Abstract
Information on unique and coordinated regulation of transcription and translation in response to stress is central to the understanding of cellular homeostasis. Here we used ribosome profiling coupled with next-generation sequencing to examine the interplay between transcription and translation under conditions of hydrogen peroxide treatment in Saccharomyces cerevisiae. Hydrogen peroxide treatment led to a massive and rapid increase in ribosome occupancy of short upstream ORFs, including those with non-AUG translational starts, and of the N-terminal regions of ORFs that preceded the transcriptional response. In addition, this treatment induced the synthesis of N-terminally extended proteins and elevated stop codon read-through and frameshift events. It also increased ribosome occupancy at the beginning of ORFs and potentially the duration of the elongation step. We identified proteins whose synthesis was regulated rapidly by hydrogen peroxide posttranscriptionally; however, for the majority of genes increased protein synthesis followed transcriptional regulation. These data define the landscape of genome-wide regulation of translation in response to hydrogen peroxide and suggest that potentiation (coregulation of the transcript level and translation) is a feature of oxidative stress.
Gene expression may be controlled at multiple levels. Globally, it is regulated by histones and satellite proteins. Locally, promoters, enhancers, and other regulatory elements are used to guide transcription. Numerous studies have yielded datasets involving the networks of transcription factors and described the associated mechanisms of transcriptional regulation. Developments in microarray technology have facilitated such studies and made them affordable for individual laboratories. Accordingly, a vast number of studies has emerged that describe transcriptional responses to various treatments, stimuli, knockouts, and other interventions. Conversely, the investigation of the regulation of gene expression at the level of translation lagged behind because of the lack of accessible high-throughput methods.
It often is assumed that changes in mRNA abundance are proportional to changes in protein synthesis in the cell, but numerous exceptions are known. One powerful approach to assess changes in protein abundance directly is the use of whole-proteome mass spectrometry, but this method is inferior to mRNA profiling in its throughput and can detect only a fraction of protein products in the cell (1). Other high-throughput approaches, such as fluorescent protein reporter libraries, are available (2–4). However, they are designed for the quantification of individual proteins rather than for addressing the details of translation. Indirect approaches, such as comparative microarray profiling of mRNAs within monosomes and polysomes, are popular as well (5–8). These methods enable estimation of the mRNA transcripts that are being translated. Recent advances in next-generation sequencing have enhanced data acquisition, improved sensitivity, and made this method superior to microarrays in its throughput (9). Importantly, it allowed mRNA abundance and protein translation to be examined in the same sample with high accuracy (with subcodon resolution) (10, 11). This experimental strategy involves deep sequencing of mRNA fragments (footprints) buried inside the actively translating ribosomes. Protein translation can be inferred from footprint abundance. Coupled with regular mRNA-sequencing (mRNA-seq) analyses, these data give information on the actual mRNA sequences that are being translated, identity of the reading frames used, and ribosomal density at each position within these mRNAs. Hereafter, we refer to this method as “ribosome profiling” or Ribo-seq. Another promising application of Ribo-seq is measuring translational regulation by monitoring translation efficiency (TE), which is the amount of footprint normalized to underlying mRNA abundance.
In the current study, we applied Ribo-seq to investigate the fine details of Saccharomyces cerevisiae response to oxidative stress caused by hydrogen peroxide treatment. A key advantage of this method is the much higher sensitivity than obtained with microarrays. With this method we were able to detect changes in transcription and its regulation within 5 min of treatment. Oxidative stress is one of the best-studied regulators of transcription (12), but little is known about how this stress changes protein abundance and posttranscriptional regulation. Previous studies pointed to a weak correlation between transcriptional and translational gene responses, i.e., elevated mRNA transcripts in stressed cells did not match the set of proteins that changed abundance. Microarray analyses revealed that only 15% of genes involved in translational response showed the corresponding changes at the mRNA levels (6). Our study focused on using Ribo-seq to examine precisely translation and its regulation by oxidative stress.
Results
Ribo-Seq.
