Abstract
Single progenitors can give rise to any and all of the main retinal cell types: photoreceptors, interneurons (horizontal, bipolar, and amacrine cells), retinal ganglion cells (RGCs), and glia. Many of these types are divisible into multiple functionally, structurally, and molecularly distinct subtypes (e.g., ∼25 for RGCs). It remains unknown when and how progenitors become committed to generate such subtypes. Here, we determine the origin of RGCs that respond selectively to vertical motion and express cadherin 6 (cdh6). Using Cre recombinase-based lineage tracing, we show that these RGCs arise from progenitors that themselves express cdh6. These progenitors are capable of generating all major retinal cell types, but the RGCs they generate are predominantly of the single direction-selective subtype. In contrast, cdh6-positive progenitors retain the ability to generate multiple subtypes of amacrine and bipolar cells. Our results demonstrate that type and subtype specification are regulated in different ways and suggest that multipotential but fate-restricted progenitors contribute to subtype specification in retina.
A key question in developmental neuroscience is how neuroepithelial cells give rise to the hundreds of neuronal and glial cell types that comprise the adult nervous system. To what extent are progenitors initially uniform and multipotential? At what points and by what mechanisms do their fates become restricted and their progeny become specified? Are all cell fate choices determined by transcriptional codes and extrinsic factors, or are some stochastic? The retina has been widely used to study these issues (1–5).
Retinal cell types are born (i.e., become irreversibly postmitotic) in distinct albeit overlapping intervals, beginning with retinal ganglion cells (RGCs), followed by cone photoreceptors, horizontal cells, and amacrine cells, and ending with bipolar cells, rod photoreceptors, and Müller glia (1, 4). Pioneering studies in rodents and Xenopus demonstrated that single early progenitors give rise to clones that can contain any of these cell types (6–8). At later stages, progenitors appear restricted in that they give rise to clones containing only later-born retinal cell types (9, 10). Heterochronic transplantation and coculture studies indicate that the restrictions are largely intrinsic: progenitors placed in a novel environment give rise to the same cell types they would have generated if left undisturbed, rather than to those being generated by their new neighbors (4, 11–13). These and other observations led to the idea that progenitors go through a series of “competence states” in which they lose and gain abilities to generate particular cell types. The changing competence of progenitors, along with changes in their environment, act together to generate the full set of retinal cells as development proceeds (3, 4). This influential “competence model” may also apply to other regions of the central nervous system and to invertebrates (14–16).
Subsequent to the initial lineage studies, several groups used molecular methods to show that retinal progenitors are heterogeneous. For example, subsets of retinal progenitors express transcription factors such as basic helix loop helix proteins Atoh1 (Math1), Atoh7 (Math5), Olig2, AsclI (Mash1), or Ngn2 (17–20). Marking and lineage-tracing methods have been used to show that some of these subsets are restricted in their competence; for example, AsclI-positive progenitors generate few RGCs, whereas Olig2-positive embryonic progenitors give rise preferentially to cones and horizontal cells (17, 19). In addition, transcriptional profiling has revealed substantial heterogeneity among progenitors (21). These findings augment but do not fundamentally alter the competence model, in that the restricted progenitors identified appear to be “intermediate progenitors” derived from progenitors that can give rise to all retinal cell types. The properties of the earliest “totipotential” progenitors and the extent of their heterogeneity remain undetermined.
It is now apparent that most major retinal cell types can be divided into multiple subtypes (e.g., >30 amacrine and ∼25 RGC subtypes) (5). Most studies of retinal lineage were performed before markers were available for subtypes, so little is known about their origin. Here, we reexamined this issue with respect to RGCs. We recently generated several mouse lines that allow marking of specific RGC subsets (22–24). Among them are two that also permit Cre recombinase-based lineage tracing. One marks ON-OFF RGCs that express the adhesion molecule cadherin 6 (cdh6) and respond selectively to either upward or downward motion (22). The other marks OFF RGCs that express junctional adhesion molecule B (JAM-B) and respond selectively to upward motion (24). Using these lines, we show that both Cdh6- and JAM-B-positive RGCs are specified early in development. Moreover, Cdh6-positive RGCs arise from progenitors that themselves express cdh6. These progenitors are capable of generating all major retinal cell classes, but the RGCs they generate are predominantly the direction-selective RGCs that express cdh6 in adulthood. These results provide insight into both the specification of retinal subtypes and the properties of the earliest progenitors that give rise to the neural retina.
Results
Multiple RGC Subtypes Are Born During the Same Interval.
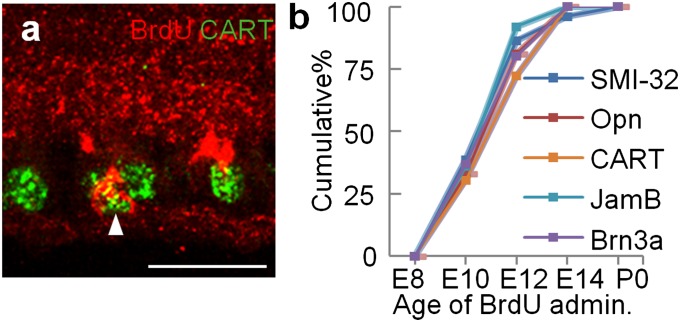
We began by asking whether RGC subtypes arise at distinct times, which would be expected if the competence model (3) applied to them. To this end, we used markers that label three nonoverlapping subsets of RGCs: ON-OFF direction-selective RGCs (ooDSGCs) express the neuropeptide CART (22), OFF direction-selective RGCs express the transmembrane adhesion molecule JAMB (24), and large α-type RGCs are rich in nonphosphorylated neurofilament heavy chains (SMI32) (25). We used antibodies to label the ooDSGCs and α-RGCs and a transgenic line described below to label the OFF direction-selective cells. We also used anti-osteopontin to label a set of large RGCs that partially overlaps the dephosphorylated neurofilament-positive group (26) and Brn3a to label RGCs broadly (∼80% of RGCs, comprising numerous subclasses; 27). Bromodeoxyuridine (BrdU) was administered to pregnant females at embryonic days (E) 8, 10, 12, 14, or 17 to label cells undergoing their final division; at postnatal day (P) 14, after all cells had acquired their type-specific markers, we determined the fraction of BrdU-positive RGCs in each class. All RGC subtypes assayed were born in the same interval, which corresponded to the interval determined with a marker of most RGCs (Brn3a) (Fig. 1). Thus, birth date does not predict RGC subtype identity.
