Abstract
Programmed −1 ribosomal frameshifting (−1 PRF) is a gene-expression mechanism used to express many viral and some cellular genes. In contrast, efficient natural utilization of −2 PRF has not been demonstrated previously in eukaryotic systems. Like all nidoviruses, members of the Arteriviridae (a family of positive-stranded RNA viruses) express their replicase polyproteins pp1a and pp1ab from two long ORFs (1a and 1b), where synthesis of pp1ab depends on −1 PRF. These polyproteins are posttranslationally cleaved into at least 13 functional nonstructural proteins. Here we report that porcine reproductive and respiratory syndrome virus (PRRSV), and apparently most other arteriviruses, use an additional PRF mechanism to access a conserved alternative ORF that overlaps the nsp2-encoding region of ORF1a in the +1 frame. We show here that this ORF is translated via −2 PRF at a conserved G_GUU_UUU sequence (underscores separate ORF1a codons) at an estimated efficiency of around 20%, yielding a transframe fusion (nsp2TF) with the N-terminal two thirds of nsp2. Expression of nsp2TF in PRRSV-infected cells was verified using specific Abs, and the site and direction of frameshifting were determined via mass spectrometric analysis of nsp2TF. Further, mutagenesis showed that the frameshift site and an unusual frameshift-stimulatory element (a conserved CCCANCUCC motif 11 nucleotides downstream) are required to direct efficient −2 PRF. Mutations preventing nsp2TF expression impair PRRSV replication and produce a small-plaque phenotype. Our findings demonstrate that −2 PRF is a functional gene-expression mechanism in eukaryotes and add another layer to the complexity of arterivirus genome expression.
Keywords: Nidovirales, virology, genetic recoding, overlapping gene, translation
In eukaryotes, translation initiation largely involves 5′ end-dependent scanning of mRNAs during which the small ribosomal subunit, in a complex with initiation factors, first binds to the 5′ cap structure and then scans in a 5′-to-3′ direction until it encounters the first suitable initiation codon, at which point translation commences (1). Consequently, the vast majority of cellular mRNAs are essentially monocistronic (although efficient reinitiation can occur after translation of very short ORFs). The fact that the cellular translational machinery essentially only decodes the 5′-most long ORF of an mRNA imposes a considerable constraint on nonsegmented RNA viruses, which must express a number of enzymatic and structural proteins to complete their replicative cycle. Strategies to overcome this limitation include the synthesis of functionally monocistronic subgenomic mRNAs, the production of precursor polyproteins that are subsequently cleaved by virus- and/or host-encoded proteases, and the use of noncanonical translational mechanisms (such as internal ribosomal entry, leaky scanning, ribosomal frameshifting, and stop codon readthrough) (2) by which additional ORFs may be translated from polycistronic mRNAs.
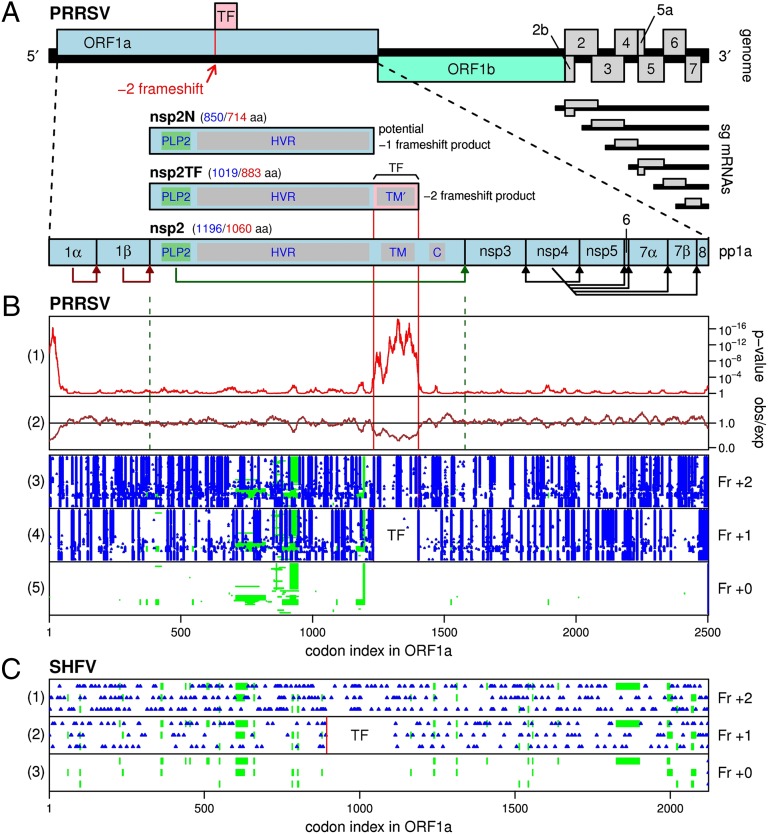
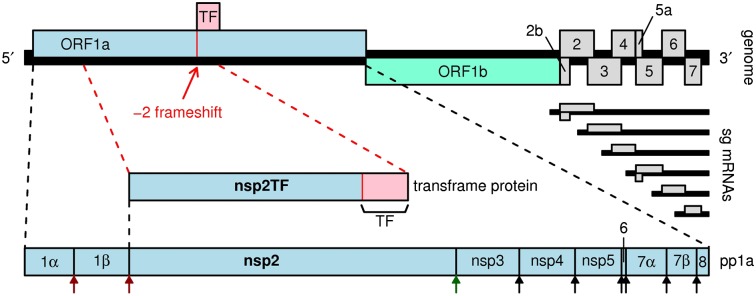
Members of the order Nidovirales (Arteriviridae, Coronaviridae, and Roniviridae), which includes the RNA viruses with the largest genomes currently known, use many of the above strategies (including polyprotein expression, subgenomic mRNA synthesis, ribosomal frameshifting, and leaky scanning) (Fig. 1A) to organize one of the most complex RNA virus replication cycles described to date (3–5). The replicative enzymes of nidoviruses are encoded in ORF1a and ORF1b, which occupy the 5′-proximal three quarters of their positive-stranded RNA genome. ORF1a and ORF1b encode two large replicase precursor polyproteins, pp1a and pp1ab, with expression of the latter depending on a −1 ribosomal frameshift in the short ORF1a/ORF1b overlap region (6, 7). Following their synthesis from the genomic mRNA template, pp1a and pp1ab are processed into at least 13–16 functional nonstructural proteins (nsps) by a complex proteolytic cascade that is directed by two to four ORF1a-encoded proteinase domains (Fig. 1A) (8–9). The 3′-proximal region of the nidovirus genome contains the genes encoding the viral structural proteins, which are translated from a nested set of 5′- and 3′-coterminal subgenomic mRNAs (10).
Fig. 1.
