doi: 10.1016/j.tics.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Chung JR, Choe Y. 2009. Emergence of memory-like behavior in reactive agents using external markers. IEEE 21st International Conference on Tools with Artificial Intelligence (IEEE), pp 404–408, 10.1109/ICTAI.2009.116.
- 6.Chung JR, Kwon J, Choe Y. 2009. Evolution of recollection and prediction in neural networks. IEEE International Joint Conference on Neural Networks, (1–6):3363–3369. Available at http://paws.kettering.edu/∼jkwon/publications/files/chung.ijcnn09.pdf.
- 7.Chatterjee R, Matsuno F. Use of single side reflex for autonomous navigation of mobile robots in unknown environments. Robot Auton Syst. 2001;35:77–96. [Google Scholar]
- 8.Balch T, Arkin R. 1993. Avoiding the past—a simple but effective strategy for reactive navigation. Proceedings: IEEE International Conference on Robotics and Automation, (1–3):678–685. Available at ftp://ftp.cc.gatech.edu/pub/people/arkin/archive/00292057.pdf.
- 9.Luh G-C, Liu W-W. An immunological approach to mobile robot reactive navigation. Appl Soft Comput. 2008;8:30–45. [Google Scholar]
- 10.Zou X-Y, Zhu J. Virtual local target method for avoiding local minimum in potential field based robot navigation. J Zhejiang Univ Sci. 2003;4:264–269. doi: 10.1631/jzus.2003.0264. [DOI] [PubMed] [Google Scholar]
- 11.Durham ACH, Ridgway EB. Control of chemotaxis in Physarum polycephalum. J Cell Biol. 1976;69:218–223. doi: 10.1083/jcb.69.1.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueda T, Hirose T, Kobatake Y. Membrane biophysics of chemoreception and taxis in the plasmodium of Physarum polycephalum. Biophys Chem. 1980;11:461–473. doi: 10.1016/0301-4622(80)87023-2. [DOI] [PubMed] [Google Scholar]
- 13.Latty T, Beekman M. Irrational decision-making in an amoeboid organism: Transitivity and context-dependent preferences. Proc Biol Sci. 2011;278(1703):307–312. doi: 10.1098/rspb.2010.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henney HRJ, Jr, Asgari M. The function of slime from Physarum flavicomum in the control of cell division. Can J Microbiol. 1975;21:1866–1876. doi: 10.1139/m75-270. [DOI] [PubMed] [Google Scholar]
- 15.McCormick JJ, Blomquist JC, Rusch HP. Isolation and characterization of an extracellular polysaccharide from Physarum polycephalum. J Bacteriol. 1970;104:1110–1118. doi: 10.1128/jb.104.3.1110-1118.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kincaid RL, Mansour TE. Measurement of chemotaxis in the slime mold Physarum polycephalum. Exp Cell Res. 1978;116:365–375. doi: 10.1016/0014-4827(78)90460-3. [DOI] [PubMed] [Google Scholar]
- 17.Kincaid RL, Mansour TE. Chemotaxis toward carbohydrates and amino acids in Physarum polycephalum. Exp Cell Res. 1978;116:377–385. doi: 10.1016/0014-4827(78)90461-5. [DOI] [PubMed] [Google Scholar]
- 18.Knowles DJC, Carlile MJ. The chemotactic response of plasmodia of the myxomycete Physarum polycephalum to sugars and related compounds. J Gen Microbiol. 1978;108:17–25. doi: 10.1099/00221287-108-1-17. [DOI] [PubMed] [Google Scholar]
- 19.Nakagaki T, Yamada H, Tóth A. Maze-solving by an amoeboid organism. Nature. 2000;407:470. doi: 10.1038/35035159. [DOI] [PubMed] [Google Scholar]
- 20.Nakagaki T, Yamada H, Tóth A. Path finding by tube morphogenesis in an amoeboid organism. Biophys Chem. 2001;92:47–52. doi: 10.1016/s0301-4622(01)00179-x. [DOI] [PubMed] [Google Scholar]
- 21.Nakagaki T, et al. Minimum-risk path finding by an adaptive amoebal network. Phys Rev Lett. 2007;99:068104. doi: 10.1103/PhysRevLett.99.068104. [DOI] [PubMed] [Google Scholar]
- 22.Nakagaki T, Kobayashi R, Nishiura Y, Ueda T. Obtaining multiple separate food sources: Behavioural intelligence in the Physarum plasmodium. Proc Biol Sci. 2004;271:2305–2310. doi: 10.1098/rspb.2004.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakagaki T, Yamada H, Hara M. Smart network solutions in an amoeboid organism. Biophys Chem. 2004;107:1–5. doi: 10.1016/S0301-4622(03)00189-3. [DOI] [PubMed] [Google Scholar]
- 24.Wehner R, Michel B, Antonsen P. Visual navigation in insects: Coupling of egocentric and geocentric information. J Exp Biol. 1996;199:129–140. doi: 10.1242/jeb.199.1.129. [DOI] [PubMed] [Google Scholar]
- 25.Wittlinger M, Wehner R, Wolf H. The ant odometer: Stepping on stilts and stumps. Science. 2006;312:1965–1967. doi: 10.1126/science.1126912. [DOI] [PubMed] [Google Scholar]
- 26.Dyer FC. Spatial memory and navigation by honeybees on the scale of the foraging range. J Exp Biol. 1996;199:147–154. doi: 10.1242/jeb.199.1.147. [DOI] [PubMed] [Google Scholar]
- 27.Zeil J. Orientation flights of solitary wasps (Cerceris, Sphecidae, Hymenoptera) 1. Description of flight. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1993;172:189–205. [Google Scholar]
- 28.Wystrach A, Graham P. What can we learn from studies of insect navigation? Anim Behav. 2012;84:13–20. [Google Scholar]
- 29.Bernstein C, Driessen G. Patch-marking and optimal search patterns in the parasitoid Venturia canescens. J Anim Ecol. 1996;65:211–219. [Google Scholar]
- 30.Armstrong DP, Gass CL, Sutherland GD. Should foragers remember where they've been? Explorations of a simulation model based on the behavior and energetics of territorial hummingbirds. In: Kamil AC, Krebs JR, Pulliam HR, editors. Foraging Behavior. New York: Plenum; 1987. [Google Scholar]
- 31.Arkin RC, Diaz J. 2002. Line-of-sight constrained exploration for reactive multiagent robotic teams. IEEE Seventh International Workshop on Advanced Motion Control (IEEE), pp 455–461, 10.1109/AMC.2002.1026963.
- 32.Arkin RC, Mackenzie D. Temporal coordination of perceptual algorithms for mobile robot navigation. IEEE T Robotic Auton. 1994;10:276–286. [Google Scholar]
- 33.Arai T, Pagello E, Parker LE. Guest editorial—Advances in multirobot systems. IEEE T Robotic Auton. 2002;18:655–661. [Google Scholar]
- 34.Trianni V, Dorigo M. Self-organisation and communication in groups of simulated and physical robots. Biol Cybern. 2006;95:213–231. doi: 10.1007/s00422-006-0080-x. [DOI] [PubMed] [Google Scholar]
- 35.Quinn RD, Nelson GM, Bachmann RJ, Ritzmann RE. Toward mission capable legged robots through biological inspiration. Auton Robots. 2001;11:215–220. [Google Scholar]
- 36.Rasband WS. 1997–2011. ImageJ (National Institutes of Health, Bethesda, Maryland). Available at http://imagej.nih.gov/ij/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.