Fig. 1.
Birthdays of RGC subsets. (A) Double labeling for a subtype marker (CART, red) and BrdU (green) allows birth dating of double-labeled cells (arrowhead). (B) Fraction of RGC subtypes born by a given age, shown as a cumulative plot. Counts are from ≥3 mice, ≥16 retina sections for each cell type at each age. (Scale bar, 30 μm.)
ON-OFF Direction-Selective RGCs Are Specified in Embryos.
Next, we asked whether subtypes might be specified late in retinal development, perhaps as a result of synaptic interactions. To this end, we focused on RGCs that express cdh6. This gene is selectively expressed by a subset of ooDSGCs comprising ∼8% of all RGCs (22). ooDSGCs can be recognized by their bistratified dendritic arbors that costratify with cholinergic starburst amacrine cells in two sublaminae of the inner plexiform layer (22) and, as noted above, by their expression of CART. No other bistratified RGCs have been described that costratify with starburst amacrines (28), and all CART-positive RGCs are ooDSGCs (22).
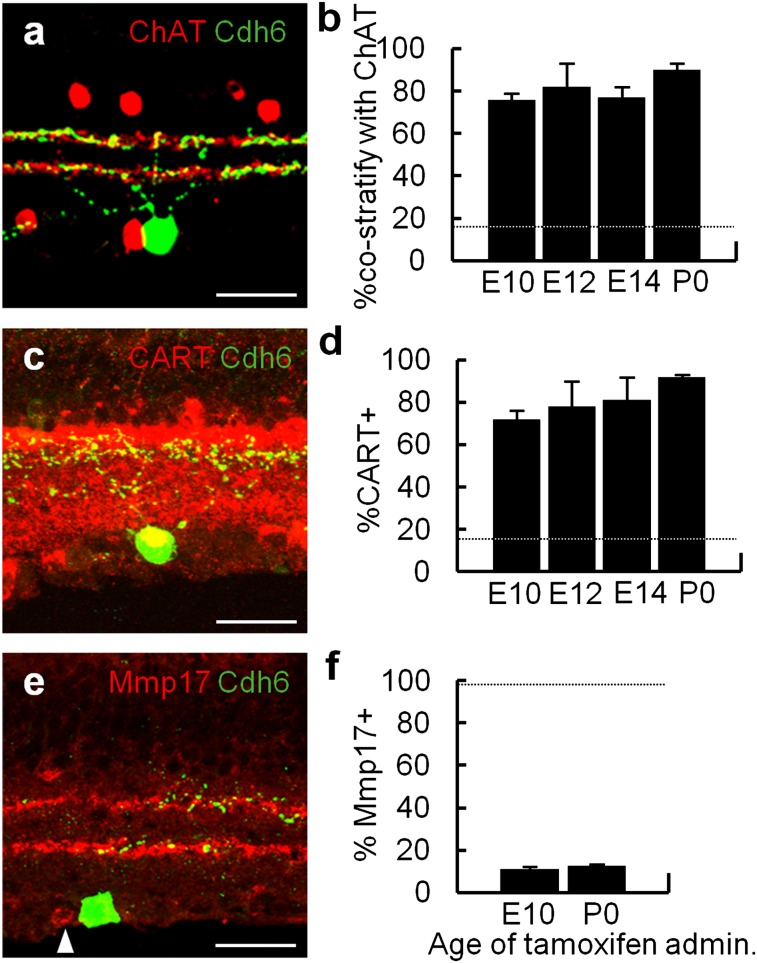
To ask when cdh6-expressing ooDSGCs become specified, we used a mouse line in which tamoxifen-activated Cre recombinase (CreER) was inserted into the cdh6 locus (22). Administration of tamoxifen to Cdh6-CreER mice bearing a Cre-dependent reporter (STOP-YFP; 29) indelibly marks cells expressing CreER within ∼1 d following administration (30). When tamoxifen is delivered to neonates, and mice are examined >3 wk later, >95% of labeled RGCs are ooDSGCs (22). We administered tamoxifen at E10 or E12, during the peak of RGC birth, or at E14, when most RGCs have been born (Fig. 1). In all cases, ∼80% of labeled RGCs were ooDSGCs as judged by either of two criteria: bistratified dendrites that costratified with processes of starburst amacrine cells (Fig. 2 A and B) and expression of CART (Fig. 2 C and D). Thus, ooDSGCs are specified at, or shortly after, their birth, before they receive or form synapses, and before most other retinal cell types are generated.
Fig. 2.
Early specification of Cdh6-RGCs. Cells were labeled by administration of tamoxifen to Cdh6-CreER;STOP-YFP mice at indicated ages, then retinas were analyzed at P20–60. (A and B) Fraction of YFP-positive RGCs that were ooDSGCs as judged by costratification with starburst amacrine cells (ChAT, red in A). (C and D) Fraction of YFP-positive RGCs that were ooDSGCs as judged by coexpression of CART (red in C). (E and F) Fraction of YFP-positive RGCs that prefer posterior motion (22), as judged by coexpression of MMP17 (red in E). Arrowhead in E indicates an MMP17-positive cell. Dotted lines in B and D indicate fraction of all RGCs that are ooDSGCs. Dotted line in F indicates fraction of posterior motion-preferring ooDSGCs that express MMP17. Each bar represents counts from ≥2 mice, ≥16 retina sections. Error bars represent SDs. (Scale bars, 30 μm.)
Although most cdh6-positive RGCs are ooDSGCs, only ∼50% of ooDSGCs are cdh6-positive (22). Physiological studies have revealed the existence of four ooDSGC subsets, responsive to upward, downward, nasal, and temporal motion (31). These subsets are morphologically similar but molecularly distinct: cdh6 is expressed primarily by ooDSGCs selective for vertical motion, whereas ooDSGCs sensitive to temporal motion express MMP17 (22). We therefore used MMP17 to determine whether cdh6 is initially expressed by ooDSGCs that eventually acquire sensitivity to all or only to some directions of motion. Fewer than 10% of YFP-positive RGCs labeled by administration of tamoxifen at either E10 or P0 were MMP17-positive (Fig. 2 E and F), indicating that not only the morphology of RGCs but also their precise physiological properties are specified early in development.
J-RGCs Are Specified in Embryos.