Arterivirus genome organization and expression mechanisms. (A) Map of the ∼15-kb PRRSV genome. Two long 5′ ORFs encode nonstructural polyproteins, and at least eight shorter 3′ ORFs encode structural proteins. The 3′ ORFs are translated from a nested set of 3′-coterminal subgenomic mRNAs, two of which are bicistronic. ORF1a and ORF1b are translated from the genomic RNA, where translation of ORF1b depends on −1 PRF at the end of ORF1a. The newly described TF ORF overlaps a central region of ORF1a in the +1 reading frame and is accessed via −2 PRF. Domains in nsp2/nsp2TF are annotated as PLP2 (papain-like protease), HVR (hypervariable region), TM/TM′ (putative TM domains), and C (Cys-rich domain). Predicted sizes (in aa) for nsp2-related products are shown for GenBank sequences NC_001961 (blue) and DQ489311 (red; the isolate SD01-08 used in this study). (B) Bioinformatic analysis of PRRSV ORF1a. Panels 1 and 2 depict the conservation at ORF1a-frame synonymous sites in an alignment of 212 PRRSV sequences using a 25-codon sliding window. Panel 2 shows the ratio of the observed number of substitutions to the number expected under a null model of neutral evolution at synonymous sites. Panel 1 shows the corresponding P value. Summed over the whole TF ORF, the corresponding P value is 5.7 × 10−65. To map the conservation statistic onto the coordinates of a specific sequence, all alignment columns with gaps in a chosen reference sequence, NC_001961, were removed (note that the original alignment is gap-free within the TF ORF itself). Panels 3–5 show the positions of stop codons (blue) in the three possible reading frames, and alignment gaps (green) in all 212 aligned sequences. Note the conserved absence of stop codons in the +1 reading frame in the TF region. (C) Positions of stop codons (blue) in an alignment of the three available SHFV sequences. The vertical red line indicates the location of the G_GUU_UUU/G_GUC_UCU motif. Note the conserved absence of stop codons in the +1 reading frame for 220 codons immediately following this site.
As indicated above, the genomes of a variety of viruses, including all currently known nidoviruses, harbor sequences that induce a proportion of translating ribosomes to frameshift −1 nt and continue translating in an alternative reading frame (6, 7, 11–13). Where functionally important, this process may be termed “−1 programmed ribosomal frameshifting” (−1 PRF). The eukaryotic −1 frameshift site typically consists of a “slippery” heptanucleotide fitting the consensus motif X_XXY_YYZ, where XXX normally represents any three identical nucleotides (although certain exceptions have been found); YYY represents strictly AAA or UUU; and Z represents A, C, or U, and underscores separate zero-frame codons. This consensus motif generally is followed by a stimulatory element that comprises a stable RNA secondary structure, such as a pseudoknot or stem-loop, beginning 5–9 nt downstream of the shift site (11, 13). In contrast, very little is known about the utilization of ‒2 programmed ribosomal frameshifting (‒2 PRF) in eukaryotic systems, including the potential shift sites and stimulatory elements.
Here we describe the identification of a short arterivirus ORF (TF) that is translated via efficient −2 PRF. In porcine reproductive and respiratory syndrome virus (PRRSV), frameshifting (with an efficiency of about 20%) occurs at a conserved G_GUU_UUU sequence (underscores separate ORF1a codons) in a central region of ORF1a and results in the expression of a transframe protein, nsp2TF, that comprises the N-terminal two thirds of nsp2 fused to a 169-aa C-terminal region encoded by the TF ORF. Mutations that prevent expression of nsp2TF seriously impair PRRSV replication in cell culture. Because ribosomes translating the TF ORF will not decode the remainder of the replicase gene, the combination of this −2 PRF mechanism and the downstream ORF1a/ORF1b −1 PRF results in differential expression of three replicase gene segments, suggesting that the regulation of genome translation plays a key role in fine-tuning the replicative cycle of arteriviruses, including PRRSV, one of the economically most important swine pathogens.
Results
Computational Analysis Reveals a Conserved ORF Overlapping the Arterivirus nsp2-Coding Sequence.
Arteriviruses comprise a family of small, enveloped positive-stranded RNA viruses that currently includes PRRSV, equine arteritis virus (EAV), lactate dehydrogenase-elevating virus (LDV), and simian hemorrhagic fever virus (SHFV) (5). The ORF1a sequences of 212 PRRSV isolates available in GenBank as of February 26, 2012 were extracted, translated, aligned, and back-translated to a nucleotide sequence alignment. Both European (EU; type I) and North American (NA; type II) genotype isolates, which typically share 50–55% amino acid identity within pp1a, were included. Next, the alignment was analyzed for conservation at ORF1a-synonymous sites, as described previously (14). The analysis revealed a striking and highly statistically significant (P < 10−64) increase in synonymous-site conservation in a region covering around 170 codons toward the 3′ end of the nsp2-encoding sequence (Fig. 1B). Within this region the mean synonymous substitution rate was reduced to 47% of the ORF1a average. Such peaks in synonymous-site conservation generally are indicative of functionally important overlapping elements, either coding or noncoding (14–16). In this case, inspection of the +1 and +2 reading frames relative to ORF1a, in all 212 sequences, revealed an almost complete absence of stop codons in the +1 reading frame in a region corresponding precisely to the region of enhanced conservation (Fig. 1B). This finding suggested an overlapping ORF in the +1 reading frame as a possible explanation for the enhanced conservation at ORF1a synonymous sites.
An inspection of other arterivirus genomes revealed further evidence for a +1 frame ORF overlapping the equivalent region of ORF1a. Currently, three SHFV sequences are available (17). With pairwise amino acid identities within pp1a of just 35–37%, these sequences are too divergent for the analysis of synonymous-site conservation. However, the conserved presence of a 219–225 codon ORF in the +1 frame in such divergent sequences is, in itself, statistically significant (P < 10−10) (Fig. 1C; also see SI Materials and Methods). A 169-codon ORF also is present in one (LDV-P) of two published LDV sequences, but in the second sequence (LDV-C) the ORF is disrupted by a single stop codon. To assess the likeliness of a sequencing error in the LDV-C sequence, we sequenced the relevant region of an additional LDV isolate (795 nt, including the last 158 codons of the ORF; GenBank accession no. JX258842). This sequence is divergent from both LDV-P and LDV-C (locally 83–90% nucleotide identity) and lacks the interrupting stop codon that is present in LDV-C. Remarkably, no evidence for a corresponding ORF was found in EAV. In fact, in this part of the genome, EAV is highly divergent from other arteriviruses, and the nsp2 region is greatly reduced in size.
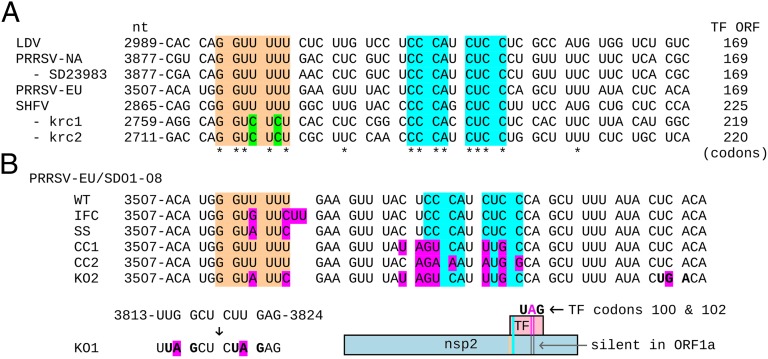
At the 5′ end of the conserved ORF, there is a G_GUU_UUU sequence that is present in 206/212 PRRSV sequences (4/212 have G_GUU_UUC), both LDV sequences, and one SHFV sequence. The other two SHFV sequences have G_GUC_UCU at the corresponding position. Significantly, the G at position 1 is conserved although the corresponding ORF1a codon is CAG (Gln), UGG (Trp), or CGG (Arg) in different PRRSV isolates (Fig. 2A). We hypothesized that this motif could facilitate −2 PRF from ORF1a into the overlapping ORF. We were not able to predict convincing downstream RNA secondary structures at a distance of 5–9 nt that might (by analogy to −1 PRF sites) be expected to be present, nor could we definitively rule out the existence of such structures. On the other hand, we did observe a highly conserved CCCANCUCC motif beginning 11 nt downstream of the G_GUU_UUU sequence. This motif is present in both LDV, all three SHFV, and 211/212 PRRSV sequences. Such high conservation could reflect protein sequence constraints (in two overlapping reading frames) but also might be part of a frameshift-stimulatory RNA sequence.