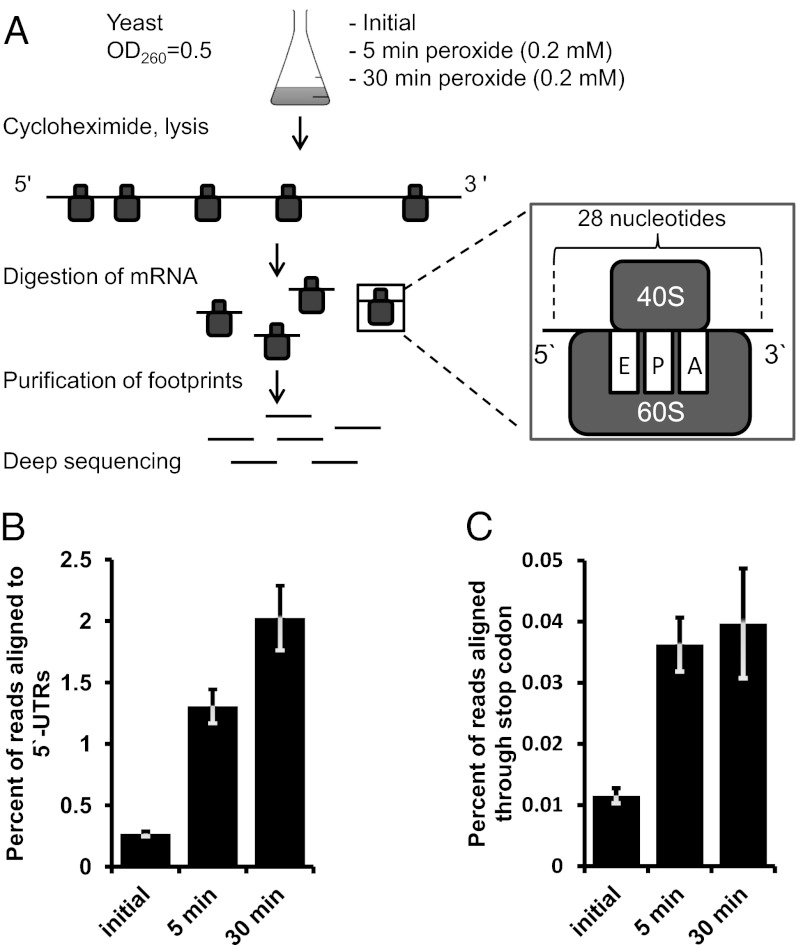
An overview of the Ribo-seq method that we used to examine the regulation of translation by oxidative stress is given in Fig. 1A. Each translating ribosome protects ∼28 nucleotides on the translated mRNA, and the unprotected regions are removed by subjecting mRNAs to RNase I digestion. The protected mRNA pieces (footprints) are extracted and analyzed by deep sequencing. Because their length is known, the exact codons that occupy the A and P sites of the ribosome can be determined. This information is used to identify frameshifts, read-through events, and altered codon use. Additionally, quantification of footprints provides an opportunity to estimate changes in translation for every mRNA species.
Fig. 1.
Oxidative stress affects the fidelity of translational machinery. (A) Design of the experiment. See text for details. (B) Hydrogen peroxide treatment leads to an increase in 5′-UTR translation. Yeast cultures were treated with 0.2 mM hydrogen peroxide for 5 or 30 min. Untreated cells served as control. A fivefold increase in net translation of 5′ UTRs occurred after 5 min of incubation. Incubation with hydrogen peroxide for 30 min further increased 5ʹ-UTR translation. (C) Oxidative stress leads to translation read-through events at stop codons. Experimental conditions are as in B. Error bars indicate SEM. Measurements from biological replicates are shown.
A key factor that decreases throughput of this method is that only 5% of total yeast RNA consists of mRNA in rapidly growing yeast cells (13). Previously, contamination was eliminated during footprint preparation by ultrafiltration, which is not very efficient; i.e., the fraction of ribosomal RNA fragments in sequencing libraries approached 80%, with an average value of about 60%, as observed in previous studies (10) and our own pilot experiments. To improve the throughput of the method, we examined the content of contaminating rRNA fragments. In our footprint samples a particular fragment of the 28S ribosomal subunit was responsible for 90% of contamination. An additional step of subtractive hybridization allowed us to get rid of this specific fragment, and 95% of the resulting library consisted of mRNA footprints (Tables S1 and S2). Such high purity made possible sample multiplexing, which increased throughput and decreased cost.
Oxidative Stress Increases Ribosome Occupancy of Upstream ORFs.
Upstream ORFs (uORFs), short ORFs immediately upstream of the main gene sequence, are known to modulate gene expression in response to amino acid depletion and other types of stress. One of the best-studied examples is the regulation of GCN4, which has multiple uORFs that block its translation when sufficient levels of amino acids are present but allow translation when amino acids are depleted (14). Precise mapping and thorough characterization of such uORFs have been complicated because of the lack of sensitive methods. Bioinformatics analysis and modeling were used instead (15). Ribo-seq overcomes this challenge, detecting uORFs quantitatively and mapping them to the mRNA at a single-nucleotide resolution (10).
We first used Ribo-seq to examine if oxidative stress caused by hydrogen peroxide treatment affects the diversity and abundance of uORFs. We used annotated 5ʹ UTRs from the yeast transcriptome-sequencing study (16). Among them, surprisingly many UTRs (1,800 genes) showed detectable presence of translating ribosomes at the uORFs. These uORFs often overlapped with each other and frequently lacked AUG start codons. In many cases, this observation complicated the analysis of individual uORFs; i.e., often it was unclear if a single uORF or several adjacent uORFs were present in the gene. uORFs are thought to be short, but when clustered they may occupy long sequences upstream of actual ORFs. Thus, we call such regions “upstream translation islets.” They can be short or long, represent a single uORF or an uORF cluster, and change their length and composition in response to various treatments. To quantify the translation events within 5ʹ UTRs, we assigned sequencing reads to the entire 5ʹ UTRs rather than attempting to separate them into individual uORFs.