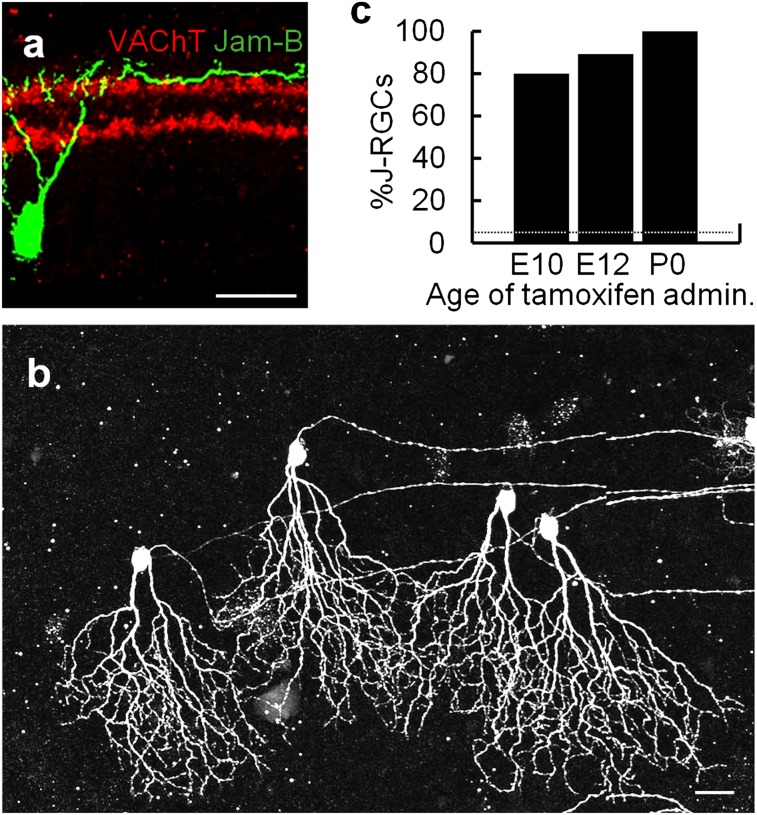
To ask whether early specification is a peculiar feature of cdh6-positive ooDSGCs, we repeated these experiments targeting RGCs that express JAM-B. This gene marks a small subset of RGCs called J-RGCs (∼4% of all RGCs) that can be identified by markedly asymmetric dendritic arbors confined to the outermost edge of the inner plexiform layer (24). We previously generated a JAM-B-CreER line that marks J-RGCs (24). We crossed this line to the STOP-YFP Cre-dependent reporter described above and administered tamoxifen to pregnant females at E10 or E12. More than 70% of labeled RGCs were J-RGCs following administration of tamoxifen to JAM-B-CreER;STOP-YFP mice at E10 or E12, judged either by sublaminar stratification of dendrites viewed in sections or asymmetric dendritic arbors viewed in whole mounts (Fig. 3). This is ∼20-fold more than would be expected by chance. Thus, early subtype specification may be a general feature of RGC development.
Fig. 3.
Early specification of J-RGCs. Cells were labeled by administration of tamoxifen to JAM-B-CreER;STOP-YFP mice at indicated ages, then RGCs were categorized at P20-60. (A) Section showing RGC labeled by tamoxifen at E12 with dendrites arborizing in the innermost sublmina, S1. Starburst amacrines, labeled with anti-ChAT, mark borders of S1/S2 and S3/S4. (B) Whole mount showing RGC labeled by tamoxifen at E12 with asymmetric dendrites pointing ventrally. (C) Fraction of YFP-positive RGCs that were J-RGCs as judged by dendritic stratification and asymmetry; data obtained from sections and whole mounts is combined. Each bar represents counts from ≥2 mice, ≥16 retina sections, and ≥2 whole mounts. [Scale bars, 100 μm (A) and 25 μm (B).]
Direction-Selective cdh6-Positive RGCs Arise from Committed Progenitors.
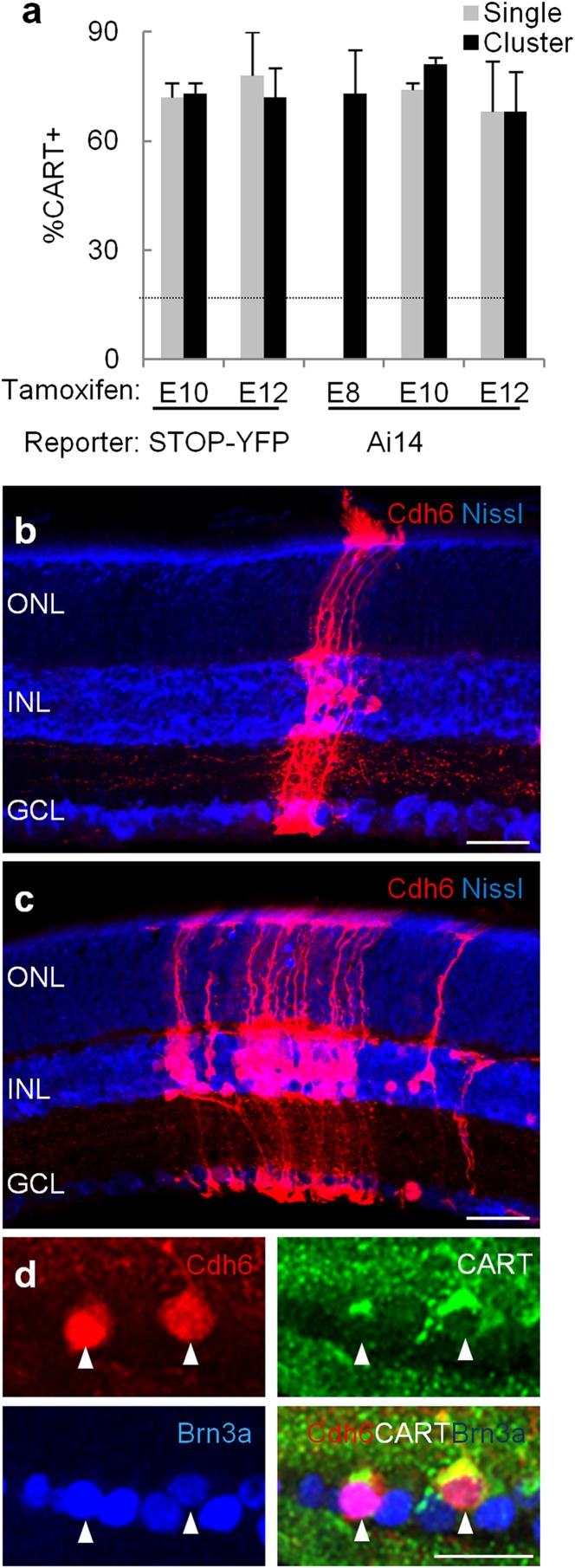
In Cdh6-CreER;STOP-YFP mice that had been administered tamoxifen at E10 or E12 (but not in JAM-B-CreER;STOP-YFP mice), we observed that some labeled RGCs were closely associated with labeled interneurons (Fig. S1). Staining with CART demonstrated that >70% of these RGCs were ooDSGCs (Fig. 4A). Because the STOP-YFP line is unable to mark photoreceptors, Müller glia, and many interneurons, we repeated the analysis using another Cre-dependent reporter called Ai14 that is capable of marking all retinal cell types (32). Administration of tamoxifen to Cdh6-CreER;Ai14 mice at E10 or E12 led to labeling of radial clusters that spanned the entire retina and contained photoreceptors (cells in the outer nuclear layer) and interneurons (cells in the inner nuclear layer) as well as cells in the ganglion cell layer (Fig. 4B). More than 70% of the RGCs in these clusters (identified by immunostaining with the RGC marker Brn3a) were positive for CART, which is expressed by ooDSGCs. Thus, RGCs in labeled clusters are predominantly ooDSGCs.