Fig. 2.
Nucleotide sequences in the vicinity of the TF frameshift site. (A) Sequences from representative arteriviruses [GenBank accession numbers: NC_001639, LDV; NC_001961, PRRSV NA (type II); JX258843, PRRSV NA (type II) isolate SD23983; DQ489311, PRRSV EU (type I) isolate SD01-08; NC_003092, SHFV; HQ845737-8. SHFV strains krc1-2]. The proposed frameshift site (confirmed in SD01-08 and SD23983) is highlighted in orange, and nucleotide variations in SHFV isolates krc1-2 are indicated in green. The highly conserved downstream CCCANCUCC motif is highlighted in cyan. Spaces separate ORF1a codons. The length of the +1 frame TF ORF is indicated at the right. (B) Overview of mutants used to investigate TF expression and function by ORF1a expression and reverse genetics. Non-WT nucleotides are shown in pink. Coordinates of terminal nucleotides refer to sequence DQ489311. IFC, in-frame control; SS, shift-site mutant; CC1 and CC2, disrupted CCCANCUCC motif; KO1, knockout mutant 1 (premature termination codons in TF); KO2, knockout mutant 2 (premature termination codon and disrupted frameshift cassette). Only the IFC and CC2 mutations are nonsynonymous with respect to the nsp2 amino acid sequence.
In PRRSV, −2 frameshifting at the G_GUU_UUU sequence would produce a transframe fusion protein comprising the N-terminal 65–72% (typically 714–850 amino acids, depending on isolate) of nsp2 fused to the 169 amino acids encoded by the overlapping ORF (Fig. 1A). We refer to the overlapping ORF as “TF ORF” and to the predicted transframe fusion protein as “nsp2TF.” In PRRSV, LDV, and SHFV, nsp2TF is 14–19% shorter than full-length nsp2, and the TF ORF overlaps the part of ORF1a that encodes the predicted nsp2 transmembrane (TM) domain. The TF ORF appears to encode an alternative TM domain containing four or more potential TM regions, depending on species and isolate, but no other conserved amino acid motifs that might give further indications as to protein function were identified.
Immunodetection of nsp2TF in PRRSV-Infected Cells.
To confirm expression of the predicted nsp2TF frameshift product in PRRSV-infected cells, a polyclonal Ab (pAb-TF) was raised against the C-terminal peptide (CPKGVVTSVGESV) of nsp2TF of PRRSV type I isolate SD01-08 (18). We also used mAbs 36-19 and 58-46, raised against the N-terminal 436 amino acids of SD01-08 nsp2 (18, 19) and therefore expected also to recognize nsp2TF, which (in SD01-08) would share its N-terminal 714 amino acids with nsp2.
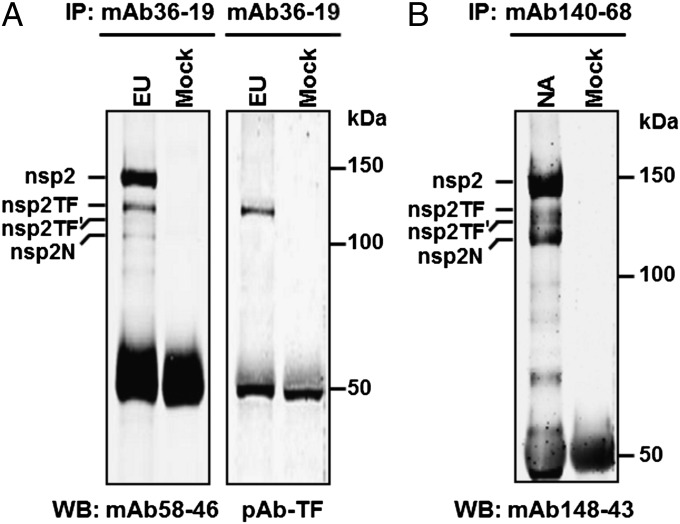
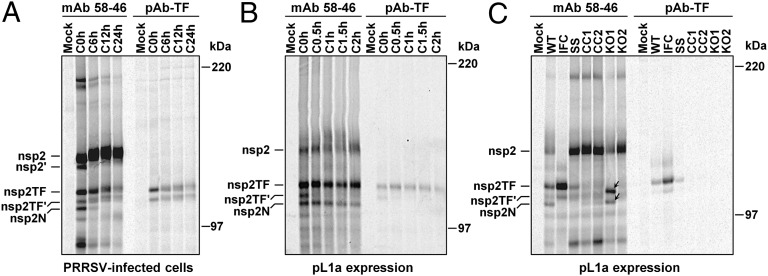
Expression of nsp2TF was first analyzed by immunoprecipitation (IP) and Western blot analysis. Lysates of SD01-08–infected MARC-145 cells were harvested at 48 h postinfection (p.i.). PRRSV proteins were immunoprecipitated using mAb36-19 and were separated by SDS/PAGE. Four high-molecular-mass bands with apparent masses between 100–150 kDa were detected by Coomassie Brilliant Blue staining, with the two smallest products clearly being less abundant. In addition, various lower-molecular-mass products were observed (Fig. S1A). To confirm that the high-molecular-mass bands represented nsp2-related products, Western blot analysis was performed using anti-nsp2 mAb58-46 and pAb-TF. All four high-molecular-mass products were specifically recognized by mAb58-46 (Fig. 3A), indicating that they must share N-terminal sequences. However, only the second-largest protein labeled nsp2TF was recognized by pAb-TF. The third protein labeled nsp2TF′ was not detected in a Western blot using pAb-TF, possibly because of its lower abundance and/or lower affinity for the Ab; this product was, however, detected in IP using pAb-TF (see below). We speculate that nsp2TF′ could be either a precursor to or a modified form of nsp2TF. The smallest product of the four proteins labeled nsp2N might derive from a −1 frameshift at the same conserved G_GUU_UUU sequence. Such a frameshift would lead to an immediate termination, because there is a −1/+2 frame stop codon adjacent to the shift site (Fig. 2A; see also Fig. S1D and Discussion).
Fig. 3.
Analysis of nsp2 and nsp2TF expression in PRRSV-infected cells. MARC-145 cells were infected with type I (EU) PRRSV isolate SD01-08 (A) or type II (NA) PRRSV isolate SD23983 (B) or were mock infected. Proteins were immunoprecipitated with mAbs specific for the common N-terminal domain of nsp2 and nsp2TF, separated by SDS/PAGE, and probed by Western blotting using an alternative mAb recognizing the common N-terminal domain or a TF-specific pAb (SD01-08 only), as indicated at the bottom. Size markers and putative PRRSV proteins are indicated.