We next compared yeast cells treated with 0.2 mM hydrogen peroxide for 5 or 30 min with corresponding untreated cells. Even short (5-min) incubation resulted in a fivefold increase in the ribosomal footprints aligning to the 5ʹ UTRs (Fig. 1B). We detected 847 5ʹ UTRs whose coverage by footprints increased more than 2.6-fold under these conditions, and the 30-min treatment increased this number to 1,217 UTRs. Interestingly, the changes in 5′-UTR utilization generally were more pronounced than those of downstream genes and occurred at an earlier time point. In addition, the majority of uORFs initiated translation at non-AUG codons under both normal conditions and oxidative stress, as is seen in cells under conditions of amino acid depletion (10). Interestingly, translation of 5ʹ UTRs increased uniformly during stress, and no 5ʹ UTR was down-regulated under these conditions.
Many Genes Show Translation Immediately Upstream of Their Known Start Codons.
Analyzing uORF distribution, we observed multiple translation events immediately upstream (i.e., within 45 nt) of their AUG start codons, and oxidative stress increased these events significantly. Elevated ribosome occupancy at uORFs may be caused by slower elongation or, conversely, by increased translation. Up-regulated translation can lead to one of two possible outcomes. First, the translation upstream of AUG may correspond to the N-terminal extensions of some proteins. Second, uORFs in the vicinity of start codons could influence the translation of downstream genes. They may facilitate reinitiation of the ribosome at a downstream AUG codon because the distance between the uORF’s stop codon and the following start codon is short (10–15 nt on average). On the other hand, dissociation of the ribosome complex at the uORF stop codon could prevent translation of the main gene. Supporting the first possibility, our analysis revealed five strong candidates with N-terminal extensions in untreated samples, 13 in samples treated with peroxide for 5 min, and 32 in samples treated for 30 min (Table S3). These peptides were translated in the same reading frame as the downstream gene and usually started with a non-AUG codon. Fig. S1 features proteins selected to represent different scenarios of the N-terminal extension/ORF interplay. The only two known yeast proteins with N-terminal non-AUG extensions, ALA1 and GRS1 tRNA synthetases (17, 18), were among our identified proteins. In these two proteins, N-terminal sequences serve as signal peptides, directing a fraction of these proteins to mitochondria. We examined the subcellular localization of our detected protein candidates using Gene Ontology (GO) annotation of the SGD database. Twenty-one of 32 proteins had experimentally verified localization in both cytosol and another compartment, such as mitochondria, Golgi, vacuoles, and membranes. Such an enrichment of GO terms supports the idea of regulation by targeted protein localization in response to oxidative stress.
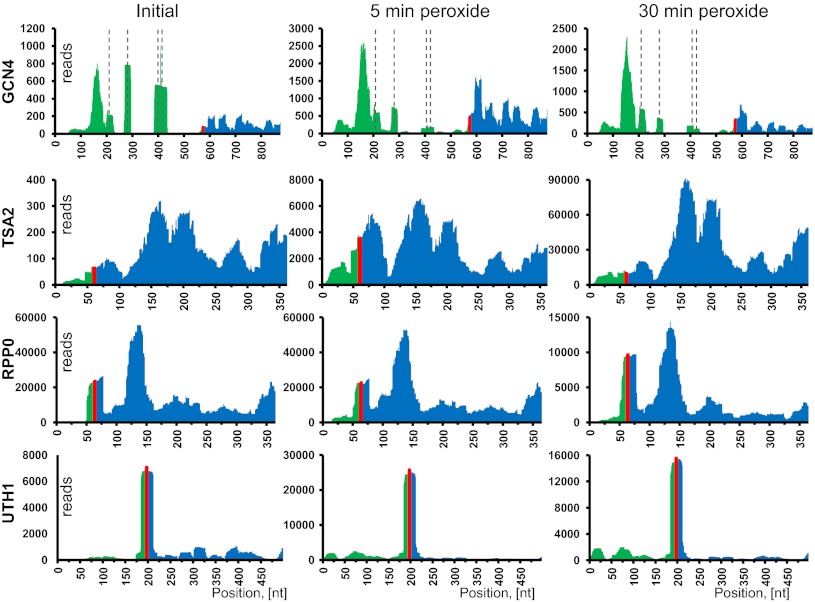
At the genome-wide level, the majority of 5ʹ UTRs supported uORF translation rather than N-terminal protein extensions. We observed intricate and widespread translation of 5ʹ UTRs under conditions of oxidative stress. Some common cases are shown in Fig. 2, illustrated by four representative proteins. Remarkably, the coverage profiles for every gene were alike in different experimental conditions and were nearly identical in replicates.
Fig. 2.
Examples of 5′-UTR translation during oxidative stress. Ribosome footprint coverage for four different mRNAs discussed in the text illustrates various patterns of translation. Panels show the footprint coverage of certain mRNAs with no in-frame stop codons upstream of annotated genes. For each mRNA, translation following 5- and 30-min hydrogen peroxide treatment is given. Untreated yeast cells served as a control. The entire 5′ UTR and 300 nt of the gene sequence were used to generate the coverage density map. The 5′-UTR part of the mRNA is shown in green, the AUG start codon in red, and the annotated gene in blue. Dashed lines in GCN4 graphs indicate positions of known uORFs.