Fig. 4.
RGCs generated by cdh6-positive progenitors are ooDSGCs. (A) Cells were labeled by administration of tamoxifen to Cdh6-CreER;STOP-YFP or Cdh6-CreER; Ai14 mice at indicated ages, then RGCs were categorized at P20–60. Graph shows fraction of labeled RGCs that are CART-positive, indicating that they are ooDSGCs. Fraction is similar for isolated RGCs (gray, replotted from Fig. 2) and RGCs in clones (black) using either reporter (STOP-YFP or Ai14). Each bar represents counts from ≥2 mice, ≥16 retina sections. Error bars represent SDs. Dotted line indicates fraction of all RGCs that are CART+. (B and C) Radial clones marked by tamoxifen administration at E10 (B) or E8 (C) in Cdh6-CreER;Ai14 retina (red). Blue, fluorescent Nissl stain. (D) High-power image of RGCs from a clone similar to that in C but labeled with Brn3a and CART to show that the Cdh6-positive cells in the ganglion cell layer (arrowheads) are CART-positive RGCs. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. (Scale bars, 30 μm.)
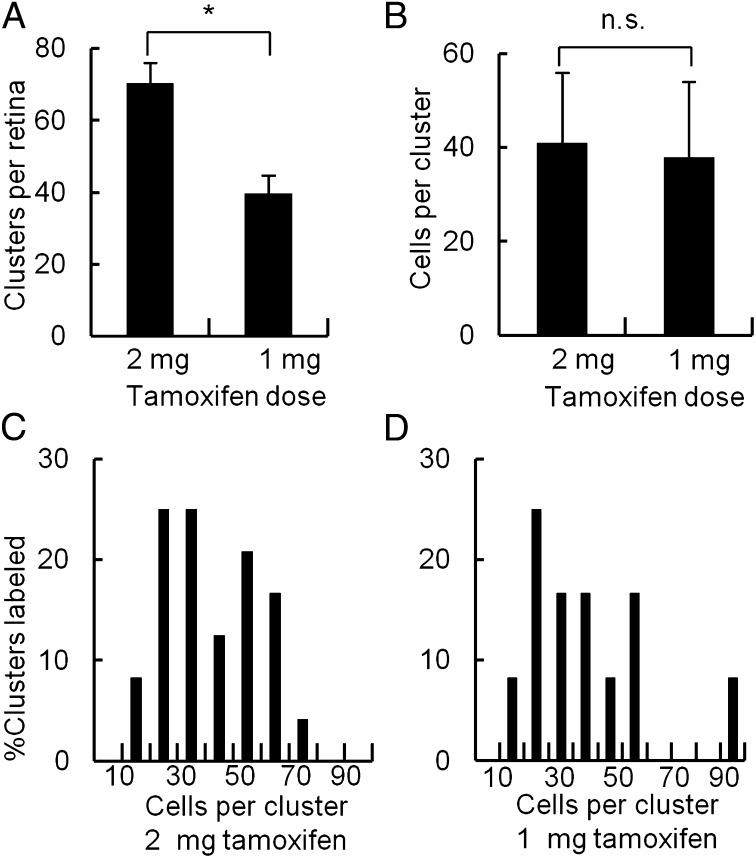
Together, these results raised the possibility that ooDSGCs are generated by multipotential progenitors, capable of giving rise to many retinal cell types but specified as to the RGC subtype they produce. To test this idea, we performed three further experiments. First, we asked whether the clusters of labeled cells are in fact clones. We used a test initially used to analyze clusters of retrovirally marked cells in retina and optic tectum (7, 33). If the clusters were clones, the number of cells per cluster would not vary among retinas bearing different numbers of clusters following infection with different amounts of retrovirus. In contrast, if clusters comprised groups of clonally unrelated cells labeled separately, both the number of clusters and the average number of cells per cluster should vary with viral titer. Here, we similarly asked whether cluster number or size varied with the tamoxifen dose. Cluster number varied with the tamoxifen dose but cluster size did not, indicating that most clusters were clones (Fig. 5 and Fig. S2).
Fig. 5.
Labeled clusters in Cdh6-CreER;Ai14 retina are clones. Graphs show number and size of RFP-positive clusters in central retina after tamoxifen administration at E10. If the clusters are clones, the number of cells per cluster should not vary between retinas bearing different numbers of clusters following administration of different doses of tamoxifen. In contrast, if clusters represent groups of cells labeled separately, both the number of clusters and the average number of cells per cluster should vary with the tamoxifen dose (7, 33). (A) The number of labeled clusters per retina varies with tamoxifen dose (*P < 0.01) of 2 mg or 1 mg. (B) The number of cells per labeled cluster does not change with tamoxifen dose (P = 0.4). (C and D) Similar distributions of cluster sizes are seen labeled in the central retina after the two different doses of tamoxifen. n = 4 mice per dose; error bars in A and B represent SDs.