A similar analysis for the PRRSV type II isolate SD23983 also revealed multiple products in the 100–150 kDa range (Fig. 3B). The available SD01-08 pAb-TF did not cross-react with any of these bands, but this result was not surprising, because the sequence of the 13-aa peptide used to produce this Ab is not conserved in type II viruses.
Mass Spectrometric Analysis of the Site and Direction of Frameshifting.
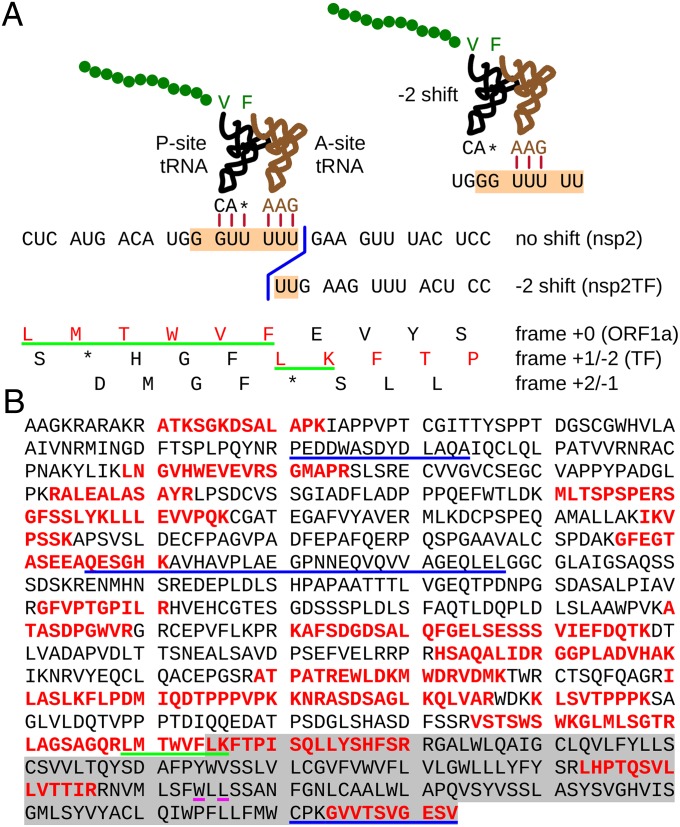
Although bioinformatic analysis suggested the site (G_GUU_UUU) and direction (−2) of frameshifting, we sought to confirm both predictions with direct protein sequence analysis. To this end, proteins from SD01-08–infected and mock-infected MARC-145 cell lysates were immunoprecipitated, resolved by SDS/PAGE, and stained with Coomassie Brilliant Blue (Fig. S1A). The gel slice containing the putative nsp2TF band was analyzed by LC/MS/MS. Four peptides specific for the 169-aa TF region were identified in addition to many peptides from the common N-terminal domain of nsp2 and nsp2TF (Fig. 4B). One of the peptides, LMTWVFLK, spanned the frameshift site itself (bold), and its sequence is fully compatible with −2 PRF (after decoding GUU_UUU as VF) (Fig. 4A and Fig. S1C) but not with +1 PRF, which would produce a shift-site peptide that is shorter by one amino acid (e.g., LMTWVFK). To verify the correct identification of the frameshift peptide, a synthetic version of the peptide was subjected to the same LC/MS/MS analysis. The tandem mass spectrum of the synthetic peptide was identical to that of the peptide derived from the gel slice, confirming that nsp2TF is indeed translated via −2 PRF at the G_GUU_UUU motif (Fig. S1B). Again, the analysis was repeated for the type II isolate SD23983, for which a −2 frameshift tryptic peptide with a different sequence (QVFLTSSPISLFSSHAFSTR; shift site-encoded amino acids in bold) was predicted. Likewise, this sequence was identified in mass spectrometric analysis of the presumed nsp2TF band excised from an SDS/PAGE gel (Fig. S2).
Fig. 4.
Mass spectrometric analysis of nsp2TF purified from MARC-145 cells infected with type I PRRSV isolate SD01-08. (A) Nucleotide sequence in the vicinity of the TF shift site G_GUU_UUU, with conceptual amino acid translations in all three reading frames shown. The product of −2 frameshifting is indicated in red. Consecutive tryptic peptides covering these amino acids were detected by mass spectrometric analysis. The peptide underlined in green, which spans the shift site, is compatible with −2 but not +1 frameshifting. Stylized P- and A-site tRNAs illustrate expected codon:anticodon duplexes before and after frameshifting (see also main text). In eukaryotes, GUU is expected to be decoded by the valine tRNA with anticodon 5′-IAC-3′ (I = inosine), but it is possible that it also is decoded by the valine tRNA with anticodon 5′-ncm5UAC-3′ (ncm5U = 5-carbamoylmethyluridine). Because currently it is not known whether one or both of these, or perhaps a hypomodified form, are compatible with frameshifting, the valine anticodon is indicated by 3′-CA*-5′ in the schematic. (B) Complete amino acid sequence of nsp2TF, with peptides identified by mass spectrometry indicated in red. The C-terminal 169 amino acids encoded by the +1 reading frame are highlighted in gray. The N-terminal 714 amino acids are shared with nsp2. The epitopes recognized by mAbs 36-19 and 58-64 and pAb-TF, in order, are underlined in blue. Pink underlining indicates the locations of the premature termination codons of mutant KO1 (Fig. 2B).
Estimation of the Frameshifting Efficiency.
The frameshifting efficiency and turnover of nsp2 and nsp2TF were investigated in a pulse-chase labeling experiment in SD01-08–infected MARC-145 cells. After a 1-h pulse labeling with 35S-labeled amino acids, the incorporated label was chased for various periods (up to 24 h), and proteins were immunoprecipitated with nsp2- and nsp2TF-specific Abs. As shown in Fig. 5A, this analysis revealed the existence of two smaller products (labeled “nsp2′” and “nsp2TF′”) that apparently are about 10 kDa smaller than nsp2 and nsp2TF. Its disappearance during the chase suggested that nsp2′ is a direct precursor of nsp2, although we cannot rule out the possibility that it may be a degradation product of nsp2. Nsp2 itself also appeared to be subject to further modification during the chase period, as indicated by its slight size increase and more heterogeneous migration in the gel (Fig. 5A; compare C0h with later time points). The possible precursor status of nsp2TF′ was less obvious, because the amount of this product was more or less stable throughout the chase period, during which both the nsp2TF and nsp2TF′ bands also appeared to convert into doublets. In terms of protein turnover, the amount of nsp2TF declined much more rapidly than that of nsp2 during the chase period. This difference in turnover was even more pronounced for nsp2N, the putative −1 frameshift product (see above), which seems to be the least stable of the various nsp2 forms described here.
Fig. 5.
Analysis of nsp2-related products in PRRSV SD01-08–infected cells and transient ORF1a expression. Following metabolic labeling with 35S, proteins were immunoprecipitated with mAb58-46 or pAb-TF and were analyzed by SDS/PAGE and autoradiography. Size markers and putative nsp2-related products are indicated at the side of each panel. (A) Pulse-chase experiment with PRRSV-infected MARC-145 cells, using a 1-h pulse labeling and various chase periods as indicated at the top. (B) Pulse-chase analysis of transient ORF1a expression in the recombinant vaccinia virus/T7 RNA polymerase system. RK-13 cells were infected with recombinant vaccinia virus and either were mock transfected or were transfected with pL1a plasmid DNA. At 5 h post vaccinia virus infection, protein synthesis was labeled for 30 min and chased for up to 2 h. (C) Analysis of −2 PRF and nsp2TF mutants expressed using pL1a and the recombinant vaccinia virus/T7 RNA polymerase expression system as described for B. Arrows point to the C-terminally truncated nsp2TF products from mutant KO1.