Oxidative Stress Induces Translational Read-Through of Stop Codons and Frameshifting.
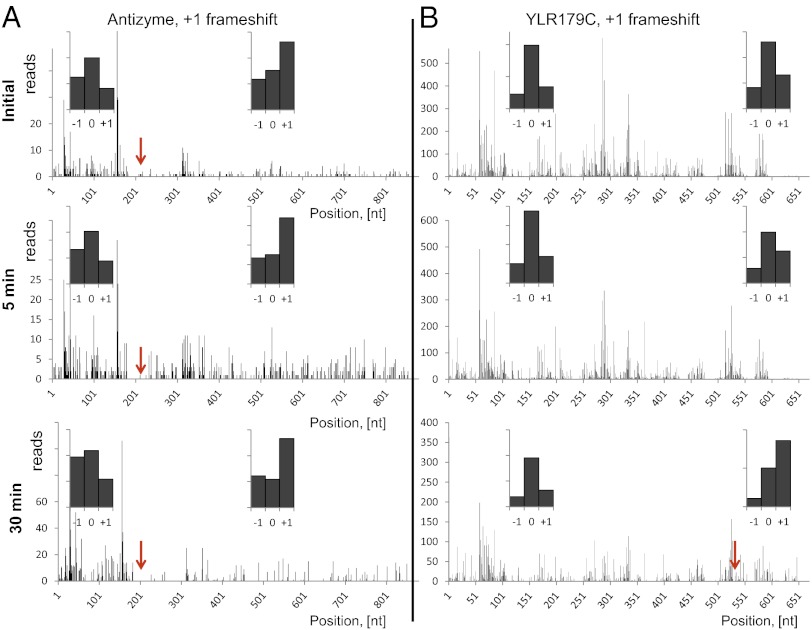
Oxidative damage is known to impact ribosomal proteins and translation factors. We examined the rate of read-through events at stop codons. Termination of translation appeared to be very efficient in the control sample, based on poor read coverage of 3ʹ UTRs immediately downstream of stop codons (Fig. 1C). Oxidative stress increased read-through events threefold in both 5- and 30-min samples. We also developed a simple method for frameshift search and validation that is technically similar to the search for N-terminal extensions. A short region downstream of the stop codon for each annotated gene was examined for the presence of ribosomal footprints with coverage comparable to the gene itself. A handful of candidates were confirmed manually. For validation, the 5ʹ ends of footprints aligned to the regions upstream or downstream of the known frameshift were quantified and assigned to the matching reading frame. The frame with the highest count would correspond to the actual ORF. This approach is shown in Fig. 3 and Fig. S2 for two known frameshifts in S. cerevisiae, antizyme and protein ABP140, respectively (19). Further analysis of genes for read-through of annotated stop codons yielded four additional genes with +1 frameshifts (i.e., ribosome slipping one nucleotide towards 3′ end) (Table S4). An example is shown in Fig. 3B. All these frameshifts were detected under conditions of oxidative stress.
Fig. 3.
Ribo-seq allows identification of frameshifts (red arrows). (A) Validation of the known frameshift in the antizyme gene. (B) Oxidative stress leads to a frameshift in the product of the YRL179C gene. We observed a change of frame, leading to translation of a longer protein in the 30-min peroxide treatment sample. The 5′ ends of footprints were mapped to the genomic sequence of YRL179C. (Insets) Histograms show the count of footprints, matching one of three possible frames either to the left or to the right of the frameshift. The “0” frame is the one with the annotated start codon. The highest count of footprints matched the “0” frame before the frameshift and the “+1” frame after the frameshift.
Correlation Between Transcriptional and Translational Responses to Oxidative Stress.
In S. cerevisiae, ∼1,700 genes are regulated by hydrogen peroxide at the level of transcription, including ∼900 genes of the environmental stress response cluster, which encompasses genes regulated in response to various stresses such as heat shock, starvation, and oxidative stress (12). Next-generation sequencing technologies can improve the sensitivity and dynamic range of gene-expression analysis significantly. We found that after 5-min treatment with hydrogen peroxide transcriptional changes were observed for 116 genes, of which 10 were down-regulated and 106 were up-regulated. The 30-min treatment yielded transcriptional changes in 1,497 genes (529 down-regulated and 968 up-regulated) with the threshold of 2.6-fold (see Datasets S1 and S2 and Fig. S3A for comparison of mRNA-seq with microarrays from ref. 12).
One of our major goals was to examine genome-wide translational changes and posttranscriptional regulation of translation in response to oxidative stress. Sequencing of ribosomal footprints enabled direct and absolute quantification of mRNAs undergoing translation. It should be noted that Ribo-seq does not provide protein concentrations but instead estimates the relative translation for a given protein. Using this method, we showed that protein synthesis cannot be inferred securely from mRNA abundance. There were genes whose translation did not correlate with mRNA abundance (Fig. S4E). In addition, a significant fraction of genes showed essentially no translation, although their mRNAs were present. We detected translational response for 97 genes after the 5-min hydrogen peroxide treatment. Only four genes showed decreased protein synthesis at this time point. After 30 min, relative protein synthesis was decreased in 593 genes and increased in 766 (Dataset S2). Some proteins increased expression between 5 and 30 min, some reached a plateau at 5 min, and others declined during the longer treatment time.