Second, we asked whether cdh6 is expressed by retinal progenitors. In situ hybridization showed that cells in the neuroblast layer, which contains mitotically active progenitors, expressed cdh6 at E10 and E12 (Fig. S3). Although the small size of the cells and the low levels of cdh6 made it infeasible to count cdh6-positive cells, it was apparent that only a subset of cells expressed detectable levels of cdh6, as expected if cdh6 marks a defined subset of progenitors. Only a few cells were cdh6-positive in the region of postmitotic cells internal to the neuroblast layer; we presume that these are newborn ooDSGCs.
Third, we asked whether we could label ooDSGC-containing clones by activating CreER at E8, as the optic vesicle forms (34) and before any RGCs are born (Fig. 1B). Activation at E8 led to labeling of large clones (Fig. 4C) but few isolated cells. At least 70% of the RGCs in these clones were ooDSGCs as indicated by staining with antibodies to CART and Brn3a (Fig. 4 A and D). Together, these results indicate that at least one RGC subtype is generated by committed but multipotential progenitors. In contrast, no cells were labeled following CreER activation at E7, before formation of the retina. This result is consistent with the idea that commitment occurs as the retina forms.
Clonal Relatives of Direction-Selective RGCs.
To determine the identity of cells in the clones that arose from cdh6-positive progenitors, we labeled them with cell type-specific markers (35, 36). Although it was not feasible to assess the full composition of individual clones, it was clear that at least some contained amacrine cells (syntaxin-1-positive), bipolar cells (Chx10-positive), horizontal cells (calbindin-positive cells at the outer margin of the inner nuclear layer), and Müller glia (Sox9-positive) as well as RGCs (Fig. S4). Most clones also contained cells in the outer nuclear layer (Fig. 4), which is composed entirely of rod and cone photoreceptors. Thus, cdh6-positive progenitors can give rise to all of the major retinal cell types.
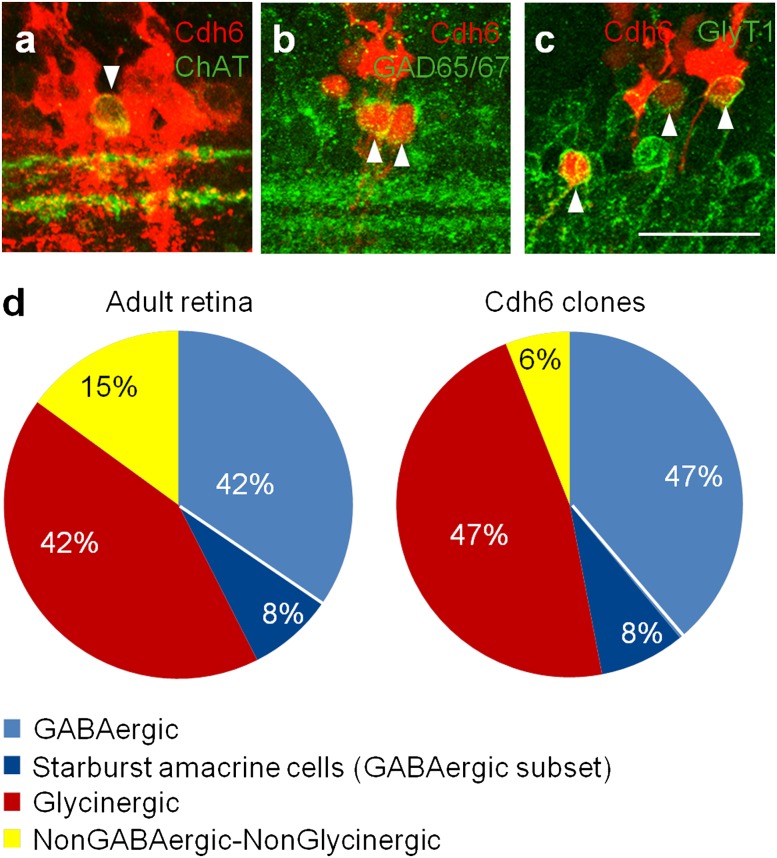
Finally, we asked whether the amacrine and bipolar cells derived from cdh6-positive progenitors were of specific subtypes. In the cortex, clonal relatives are often synaptic partners (37). In the retina, starburst amacrine cells provide a major input to ooDSGCs, but to few other RGC subtypes (5, 31). We therefore asked whether ooDSGCs-containing clones included a disproportionate number of starburst amacrines. The fraction of starburst amacrines (choline acetyltransferase-positive) among all amacrines (syntaxin-positive) in clones was the same as the fraction in the retina generally (Fig. 6 A and D). Moreover, the distribution of the major amacrine classes (glycinergic, GABAergic and neither glycinergic nor GABAergic) in clones reflected their proportion in the whole retina (38) (Fig. 6 B–D). Likewise, two bipolar subsets, Types 1+2 and rod (36), were both present in clones (Fig. S5), even through rod bipolars provide no direct input to ooDSGCs. Thus, Cdh6-positive progenitors are biased to generate a specific RGC subtype but can give rise to many and perhaps all interneuronal subtypes.
Fig. 6.
Amacrine cell subtype composition of Cdh6-marked clones. (A–C) Clones (red) contain starburst amacrine cells (ChAT-positive, green in A), GABAergic amacrine cells (Gad65/67-positive, green in B), and glycinergic amacrine (GlyT1-positive, green in C). Arrowheads indicate double-positive cells. (D) The main amacrine subtypes are found in clones (Right) in about the same proportions as occur in the whole retina (Left; replotted from ref. 38). (Scale bar, 30 μm.)
Discussion
We have identified a unique class of progenitor cell that can give rise to all of the main retinal cell types but is strongly biased toward generating a particular RGC subtype: ooDSGCs that respond preferentially to vertical motion and express cdh6. These progenitors differ from previously described restricted retinal progenitors (17, 19, 39, 40), which are biased toward generation of specific cell types; in none of these cases has the range of subtypes present in cloned been analyzed. Although completely consistent with previous results on retinal lineage (1–10), our observations suggest that early retinal progenitors are molecularly heterogeneous and committed to extents that had not been appreciated.