Several smaller nsp2-specific products were observed in the C0h sample, including a prominent band migrating at around 90 kDa, a fainter band migrating slightly behind the 90-kDa band, and a faint band migrating at around 98 kDa (Fig. 5A). The prominent 90-kDa product was not detected by pAb-TF and most likely derives from internal cleavage of nsp2. Two additional nsp2-specific products were observed to migrate at around 200 kDa. Based on their size, these may represent nsp2–8 and a modified or precursor form of nsp2–8. The definitive identity of these products cannot be explained at this point in time, and they add to the puzzling complexity of nsp2 expression and posttranslational modification.
To assess the frameshifting efficiencies in the context of PRRSV infection, we measured the radioactive incorporation into the nsp2+nsp2′, nsp2TF+nsp2TF′, and nsp2N bands directly after the pulse labeling (Fig. 5A; lane C0h) and corrected these numbers for the Met and Cys content of the different proteins (Table S1). Notwithstanding potential differences in Ab affinity for the different nsp2-related products and other potential confounding factors, these measurements suggested −2 and (putative) −1 frameshifting efficiencies of ∼20% and 7%, respectively. Because the presence and identity of the minor products, including those migrating at around 90, 98, and 200 kDa, could affect the calculated frameshifting efficiencies to a limited extent, we repeated the calculations under the conservative assumption that these minor products are all derived from nonframeshift products. These results, combined with those above, indicate frameshifting efficiencies in the range of 16–20% and 6–7% for −2 and −1 frameshifting, respectively.
The G_GUU_UUU and CCCANCUCC Motifs Are Required for Efficient Frameshifting.
The transient expression of ORF1a in the recombinant vaccinia virus/T7 polymerase expression system (19) was used to develop an assay for PRRSV −2 PRF in uninfected cells. A T7 promoter-driven, full-length ORF1a expression vector (pL1a) was constructed, and the synthesis of nsp2, nsp2TF, and nsp2N was monitored by radiolabeling (30-min pulse, 0- to 120-min chase) and IP (Fig. 5B). With the exception of nsp2′, all products immunoprecipitated from PRRSV-infected cell lysates (Fig. 5A) could be identified. Although differences in abundance, time of appearance, and stability were observed, these data clearly demonstrated that translation of the PRRSV ORF1a sequence is sufficient to allow efficient −2 frameshifting. In fact, the efficiency of frameshifting was estimated to be even higher in this system than in virus-infected cells (with an estimated −2 shift efficiency of around 50%).
Subsequently, using this ORF1a expression system, we investigated the requirement for an intact shift site and downstream elements for efficient frameshifting. First we engineered a shift-site mutant (SS) (Fig. 2B) that contains two mutations in the shift site (G_GUU_UUU to G_GUA_UUC) and therefore is expected to express only nsp2. To mark the position at which WT nsp2TF migrates in gels, an in-frame control (IFC) was constructed in which the shift site was mutated synonymously and an extra 2 nt were inserted to force expression of the TF reading frame (G_GUU_UUU to G_GUG_UUC_UU) (Fig. 2B). As shown in Fig. 5C, WT pL1a produced both nsp2 and nsp2TF, and expression of the latter protein was confirmed by IP with pAb-TF. As expected, the IFC mutant produced only nsp2TF, as detected by both pAb-TF and mAb58-46. For the SS mutant, production of nsp2TF was greatly reduced, although, unexpectedly, frameshifting was not totally inhibited (Fig. 5C). Next we investigated whether the conserved CCCANCUCC motif (see above and Fig. 2A) is involved in the stimulation of frameshifting at the G_GUU_UUU shift site by constructing two additional mutants in which the CCCANCUCC motif was disrupted (CC1 and CC2) (Fig. 2B). The CC1 mutations are synonymous in the nsp2 frame; the CC2 mutations disrupt the motif more thoroughly but include substitutions that are not synonymous with respect to nsp2. The CC1 and CC2 mutants produced nsp2, but no nsp2TF was detected (Fig. 5C). These results indicate that both the G_GUU_UUU shift site and the downstream CCCANCUCC motif are required for ribosomal frameshifting at the WT efficiency. When frameshifting is prevented (in the mutants SS, CC1, and CC2), expression of nsp2 is expected to increase substantially, because ribosomes no longer are being diverted into the alternative reading frame (Fig. 5C).
Inactivation of nsp2TF Expression Affects PRRSV Replication.
To investigate whether nsp2TF expression and/or the diversion of a proportion of the ribosomes out of ORF1a are relevant for virus replication, two mutants were generated (KO1 and KO2) (Fig. 2B) in which nsp2TF expression was partially or completely knocked out with mutations that are translationally silent with respect to ORF1a. KO1 makes a truncated nsp2TF protein because of the mutagenesis of codons 100 and 102 of the 169-codon TF ORF into stop codons (nsp2-frame UUG_GCU_CUU_GAG to UUA_GCU_CUA_GAG; stop codons indicated in bold) (Figs. 2B and 4B). Consequently, the truncated nsp2TF lacks the C-terminal pAb-TF epitope. KO2 contains nine mutations that disrupt the frameshift site and the downstream CCCANCUCC motif, besides introducing a stop codon into the TF ORF (Fig. 2B). This mutant was intended to knock out the frameshift signal completely and to express only nsp2. KO1 and KO2 differ from WT virus by two and nine nucleotide substitutions, respectively, none of which affect the encoded nsp2 amino acid sequence (Fig. 2B).
The mutants were tested in the ORF1a expression system to verify synthesis of the expected proteins. As expected, the KO1 frameshift product could not be detected using pAb-TF because of the truncation of the C-terminal epitope region. However, IP with mAb58-64 revealed the synthesis of truncated forms of nsp2TF and nsp2TF′ (Fig. 5C, arrows) and a ratio between full-length nsp2 and the frameshift product similar to that in WT. On the other hand, the mutations introduced in KO2 indeed were found to eliminate frameshifting, and, as anticipated, a much larger amount of full-length nsp2 was produced (Fig. 5C).