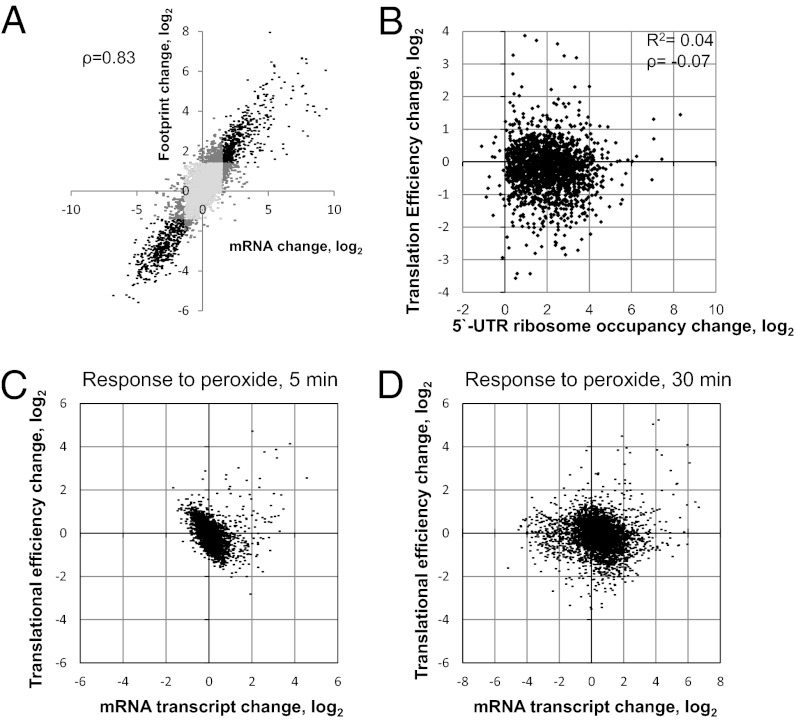
Interestingly, the values of translation change in response to hydrogen peroxide did not match those for mRNA transcripts exactly, even if we only consider coregulated genes (Fig. 4A, black dots), although in most cases the changes in values are in the same direction. For instance, the footprint density of a representative protein increased 10-fold, but its mRNA expression increased only twofold. These data suggest a specific posttranscriptional control of protein expression. Indeed, by comparing changes in TE with changes in mRNA transcripts, we observed multiple proteins in which translational regulation was greater than transcriptional regulation (Fig. 4 C and D).The TE is the ratio of Ribo-seq read counts to mRNA-seq read counts, and it describes the propensity of mRNA to undergo translation. The higher the TE, the better is the mRNA translated. Posttranscriptional regulation can be simply permissive, allowing an mRNA transcript to be translated under stress conditions. However, based on our analysis, posttranscriptional regulation usually makes an addition to transcription changes, modulating protein synthesis (see Fig. S3B for the TE error rate). Because we observed an immediate increase in uORF footprint density in response to hydrogen peroxide treatment, we further examined a possible effect on the TE of downstream genes. In our reference database, 3,830 genes had annotated 5ʹ UTRs with an unambiguous sequence longer than 23 nt. Among them, nearly 1,800 were covered by ribosomal footprints in at least one of the samples, and 1,217 had increased footprint density after the 30-in peroxide treatment. We analyzed the potential coregulation of translation and increased ribosomal density at 5ʹ UTRs in these 1,800 genes and found that, on a genome-wide scale, ORF translation and ribosomal density at uORFs were mostly independent under oxidative stress conditions (Fig. 4B).
Fig. 4.
Interplay between translation and transcription. (A) Correlation between changes in footprint and transcript abundances in response to hydrogen peroxide. Light gray dots represent genes whose footprint count and mRNA count were not affected by peroxide treatment; dark gray dots represent genes with only the footprint or mRNA affected; and black dots represent coaffected genes. Changes in transcript and in footprint abundance between the initial and the 30-min peroxide samples are plotted on the axes (for further details see SI Materials and Methods). (B) Increased ribosomal occupancy at the 5ʹ UTR does not affect the TE of a downstream gene. (C) Relationship between change in TE and change in mRNA transcript change after 5-min incubation with peroxide. (D) Relationship between change in TE and change in mRNA transcript after 30-min incubation with peroxide.
Oxidative Stress Regulates Translation Elongation.