In considering how RGCs become diversified into ∼25 subtypes, we first asked whether they arise at distinct times, and might therefore be determined by the changing competence of their progenitors, as is the case for the main retinal types, and for distinct neuronal populations in other systems, including cerebral cortex and Drosophila motor neurons (4, 14–16). In fact, no significant differences were observed among the RGC subtypes we assayed. We then asked whether RGCs might acquire subtype-specific properties late in development. If this were the case, they might be shaped by synaptic partners or patterns of activity, as is the case for diversification of slow and fast muscle fiber subtypes or cholinergic and adrenergic sympathetic neuronal subtypes (41, 42). However, Cdh6-RGCs and J-RGCs are specified at or soon after the time they are born and, for Cdh6-RGCs, specification clearly occurs at the progenitor stage. Thus, specification of at least some RGC subtypes may result from differences among their progenitors.
An important unanswered question is when progenitors become specified to generate cdh6-positive RGCs. At one extreme, expression of cdh6 might commence only shortly before the RGC is born, in which case progenitors might be uniform and unspecified until around the time of the mitosis that generates the cdh6-positive RGC. At the other extreme, specification could occur two or more cell divisions before the RGCs themselves are born. Further lineage analysis may be helpful in distinguishing these alternatives. For example, if specification occurs early, some clones marked in the Cdh6-CreER line would be expected to contain multiple cdh6-positive RGCs but few, if any, cdh6-negative RGCs.
In contrast to the RGC subtype we describe, other retinal subtypes may be specified at later stages. For example, recent studies have shown that amacrine subtypes arise at distinct times, as predicted by the competence model (38, 43), and that further distinctions among them may result from postmitotic fate choices (44). Similarly, analysis of clones containing late-born retinal cell types (rods, amacrines, bipolars, and Müller glia) indicates that stochastic mechanisms acting on a homogeneous population of progenitors can account for the much of the diversity observed (1, 45). The observation that cdh6-positive progenitors produce multiple subtypes of bipolar and amacrine cells could also be explained by stochastic mechanisms acting to produce the diversity of later-born cell types.
In view of this difference, it is interesting to consider why RGC subtypes might be specified by a deterministic mechanism acting at the progenitor stage. The difference may reflect the fact that RGCs are the sole “output” neurons of the retina. Specification at the progenitor stage may ensure that that these feature detectors, which send ∼25 channels of visual information to the brain, are present in correct proportions and properly distributed through the visual field. Moreover, RGCs are the first-born retinal neurons; once they are specified, they can participate in organizing the cells and circuits that endow them with their subtype-specific functions.
Finally, it is important to consider whether other RGC subtypes also arise from committed progenitors. The two RGC subsets we have analyzed here, Cdh6-RGCs and J-RGCs, are both direction-selective cells that respond preferentially to vertical motion. This merely reflects the fact that Cdh6-CreER and JAM-B-CreER lines were the only two available to us that were suitable for these experiments, but we cannot rule out the possibility that RGC subsets with different preferences arise by other mechanisms. In addition, of these two lines, only Cdh6-CreER marked early progenitors. We note, however, that the JAM-B-CreER line was generated from a genomic fragment (24) and may therefore contain only some of the regulatory elements associated with the JAMB gene; the Cdh6-CreER line is a knock-in (22) that preserves the entire genomic region.
Taken together, our results lead to the speculation that the retina may arise from a mosaic of totipotential progenitors, each committed to generating one or a few RGC subtypes (Fig. S6). Comparing cdh6-positive and -negative progenitors may provide insights into the mechanisms by which the earliest progenitor populations acquire their distinct properties.
Materials and Methods
Animals.
Cadherin 6-CreER (Cdh6-CreER) and JAM-B-CreER mice were generated as described previously (22, 24) and maintained on a C57/B6 background. Briefly, tamoxifen-responsive Cre recombinase (CreER) cDNA was inserted at the initiation codon of the cdh6 gene by homologous recombination in embryonic stem cells. The JAM-B-CreER transgene was generated from a bacterial artificial chromosome by insertion of CreER cDNA at the initiation codon of the JAM-B gene. Cdh6-CreER heterozygote knock-in mice and JamB-CreER transgenic mice were crossed to reporter mouse lines that express a fluorescent protein following Cre-mediated excision of a stop signal. The reporter mouse lines used were Thy1-lox-STOP-lox-YFP line 15 (STOP-YFP; ref. 30), in which YFP expression is controlled by regulatory elements from the thy1 gene, and Ai14 (ref. 32; obtained from The Jackson Laboratory), in which tdTomato expression is controlled by the CAG promoter/enhancer inserted into the Rosa26 locus (17). All matings were timed; the presence of a vaginal plug marked embryonic day (E) 0.
BrdU birth dating was performed as described in ref. 38. Both CD-1 and C57/B6 mice were used in these studies, and no significant difference was detected between birthdays of RGC subsets in the two strains. To activate CreER, tamoxifen (Sigma; 20 mg/mL in sterile sunflower oil, dissolved by sonication) was administered by s.c. injection at P0 or to pregnant females by gavage. Except as otherwise indicated, doses were as follow: E8, 1 mg; E10, 2 mg; E12, 4 mg; E14, 4 mg; P0, 50 μg/g body weight. For tamoxifen administration to embryos, male double heterozygotes (JAM-B-CreER or Cdh6CreER; STOP-YFP or Ai14) were crossed to wild-type CD-1 females (Charles River Laboratories). CD-1 females were used because, in our experience, tamoxifen administration to pregnant C57/B6 females interferes with their ability to give birth. All animal procedures were approved by the Animal Care and Use Program at Harvard University and were in compliance with federal guidelines.
Tissue Processing and Immunohistochemistry.
Mice were killed at postnatal day (P) 14 (for birth dating) or P21–P60 (for all other experiments) by an overdose of pentobarbital and eyes were collected. Retinas were removed from the eye cup, fixed in 4% (vol/vol) paraformaldehyde (PFA) in PBS for 1 h on ice, cryoprotected in 30% sucrose/PBS for 1 h on ice, frozen, and sectioned at 20 μm in a cryostat. Sections were rinsed in PBS and incubated successively in blocking solution (3% donkey serum/0.5% Triton X-100/PBS) for 1 h at room temperature, and primary antibodies overnight at 4 °C. For BrdU immunostaining sections were preincubated in 2 M HCl for antigen retrieval (38), then processed as above. For whole mounts, retinas were incubated in blocking solution for 2 h followed by incubation with primary antibodies for 3 d at 4 °C. All tissues were washed in PBS, incubated with Alexa-conjugated secondary antibodies in blocking solution for 2 h at room temperature, washed with PBS, mounted on Superfrost Plus slides (Fisher) and coverslipped with Vectashield (Vector Laboratories).