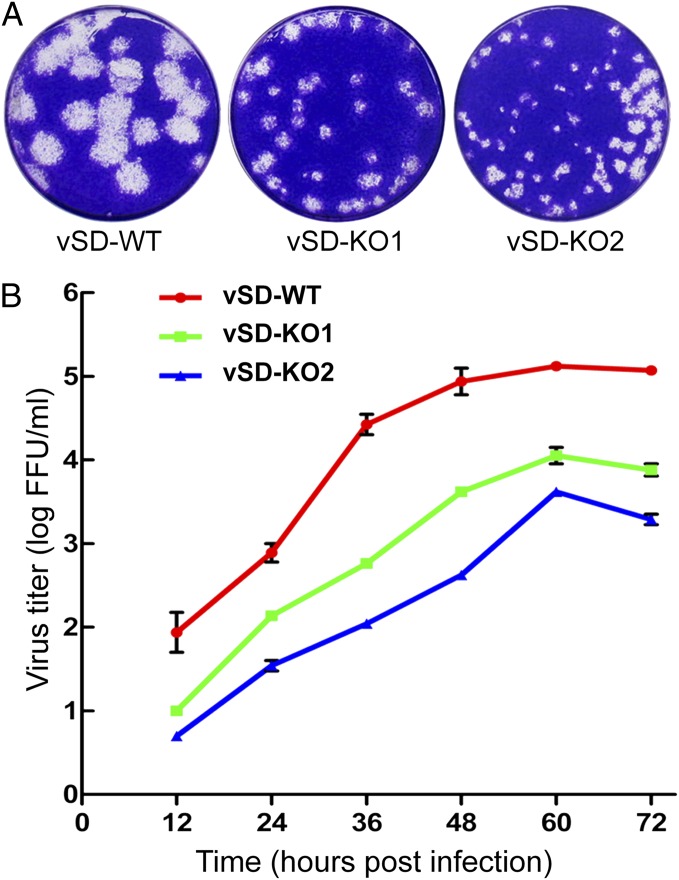
The KO1 and KO2 mutations subsequently were transferred to a PRRSV full-length cDNA infectious clone, and the resulting recombinant viruses, vSD-KO1 and vSD-KO2, were found to be viable. However, both mutants produced plaques that were clearly smaller than those of the WT virus (vSD-WT), with vSD-KO2 producing somewhat smaller plaques than vSD-KO1 (Fig. 6A). Analysis of growth kinetics consistently showed that replication of both vSD-KO1 and vSD-KO2 is seriously impaired in MARC-145 cells, with peak titers of both mutants being 50–100-fold lower than those of the WT control, and vSD-KO2 again displaying the larger reduction (Fig. 6B).
Fig. 6.
Comparison of growth characteristics of WT PRRSV and nsp2TF knockout mutants. (A) Plaque morphology of WT and nsp2TF knockout PRRSVs on MARC-145 cells. Confluent cell monolayers were infected with 10-fold serial dilutions of the virus suspension, and after 2 h an agar overlay was applied. Plaques were detected after 4 d of incubation at 37 °C and stained with 0.1% crystal violet. (B) Growth kinetics of WT and nsp2TF-knockout PRRSV on MARC-145 cells. The results shown are mean values from three replicates. Virus titers are expressed as numbers of fluorescent-focus units (FFU) per milliliter.
PRRSV nsp2 and nsp2TF Localize to Different Intracellular Compartments.
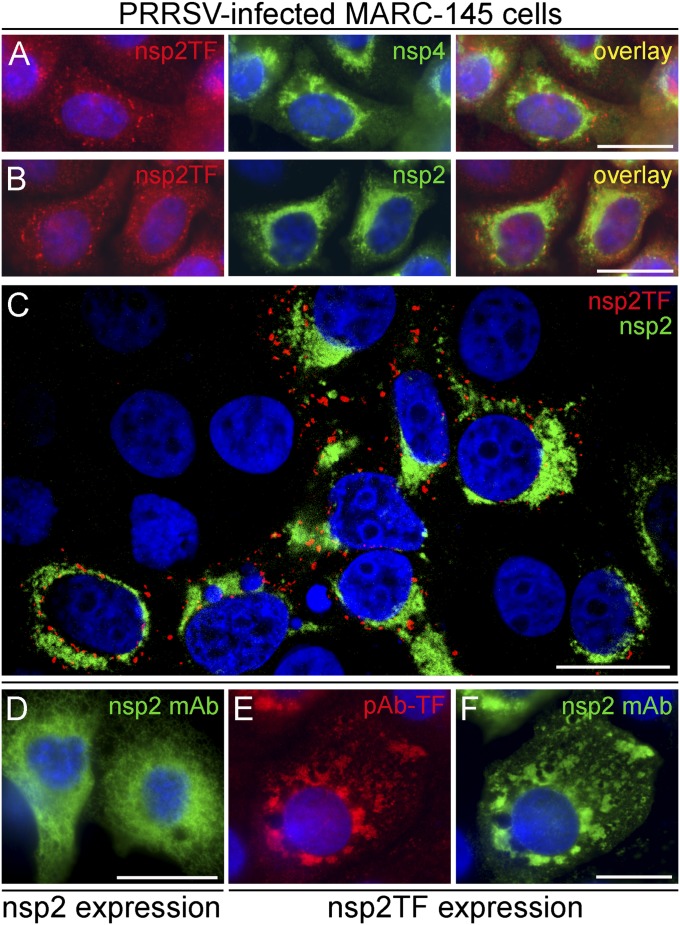
The localization of nsp2TF in PRRSV SD01-08–infected cells was investigated using immunofluorescence (IF) microscopy. As shown in Fig. 7A, pAb-TF labeled specific foci mainly localizing to the perinuclear region of infected cells. This region also is known to contain the arterivirus replication structures, modified membranes that label abundantly for most of the viral nsps (19–21). We used an mAb that recognizes nsp4 (19) to visualize these structures, which revealed a labeling pattern clearly different from that observed with pAb-TF. Surprisingly, a control labeling with nsp2-specific mAbs (36-19 and 58-46, recognizing different epitopes in the common N-terminal domain of nsp2 and nsp2TF) (Fig. 4B) revealed that the pAb-TF–specific foci were not, or were barely, recognized by nsp2-specific mAbs (Fig. 7B), even though these mAbs did detect nsp2TF convincingly in other immunoassays (Figs. 3 and 5). Apparently, this part of nsp2TF is not accessible in these formaldehyde-fixed, Triton X-100-permeabilized cells. Confocal microscopy further corroborated that the labeling patterns obtained with nsp2-specific mAbs and pAb-TF in PRRSV-infected cells did not overlap (Fig. 7C).
Fig. 7.
IF microscopy analysis of the subcellular localization of nsp2TF in PRRSV-infected MARC-145 cells at 18–24 h p.i. (A–C) and a CMV promoter-driven expression system for nsp2 and nsp2TF (D–F). (A) Infected cells were double labeled with pAb-TF and a PRRSV nsp4-specfic mAb (mAb54-19) (19), revealing that nsp2TF does not colocalize with the viral replication structures. (B) Double labeling with pAb-TF and PRRSV nsp2-specific mAb36-19 results in nonoverlapping labeling patterns suggesting that the nsp2-specific mAb cannot access the N-terminal domain of nsp2TF in fixed PRRSV-infected cells. (C) Confocal microscopy of a 0.8-μm slice through the nucleus confirming the nearly complete spatial separation of the structures labeled with pAb-TF and mAb36-19. (D) Nsp2-expressing MARC-145 cells labeled with mAb58-46. (E and F) Nsp2TF-expressing MARC-145 cells double labeled with pAb-TF (E) and mAb58-46 (F) show that, in contrast to PRRSV-infected cells, the latter antibody is able to recognize nsp2TF in this expression system. Cell nuclei were stained with Hoechst 33342. (Scale bars, 20 nm.) See also Fig. S3.