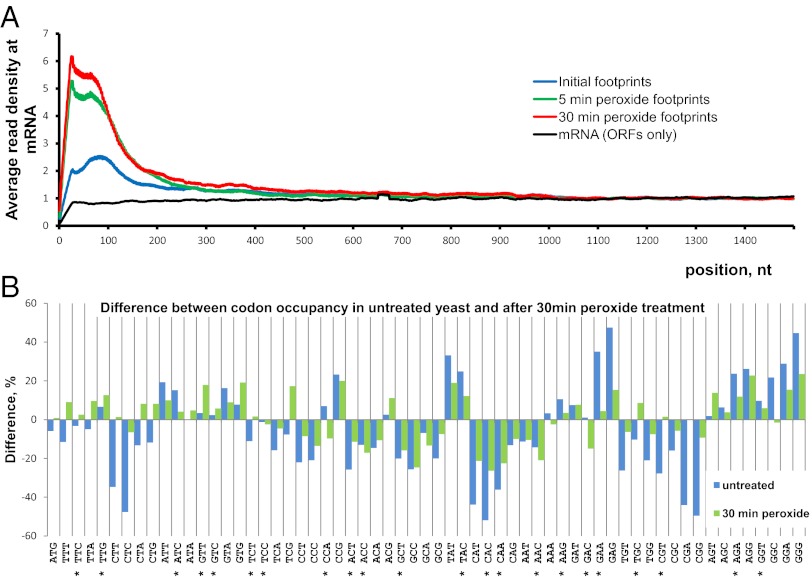
We found that the density of elongating ribosomes on the mRNAs was consistently higher within the first 100–150 nt from the start codon. This observation may be explained by codon use and the corresponding tRNA copy number (20). Hydrogen peroxide treatment caused a significant increase in ribosome occupancy and, therefore, in the density of footprint coverage within the beginning of the ORF (Fig. 5A), and this effect was similar for the 5- and 30-min treatment samples. Treatment affected transcripts regardless of their length or expression level [similar to the previous observations (10)]. The data suggest that oxidative stress influenced elongation, forcing ribosomes to spend more time at the beginning of their ORFs. Together with the increased utilization of the 5ʹ UTRs it explains the contradiction with previous experimental observations (6). The fact that ribosome density increased so rapidly upon addition of hydrogen peroxide implies a direct effect of the oxidant, which targets ribosomes and elongation factors.
Fig. 5.
Global features of translation examined by Ribo-seq. (A) Density of footprint coverage along the mRNA. Profiles of read coverage were calculated for each mRNA longer than 1,500 nt and rpkm >10. The profiles were normalized based on the average density in the region from 1,000–1,500 nt. Densities for each nucleotide position were averaged across all mRNAs. An average between the two experimental replicates is shown. (B) Ribosomal occupancy of individual codons measured in vivo. Percentage of difference is calculated between the predicted codon distribution across mRNAs and the experimental codon appearance at the ribosomal A site. For details of normalization and prediction, see SI Materials and Methods. Codon values greater than zero are encountered more often at the A site of ribosome. Asterisks mark codons with the highest RSCU values (21). Blue bars represent untreated control; green bars represent samples treated for 30 min with 0.2 mM hydrogen peroxide.
Ribo-Seq Enables Codon Occupancy Quantification in Vivo.
Because Ribo-seq can track translation at a single-nucleotide resolution, we examined the experimental relative frequency of translated codons and compared the experimental observations with the predicted values. Assuming that all codons are translated at the same rate, one would expect the distribution of codons trapped at the ribosomal A site to be identical to the frequency distribution of codons across mRNAs (normalized to expression levels). However, our experimental data showed that some codons were more enriched (Fig. 5B, bars above the baseline), meaning that they are met more frequently in ribosomes and are translated less efficiently. Codons such as CAC or GGT fit into the relative synonymous codon use (RSCU) table, which is used for calculations of the codon adaptation index (21) that rely partially on tRNA copy numbers in the yeast genome (20). The number of experimental replicates does not allow us to compare a particular codon directly in untreated and peroxide-treated yeast. Nevertheless, by analyzing the whole distribution (Fig. 5B), we observed that the difference between predicted and experimental codon occupancy was less in stressed than in unstressed yeast. In other words, untreated, logarithmically grown yeast cells have more selective pressure on translation machinery (e.g., the availability of charged tRNA.). Oxidative stress causes a rapid decrease in translation but, perhaps, less of a decrease in the pool of tRNAs and in the amount of mRNA, thus relaxing the competition of ribosomes for tRNAs. Therefore, the observed codon occupancy tends to be similar to the codon distribution of genes. Increasing the number of experimental replicates can make this method sensitive enough to detect changes in individual codon translation upon stress or any other change in condition.