Antibodies.
Primary antibodies used were rat anti-BrdU (1:200; BU1/75; Accurate), rabbit anti-CART (1:2,000; H-003–62; Phoenix Pharmaceuticals), mouse monoclonal anti-Brn3a (1:500; MAB1585; Chemicon), goat anti-Osteopontin (1:1,000; AF808; R&D Systems), mouse anti-neurofilaments (1:1,000; SMI-32P; Covance), rabbit anti-GFP (1:1,000; AB3080P; Chemicon), chicken anti-GFP (1:1,000; GFP-1020; Aves Laboratories), rabbit anti-DsRed (1:1,000; 632496; Clontech), goat anti-choline acetyltransferase (1:200; AB144P; Chemicon), goat anti-vesicular acetylcholine transporter (1:1,000; G488A; Promega), rabbit anti-glutamate decarboxylase 65/67 (1:1,000; AB1511; Chemicon), goat anti-glycine transporter 1 (1:5,000; AB1770; Chemicon), mouse monoclonal anti-syntaxin-1 (1:500; S0664; Sigma), goat anti-Chx10 (1:300; 21690; Santa Cruz), rabbit anti-Calbindin (1:4,000; CB38a; Swant), rabbit anti-Sox9 (1:1,000; AB5585; Chemicon), rabbit anti-neurokinin receptor 3 (1:5,000; 480739; Calbiochem), and mouse monoclonal anti-protein kinase C (1:500; MC5; ABCAM). Secondary antibodies conjugated to Alexa Fluor 488, 568, or 647 (Invitrogen) were diluted 1:500. Neurotrace 435/455 (Invitrogen) was used as a nuclear counterstain where indicated. Immunostaining patterns obtained with these antibodies in mouse retina are detailed in refs. 22, 25–27, 35 and 38.
In Situ Hybridization.
Timed-pregnant female mice were killed and embryos were fixed in 4% PFA in PBS at 4 °C overnight then incubated for 24 h in 30% sucrose in PBS for cryoprotection, flash-frozen, and stored at −80 °C before sectioning. Retina sections (20 μm) were mounted on Superfrost Plus slides (Fisher). Riboprobes for cdh6 were generated and sections were prepared and hybridized at 65 °C as previously described (22). Probes were detected using anti-digoxigenin antibodies conjugated to alkaline phosphotase, followed by reaction with BCIP (5-bromo-4-chloro-3-indolyl phosphate) NBT (nitro blue tretrazolium) substrate for 24 h.
Imaging and Analysis.
Samples were imaged using either a Zeiss Axio Plan 2 microscope (for in situ hybridization) or an Olympus FV1000 confocal microscope with 440, 488–515, 568, and 647 lasers with a step size of 1 μm. Images were analyzed using Imaris software.
Birth dating analysis was performed as described previously (38). Briefly, retinal sections were immunostained with antibodies against markers of interest and BrdU, and the percentage of marker-positive RGCs that were also positive for BrdU was determined for each time point. BrdU-positive nuclei indicated cells undergoing their last S-phase during the time BrdU was available (2–3 h following injection) (46). The significance of differences among subtypes at each time point was tested by Student t test for means, performed on the raw data as well as on the cumulative values.
To determine what fraction of the RGCs found in the clones marked at E8, E10, or E12 in the Cdh6-CreER;reporter retina were ON-OFF DSGCs, it was necessary to distinguish RGCs from displaced amacrine cells in the ganglion cell layer. RGCs found in clones were distinguished from displaced amacrines by immunostaining for the marker Brn3a, and the ooDSGCs were identified by coimmunostaining with CART. The percentage of ooDSGCs in clones was calculated as the fraction of Brn3a+ cells that were also CART+.
Clonal analysis was based on the dilution method described in refs. 7 and 33. In the central retina, clusters within a 500-μm radius from the optic nerve head were imaged, whereas in the peripheral retina, clusters within ∼500 μm from the ora serrata were imaged.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants NS29169 and EY019355 and a National Science and Engineering Research Council of Canada grant (to I.D.l.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215806109/-/DCSupplemental.
References
- 1.Bassett EA, Wallace VA. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012;35:565–573. doi: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Cayouette M, Poggi L, Harris WA. Lineage in the vertebrate retina. Trends Neurosci. 2006;29:563–570. doi: 10.1016/j.tins.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci USA. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: Lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 5.Sanes JR, Zipursky SL. Design principles of insect and vertebrate visual systems. Neuron. 2010;66:15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holt CE, Bertsch TW, Ellis HM, Harris WA. Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron. 1988;1:15–26. doi: 10.1016/0896-6273(88)90205-x. [DOI] [PubMed] [Google Scholar]
- 7.Turner DL, Snyder EY, Cepko CL. Lineage-independent determination of cell type in the embryonic mouse retina. Neuron. 1990;4:833–845. doi: 10.1016/0896-6273(90)90136-4. [DOI] [PubMed] [Google Scholar]
- 8.Wetts R, Fraser SE. Multipotent precursors can give rise to all major cell types of the frog retina. Science. 1988;239:1142–1145. doi: 10.1126/science.2449732. [DOI] [PubMed] [Google Scholar]
- 9.Turner DL, Cepko CL. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- 10.Wong LL, Rapaport DH. Defining retinal progenitor cell competence in Xenopus laevis by clonal analysis. Development. 2009;136:1707–1715. doi: 10.1242/dev.027607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexiades MR, Cepko CL. Subsets of retinal progenitors display temporally regulated and distinct biases in the fates of their progeny. Development. 1997;124:1119–1131. doi: 10.1242/dev.124.6.1119. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe T, Raff MC. Rod photoreceptor development in vitro: Intrinsic properties of proliferating neuroepithelial cells change as development proceeds in the rat retina. Neuron. 1990;4:461–467. doi: 10.1016/0896-6273(90)90058-n. [DOI] [PubMed] [Google Scholar]
- 13.Cayouette M, Barres BA, Raff M. Importance of intrinsic mechanisms in cell fate decisions in the developing rat retina. Neuron. 2003;40:897–904. doi: 10.1016/s0896-6273(03)00756-6. [DOI] [PubMed] [Google Scholar]
- 14.Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol. 2004;20:619–647. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- 15.Qian X, et al. Timing of CNS cell generation: A programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- 16.Shen Q, et al. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- 17.Brzezinski JA, 4th, Kim EJ, Johnson JE, Reh TA. Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina. Development. 2011;138:3519–3531. doi: 10.1242/dev.064006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brzezinski JA, 4th, Prasov L, Glaser T. Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev Biol. 2012;365:395–413. doi: 10.1016/j.ydbio.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hafler BP, et al. Transcription factor Olig2 defines subpopulations of retinal progenitor cells biased toward specific cell fates. Proc Natl Acad Sci USA. 2012;109:7882–7887. doi: 10.1073/pnas.1203138109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jasoni CL, Reh TA. Temporal and spatial pattern of MASH-1 expression in the developing rat retina demonstrates progenitor cell heterogeneity. J Comp Neurol. 1996;369:319–327. doi: 10.1002/(SICI)1096-9861(19960527)369:2<319::AID-CNE11>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 21.Trimarchi JM, Stadler MB, Cepko CL. 2008. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PloS one 3:e1588.