To investigate this phenomenon in more detail, nsp2 and nsp2TF were expressed individually in MARC-145 cells (nsp2TF was expressed using the in-frame control sequence to mimic −2 frameshifting) (Fig. 2B). In the absence of replication structures and other nsps, arterivirus nsp2 localizes to endoplasmic reticulum (ER) (Fig. 7D), as was confirmed using an Ab recognizing an ER-specific marker protein (Fig. S3F). Individual expression of nsp2TF (Fig. 7 E and F and Fig. S3E) again yielded a pattern clearly different from that observed for nsp2 expression. It resembled the foci found in infected cells, but in this case these foci could be labeled with both pAb-TF and mAbs recognizing the N-terminal domain of nsp2, suggesting a conformational difference between nsp2TF (or nsp2TF-containing structures) in infected cells and the expression system. In both systems, however, the foci labeled by pAb-TF partially overlapped with the staining for exocytic pathway markers (specifically intermediate compartment and Golgi complex) (Fig. S3 A–D), an observation that currently is being investigated in more detail. Together, our microscopy studies strongly suggest that, unlike nsp2, nsp2TF is targeted not to the arterivirus replication structures but instead to an alternative intracellular destination.
Discussion
RNA viruses of eukaryotes use a variety of noncanonical translational mechanisms to express multiple proteins from a limited number of transcripts, regulate gene expression, and otherwise manipulate the host cell translational machinery for their own specific needs. However, in contrast to −1 PRF, little was known about the potential for efficient functional utilization of −2 PRF. We now have demonstrated that PRRSV uses this mechanism to produce an nsp2-related transframe protein, nsp2TF. Bioinformatic analysis strongly suggests that such an nsp2TF product is encoded by all arteriviruses, with the striking exception of EAV, in which the relevant region of ORF1a has diverged dramatically. Efficient expression of nsp2TF in virus-infected cells was verified experimentally in multiple independent immunoassays using nsp2- and nsp2TF-specific Abs. Mass spectrometric analysis of proteins purified from infected cells further confirmed the expression of nsp2TF and the exact site and direction of frameshifting. Both nsp2TF knockout mutants exhibited a crippled phenotype with a smaller plaque size, indicating an important role for nsp2TF (KO1) and possibly also for frameshifting per se (compare KO1 and KO2) in virus replication.
Currently, there are very few, if any, other examples of natural utilization of −2 PRF in eukaryotic systems. In diverse animals and fungi, expression of the cellular gene antizyme involves +1 PRF (22). However, when mammalian antizyme is expressed artificially in the yeast Saccharomyces cerevisiae, the full-length antizyme product is expressed via −2 frameshifting (23). Here the stimulatory elements comprise a stop codon (bold, lower case) 3′ adjacent to the shift site (UGC_UCC_uga) and a 3′-proximal RNA pseudoknot structure. The −2 frameshift translation reads CSPD, and frameshifting is thought to involve mainly P-site slippage on GC_UCC with an empty A-site. Recently it was found that by artificially reducing the spacer length to the downstream stimulator (either an RNA secondary structure or an antisense oligonucleotide), the slippery site for −1 PRF in HIV, i.e., U_UUU_UUA, also could serve as a slippery site for efficient −2 frameshifting (24). In prokaryotic systems, relatively inefficient −2 PRF (about 2.2%) is used in the expression of the gpGT tail-assembly protein in phage Mu, although the majority of dsDNA phages that express gpGT via frameshifting appear to use −1 PRF instead (25). Finally, although not yet formally demonstrated, sequence analysis suggests that −2 PRF on CC_CUU_UUU is used in the expression of Gag-Pol in Trichomonas vaginalis virus 1 (TVV1) (26, 27). However, PRF in TVV1 is likely to be relatively inefficient, because the Gag-Pol:Gag ratio in virions is extremely low (e.g., 1–2%) (26), in sharp contrast to the high efficiencies observed here for PRRSV −2 PRF (Fig. 5A).
Although typical −1 PRF sites (X_XXY_YYZ) allow codon:anticodon re-pairing in both the P- and A-sites with mismatches only at the wobble positions, for −2 PRF on G_GUU_UUU perfect re-pairing is maintained only in the A-site (Fig. 4A). The nucleotide preceding the heptanucleotide is typically a G or an A in different arterivirus sequences; thus the post-shift P-site anticodon:codon duplex, if a duplex forms at all, may have multiple mismatches. However, although the integrity of the A-site duplex is strictly monitored by the translating ribosome (28), the P-site duplex is not monitored so strictly, and, even for −1 PRF, a number of variations on the canonical XXX are allowed, including UCC, GGA, GUU, and GGU (2, 7, 15). The potential −2 PRF site in TVV1 (CC_CUU_UUU) and the site of presumed −2 PRF in SHFV isolates HQ845737 and HQ845738 (G_GUC_UCU) also concur with the theme of perfect re-pairing in the A-site but reduced potential for re-pairing in the P-site.
With few exceptions, eukaryotic −1 PRF is stimulated by a 3′-proximal RNA secondary structure (11, 13). Whether such structures are sufficient and/or necessary for the stimulation of −2 frameshifting remains to be determined. However, our results indicate that −2 PRF in PRRSV, and by implication other arteriviruses, may be stimulated instead by unstructured 3′ sequence elements including a highly conserved CCCANCUCC motif. Unstructured 3′ sequences also have been implicated in the stimulation of +1 PRF in yeast (29) and −1 PRF in Semliki Forest alphavirus (30). Depending on their distance from the shift site, such sequences may exert their action via mRNA:rRNA base-pairing or interaction with other translational components, although precise mechanisms have not yet been elucidated.
A variety of nsp2-related proteins were observed in our analysis, as previously reported for type II PRRSV isolate VR2332, in which such products were assumed to derive from the use of alternative N- and/or C-terminal cleavage sites (31). At least one of these products now seems to correspond to nsp2TF; some others may correspond to the nsp2- and nsp2TF-related products observed in the pulse-chase experiments in Fig. 5. Moreover, the G_GUU_UUU sequence is also a suitable shift site for −1 frameshifting (15), and a potential −1 frameshift product (nsp2N) was observed (Figs. 3 and 5 and Fig. S1D). In the vast majority of PRRSV sequences (205/212, including isolate SD01-08), such a frameshift would result in immediate termination at a −1 frame stop codon (with G_GUU_UUU_ga, G_GUU_UUU_ag, and G_GUU_UUU_aa all found in different PRRSV isolates; stop codons in −1 frame indicated in bold). Mass spectrometry of the nsp2N band identified a peptide corresponding to the predicted C terminus of a −1 frameshift product (Fig. S1D), although formally such a peptide also could derive from internal cleavage of nsp2 or nsp2TF at this position. The identification of the nsp2N band as the −1 frameshift product was supported further by the fact that this protein was not observed upon expression of the SS mutant, in which the frameshift site is mutated (Fig. 5C). The identities and posttranslational modifications of the various nsp2-related products are currently under further investigation, but the data accumulated thus far leave no doubt that efficient −2 PRF occurs, likely accompanied by a lower level of −1 PRF at the same nucleotide sequence.