Discussion
Our data define the landscape of translational control of oxidative stress in yeast. We made several interesting observations. First, we found widespread translation of uORFs under conditions of oxidative stress. A dramatic increase in uORF ribosome occupancy occurred only 5 min after the addition of hydrogen peroxide and greatly exceeded the overall changes in protein translation. Comparisons between our study and the previously identified uORFs under conditions of starvation revealed a more extensive use of the 5ʹ UTRs under oxidative stress. Two times as many genes showed increased ribosome occupancy at their 5ʹ UTRs under oxidative stress than under starvation (Fig. S3C). The greater fraction of ribosomes bound to the 5ʹ-UTR regions may be caused by two opposite events. First, translation of these regions may be up-regulated, thus producing short, cryptic peptides. On the other hand, ribosomes may move to the 5ʹ-UTR regions slowly, accumulating footprints without affecting polypeptide yields. We think the second explanation is more likely. It agrees with the elevated density in the first 30–50 codons within the mRNA and with the reported increase in elongation time under conditions of oxidative stress (6). It also is consistent with the complex relationships between gene translation and 5ʹ UTR translation. Mass-spectrometry analyses would show the real yield of uORF-produced peptides and would be useful for the development of future Ribo-seq applications. We did not detect up- or down-regulation of known translation initiating factors at the 5-min time point, so the observed effects on the 5ʹ UTR likely were caused by posttranslational modifications of initiation factors or ribosomal proteins. Phosphorylation of eIF2, a component of the ternary initiation complex, is known to inhibit translation initiation and, consequently, protein expression (22). In some cases, this factor was shown to induce translation of proteins, such as ATF4 or GCN4, through the intricate system of translation and reinitiation events at the uORFs (14, 23, 24). An additional reason for increased ribosome occupancy at the 5ʹ UTRs may involve initiation at non-AUG codons. The majority of our detected uORFs had no AUG start codons. eIF1 and eIF5 are the factors that control the recognition of start codons during translation initiation in eukaryotes (25, 26). We suggest that hydrogen peroxide impairs the fidelity of these factors, which normally restrict initiation to AUG codons, thereby facilitating non-AUG initiation of translation as the ribosome scans the mRNA. Our observations imply a mechanism that slows down the ribosome at uORFs and the beginnings of ORFs. It can be achieved by impairing the exchange of elongation factors, incomplete dissociation of initiation factors, or binding additional stress-activated proteins. In addition, hydrogen peroxide may damage tRNAs (27), amino acids (28), and aminoacyl-tRNA synthetases (29). The exact molecular mechanism requires further studies.
Translatome and transcriptome in yeast are regulated conjointly in response to various stresses, such as amino acid depletion, osmotic shock, and sorbitol treatment (7). Thus the genes up-regulated at the level of transcription also yield more protein product as well, a process that is termed “potentiation.” However, in the response to hydrogen peroxide only ∼15% of transcriptionally regulated genes were believed to be linked by potentiation (6). Our data indicate that the overlap is greater and that oxidative stress is not unique in this respect (Fig. 4A). We compared our results directly with the published reports on the translation response to oxidative stress (6). All proteins with high scores from that study were present in our list, and the two studies also had several down-regulated proteins in common. However, about 70% of peroxide-regulated proteins from that study did not overlap with our hits, perhaps because the greatly increased ribosomal density at the 5ʹ UTRs and at the beginnings of regular ORFs, which does not reflect the actual increase of translation, compromises the microarray-based approaches. In this regard, Ribo-seq has an advantage over microarrays. Overall, our study offers a more detailed view of the translational response to oxidative stress and leads to reevaluation of many translational targets of peroxide. We also observed a significant difference between mRNA abundance and its translation (Fig. S4E). Some mRNAs were not translated at all. Several genes had remarkably permissive posttranscriptional regulation upon hydrogen peroxide treatment. For example, Srx1, coding for sulfiredoxin, is present in unstressed yeast cells as a moderately transcribed gene with no detectable ribosomal occupancy. Its translation increases immediately after the addition of peroxide, increasing the TE by orders of magnitude. Srx1 reduces cysteine-sulfinic acid, formed upon reaction with hydrogen peroxide in the active sites of peroxiredoxins. Among them, Tsa1 is one of the major proteins contributing to stress resistance (30). An opposite example is PAB1, a polyA-binding protein mediating interactions between the 5′ cap structure and the 3′ mRNA poly(A) tail and facilitating translation. Treatment with hydrogen peroxide greatly decreased the TE of PAB1, but its transcript abundance remained unchanged.
Importantly, the degree of translational response to hydrogen peroxide did not match the transcriptional response precisely. There are multiple cases of posttranscriptional regulation in addition to the general transcriptional response. For example, 5-min incubation with the oxidant increased the TE of 32 genes and decreased the TE of 13. A longer incubation up-regulated 62 genes and down-regulated 122 (Dataset S1). This finding highlights our incomplete understanding of molecular mechanisms controlling gene expression. Increasing numbers of high-throughput studies involving S. cerevisiae and mammalian cells that address an interplay between translation and transcription suggest that these processes do not correlate perfectly with each other in either single-cell or culture-wide conditions (3, 31, 32).
Ribo-seq offers an improved experimental alternative to the codon adaptation index (21). It is able to detect differences between the TEs of synonymous codons. Ribo-seq may become a valuable tool for addressing the effects of deliberate starvation and amino acid depletion on codon-specific translation. Overall, our study defined the genome-wide regulation of translation by oxidative stress.
Materials and Methods
Additional details can be found in SI Materials and Methods. Primers used in library preparation are listed in Table S5.
Yeast Strains and Growth Conditions.
One milliliter of BY4741 strain (MATa his3 leu2 met15 ura3) from a frozen stock (OD600 of 0.6 in 15% glycerol) was added to 50 mL of yeast extract-peptone-glucose(YPD) medium, and the cells were grown for 16 h at 30 °C. A 1-mL aliquot of that culture was added to 400 mL of fresh YPD and grown to an OD600 of 0.5. This culture then was used for treatments and sample collection.
Preparation of Lysates.