- 22.Kay JN, et al. Retinal ganglion cells with distinct directional preferences differ in molecular identity, structure, and central projections. J Neurosci. 2011;31:7753–7762. doi: 10.1523/JNEUROSCI.0907-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim IJ, Zhang Y, Meister M, Sanes JR. Laminar restriction of retinal ganglion cell dendrites and axons: subtype-specific developmental patterns revealed with transgenic markers. J Neurosci. 2010;30:1452–1462. doi: 10.1523/JNEUROSCI.4779-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim IJ, Zhang Y, Yamagata M, Meister M, Sanes JR. Molecular identification of a retinal cell type that responds to upward motion. Nature. 2008;452:478–482. doi: 10.1038/nature06739. [DOI] [PubMed] [Google Scholar]
- 25.Coombs J, van der List D, Wang GY, Chalupa LM. Morphological properties of mouse retinal ganglion cells. Neuroscience. 2006;140:123–136. doi: 10.1016/j.neuroscience.2006.02.079. [DOI] [PubMed] [Google Scholar]
- 26.Ju WK, et al. Ganglion cells of the rat retina show osteopontin-like immunoreactivity. Brain Res. 2000;852:217–220. doi: 10.1016/s0006-8993(99)02140-x. [DOI] [PubMed] [Google Scholar]
- 27.Xiang M, et al. The Brn-3 family of POU-domain factors: Primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J Neurosci. 1995;15:4762–4785. doi: 10.1523/JNEUROSCI.15-07-04762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Völgyi B, Chheda S, Bloomfield SA. Tracer coupling patterns of the ganglion cell subtypes in the mouse retina. J Comp Neurol. 2009;512:664–687. doi: 10.1002/cne.21912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buffelli M, et al. Genetic evidence that relative synaptic efficacy biases the outcome of synaptic competition. Nature. 2003;424:430–434. doi: 10.1038/nature01844. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 31.Vaney DI, Sivyer B, Taylor WR. Direction selectivity in the retina: Symmetry and asymmetry in structure and function. Nat Rev Neurosci. 2012;13:194–208. doi: 10.1038/nrn3165. [DOI] [PubMed] [Google Scholar]
- 32.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray GE, Glover JC, Majors J, Sanes JR. Radial arrangement of clonally related cells in the chicken optic tectum: Lineage analysis with a recombinant retrovirus. Proc Natl Acad Sci USA. 1988;85:7356–7360. doi: 10.1073/pnas.85.19.7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaufman M. Cephalic neurulation and optic vesicle formation in the early mouse embryo. Am J Anat. 1979;155:425–443. doi: 10.1002/aja.1001550403. [DOI] [PubMed] [Google Scholar]
- 35.Haverkamp S, Wässle H. Immunocytochemical analysis of the mouse retina. J Comp Neurol. 2000;424:1–23. [PubMed] [Google Scholar]
- 36.Ghosh KK, Bujan S, Haverkamp S, Feigenspan A, Wässle H. Types of bipolar cells in the mouse retina. J Comp Neurol. 2004;469:70–82. doi: 10.1002/cne.10985. [DOI] [PubMed] [Google Scholar]
- 37.Yu YC, Bultje RS, Wang X, Shi SH. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature. 2009;458:501–504. doi: 10.1038/nature07722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voinescu PE, Kay JN, Sanes JR. Birthdays of retinal amacrine cell subtypes are systematically related to their molecular identity and soma position. J Comp Neurol. 2009;517:737–750. doi: 10.1002/cne.22200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Godinho L, et al. Nonapical symmetric divisions underlie horizontal cell layer formation in the developing retina in vivo. Neuron. 2007;56:597–603. doi: 10.1016/j.neuron.2007.09.036. [DOI] [PubMed] [Google Scholar]
- 40.Rompani SB, Cepko CL. Retinal progenitor cells can produce restricted subsets of horizontal cells. Proc Natl Acad Sci USA. 2008;105:192–197. doi: 10.1073/pnas.0709979104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buonanno A, Fields RD. Gene regulation by patterned electrical activity during neural and skeletal muscle development. Curr Opin Neurobiol. 1999;9:110–120. doi: 10.1016/s0959-4388(99)80014-2. [DOI] [PubMed] [Google Scholar]
- 42.Francis NJ, Landis SC. Cellular and molecular determinants of sympathetic neuron development. Annu Rev Neurosci. 1999;22:541–566. doi: 10.1146/annurev.neuro.22.1.541. [DOI] [PubMed] [Google Scholar]
- 43.Cherry TJ, Trimarchi JM, Stadler MB, Cepko CL. Development and diversification of retinal amacrine interneurons at single cell resolution. Proc Natl Acad Sci USA. 2009;106:9495–9500. doi: 10.1073/pnas.0903264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kay JN, Voinescu PE, Chu MW, Sanes JR. Neurod6 expression defines new retinal amacrine cell subtypes and regulates their fate. Nat Neurosci. 2011;14:965–972. doi: 10.1038/nn.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gomes FL, et al. Reconstruction of rat retinal progenitor cell lineages in vitro reveals a surprising degree of stochasticity in cell fate decisions. Development. 2011;138:227–235. doi: 10.1242/dev.059683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Packard DS, Jr, Menzies RA, Skalko RG. Incorportaiton of thymidine and its analogue, bromodeoxyuridine, into embryos and maternal tissues of the mouse. Differentiation. 1973;1:397–404. doi: 10.1111/j.1432-0436.1973.tb00137.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.