The initial protocol was based on a previously described procedure (10, 11). Before the addition of peroxide, a 50-mL aliquot of culture was taken rapidly and pelleted by centrifugation for 1 min at 3,400 × g at 4 °C; then the pellet was frozen immediately in liquid nitrogen. This aliquot was used for mRNA isolation, and the rest of culture was used for footprints. The peroxide concentration used in this study was 0.2 mM with incubation times of 5 and 30 min.
Ribosome Fractionation and RNA Extraction.
A 50-U aliquot of cell extract (OD260) was used for footprints extraction. It was treated with 1,000 U of Escherichia coli RNase I (Ambion) and was incubated for 1 h at room temperature with gentle shaking to digest the mRNA. After fractionation in sucrose gradient, the monosomal fraction was collected, and footprints were isolated.
Library Construction for Footprint Sequencing.
Libraries were prepared with the strand information preserved to minimize ambiguously aligned reads. A protocol that included polyadenylation of RNA fragments and subsequent DNA circularization was used. The resulting libraries were sequenced on the Illumina GLx2 or HiSeq2000 platforms.
Bioinformatics Analyses.
In-house Perl scripts were used to prepare reference databases. Alignment of sequencing reads was performed by Bowtie software v.0.12.7 (33), allowing two mismatches per read. Because every read bears a polyA tail at the end, we omitted all “A” from the 3ʹ ends of sequences before aligning. A detailed description is given in SI Materials and Methods.
Codon Translation Analysis.
In an ideal situation, ribosomal footprints should be 28 nt in length. However, RNase I used to degrade unprotected mRNA segments occasionally left extra nucleotides or cut off extra nucleotides. By plotting a distribution of the footprint length, we found that RNase creates footprints that are mostly 27–29 nt in length (Fig. S4C). These footprints can be aligned to the reference ORFs, and the position of a footprint’s 5ʹ end relative to the reading frame can be obtained. If the 5ʹ end of a footprint matched the exact border of a codon, we considered it “ideal.” If the 5ʹ end of a footprint matched the position of a codon ±1 nt, we deleted or added the first nucleotide, respectively. Thus, we minimized the error in determining the ribosome position and defined which codon was located in the A site.
Differential Gene Translation Analysis.
All experimental samples were collected in duplicates. Based on correlation between the replicates, we set up a reads per kilobase per million mapped reads (rpkm) threshold of 10 for the genes whose translation and transcription could be determined reproducibly (Fig. S4 A and B). The gene was considered regulated if its rpkm value changed more than 2.6-fold (1.4 in log2 scale). This threshold eliminated most false-positive hits (Fig. S4D).
Supplementary Material
Acknowledgments
We thank Dr. Audrey Atkin (University of Nebraska-Lincoln) for technical support. This work was supported by National Institutes of Health Grant GM065204 (to V.N.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120799109/-/DCSupplemental.
References
- 1.Hinkson IV, Elias JE. The dynamic state of protein turnover: It’s about time. Trends Cell Biol. 2011;21:293–303. doi: 10.1016/j.tcb.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 3.Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 4.Bar-Even A, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 5.Arava Y, et al. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shenton D, et al. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem. 2006;281:29011–29021. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- 7.Halbeisen RE, Gerber AP. Stress-dependent coordination of transcriptome and translatome in yeast. PLoS Biol. 2009;7:e1000105. doi: 10.1371/journal.pbio.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arava Y, Boas FE, Brown PO, Herschlag D. Dissecting eukaryotic translation and its control by ribosome density mapping. Nucleic Acids Res. 2005;33:2421–2432. doi: 10.1093/nar/gki331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozsolak F, Milos PM. RNA sequencing: Advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingolia NT. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol. 2010;470:119–142. doi: 10.1016/S0076-6879(10)70006-9. [DOI] [PubMed] [Google Scholar]
- 12.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 14.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 15.Lawless C, et al. Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast. BMC Genomics. 2009;10:7. doi: 10.1186/1471-2164-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang KJ, Wang CC. Translation initiation from a naturally occurring non-AUG codon in Saccharomyces cerevisiae. J Biol Chem. 2004;279:13778–13785. doi: 10.1074/jbc.M311269200. [DOI] [PubMed] [Google Scholar]
- 18.Tang HL, et al. Translation of a yeast mitochondrial tRNA synthetase initiated at redundant non-AUG codons. J Biol Chem. 2004;279:49656–49663. doi: 10.1074/jbc.M408081200. [DOI] [PubMed] [Google Scholar]
- 19.Asakura T, et al. Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene. 1998;16:121–130. doi: 10.1038/sj.onc.1201487. [DOI] [PubMed] [Google Scholar]
- 20.Tuller T, et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 21.Sharp PM, Li WH. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nanda JS, et al. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J Mol Biol. 2009;394:268–285. doi: 10.1016/j.jmb.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanov IP, Loughran G, Sachs MS, Atkins JF. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1) Proc Natl Acad Sci USA. 2010;107:18056–18060. doi: 10.1073/pnas.1009269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207–218. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 29.Ling J, Söll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc Natl Acad Sci USA. 2010;107:4028–4033. doi: 10.1073/pnas.1000315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 31.Lu R, et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462:358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 33.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.