As previously described, the balance between the synthesis of the arterivirus pp1a and pp1ab replicase polyproteins is regulated by another ribosomal frameshift event, a −1 PRF (Fig. 1A), leading to an estimated pp1a:pp1ab ratio of about 4:1 (7). It now is apparent that the expression level of the different replicase proteins also is affected by efficient −2 PRF (and probably also −1 PRF) at the TF shift site, leading to a more complex series of ratios. Of the ribosomes that translate nsp1α/nsp1β, ∼20% synthesize nsp2TF, ∼7% synthesize nsp2N, the other ∼73% synthesize nsp2–8, and only ∼15% subsequently translate the ORF1b-encoded proteins nsp9–12. Interestingly, betaretroviruses and deltaretroviruses also use two ribosomal frameshifts (both −1 PRF) in the expression of their Gag-Pro-Pol polyprotein (32). Here Gag, Pro, and Pol are encoded by consecutive terminally overlapping ORFs, and, in contrast to the arteriviruses, the polymerase is translated by ribosomes that have frameshifted twice.
PRRSV nsp2, the largest replicase cleavage product, is released by the autoproteolytic activities of the upstream papain-like protease (PLPβ) in nsp1β and the PLP2 protease residing in the N-terminal domain of nsp2 (Fig. 1A) (33–34). Nsp2 is a multidomain and multifunctional protein. Besides cleaving the nsp2/3 site, nsp2 functions as a cofactor for the nsp4 serine protease during processing of the C-terminal half of pp1a (35). The C-terminal domain of nsp2, but not nsp2TF, is a highly conserved Cys-rich domain of unknown function (36). Furthermore, nsp2 is predicted to be a multispanning TM protein that contributes to the formation of the membranous structures that scaffold the assembly of the viral replication complex (20, 21). Recent studies also have implicated nsp2 in viral pathogenesis, specifically by virtue of PLP2’s deubiquitinating and deISGylating activities (37–40). The biological significance of these activities was supported by the ability of PLP2 to inhibit type I IFN activation and antagonize the antiviral effect of ISG15. Finally, certain regions of nsp2 that appear to be less or nonessential for PRRSV replication are thought to play a role in the modulation of host immune responses in vivo (41).
The nsp2TF protein adds to the complexity of functions potentially encoded in this region of the genome. Its conservation in three of four distantly related arteriviruses and our reverse-genetics studies (Fig. 6) suggest it is an important protein. The frameshift site is located just upstream of the region encoding the predicted nsp2 TM domain. Thus, nsp2 and nsp2TF share the PLP2 domain and the hypervariable region of nsp2 but have distinct C-terminal segments of different sizes that appear to constitute alternative TM domains. Strikingly, IF microscopy revealed that, in both infected cells and expression systems, nsp2TF and nsp2 are targeted to different locations. Although nsp2TF’s specific destination and function in replication or pathogenesis requires further study, these data may be a first step in understanding the presence and conservation of this additional ORF in most arteriviruses. If a fraction of the frameshifting ribosomes indeed make a −1 rather than a −2 shift (see above), the production of an nsp2 variant lacking either TM domain would add further to the complexity of nsp2 expression. Such a presumably cytosolic version of PLP2 may have major implications for the interactions of this protease with host cell targets.
Our reverse-genetics analysis suggests that, although nsp2TF is not essential for replication, it is crucial for maximum virus fitness. When nsp2TF expression was prevented (mutant KO2) or a C-terminally truncated nsp2TF was produced (mutant KO1), the virus exhibited a lower growth rate and a clearly reduced plaque size. KO1 was designed to truncate the TF ORF without disrupting frameshifting per se (Fig. 5C). That this mutation still resulted in a crippled phenotype highlights the functional importance of full-length nsp2TF. In contrast, KO2 was intended to knock out frameshifting completely. There can be multiple reasons why KO2 was more crippled than KO1. KO2 does not express nsp2TF, and the absence of nsp2TF, as demonstrated by KO1, is sufficient to impair virus replication. However, knocking out the frameshift signal also would be expected to up-regulate expression of all the downstream nsps (nsp3–12), perhaps disturbing the balance of viral protein synthesis in a way that is detrimental to virus growth. Potential disruptive effects might involve replicase proteolytic processing by nsp4 (also using nsp2 as a cofactor), the formation of replication complexes, or the overexpression of RdRp and helicase, all of which individually have the potential to affect viral RNA synthesis directly or indirectly.
Because of its highly immunogenic nature, the PRRSV nsp2 region has been explored for the development of a diagnostic assay (42). Although the immunological properties of nsp2TF have not yet been determined, it might represent a viral antigen and/or a potential target for diagnostic assay development. The nsp2-coding region also was explored for its potential application in the development of PRRSV vaccines (reviewed in ref. 8). Using reverse-genetics approaches, modified live viruses with engineered deletions and foreign inserts were created in an attempt to generate differentiable marker vaccines. In addition, mutations and deletions were introduced into certain nsp2 regions during attempts to attenuate the pathogenicity of the virus. The identification of −2 PRF and the TF ORF has important consequences for the rational design of such recombinant viruses. Engineering of the PRRSV nsp2-encoding sequence may unintentionally affect the integrity and/or expression level of nsp2TF and all downstream replicase subunits, perhaps crippling virus replication, as discussed above. On the other hand, such effects also could be useful in the context of modified live virus vaccine design.
Materials and Methods
Computational Analysis.
A total of 255 arterivirus nucleotide sequences in GenBank with full coverage of ORF1a (listed in SI Materials and Methods) were identified by applying tBLASTn (43) to the pp1a peptide sequence derived from GenBank sequences NC_001961 (PRRSV-NA), NC_001639 (LDV), NC_003092 (SHFV), and NC_002532 (EAV). Six ORF1a-defective PRRSV sequences were excluded from subsequent analysis. For each virus, the ORF1a sequences were extracted, translated, aligned, and back-translated to produce nucleotide-sequence alignments using EMBOSS and Clustal (44, 45). Synonymous-site conservation was calculated as described previously (14).
Viruses and Cells.
BHK-21, RK-13, and MARC-145 cells were cultured as described previously (18, 36). The type I PRRSV isolate, SD01-08 (GenBank accession DQ489311) (18), and type II PRRSV isolate, SD23983 (GenBank accession JX258843), were used. Recombinant vaccinia virus vTF7-3 (46) was propagated in RK-13 cells.
Immunoassays.
To detect nsp2 and nsp2TF expression in infected cells, proteins were immunoprecipitated with mAb36-19 and analyzed by Western blot as described previously (19, 47). To determine the nsp2TF subcellular localization, IF microscopy was conducted essentially as described previously (19, 21). The frameshifting efficiency and turnover of nsp2 and nsp2TF were investigated in a pulse-chase experiment using a method modified from that described by Snijder et al. (36). Transient PRRSV ORF1a expression in RK-13 cells, using plasmid pL1a and the recombinant vaccinia virus/T7 polymerase expression system, was performed as described previously (36). See SI Materials and Methods for detailed procedures.
Mass Spectrometry.
Nsp2TF was immunoprecipitated from SD01-08–infected cell lysate using mAb36-19. Proteins from IP were separated on a 6% SDS/PAGE gel, which subsequently was fixed and stained with Coomassie Brilliant Blue G-250 (Bio-Rad). The gel was destained, and the band expected to contain nsp2TF was excised. Trypsin digestion and LC-MS/MS analysis were performed as described previously (48). MS spectra were searched against a custom-made protein database containing possible nsp2 frameshift proteins.
DNA Constructs.
Fig. 2B lists the constructs used in this study. All constructs were made by standard PCR mutagenesis and recombinant DNA techniques and were verified by DNA sequencing. Further details are given in SI Materials and Methods.
Rescue and Characterization of Recombinant PRRSVs.
Recombinant PRRSVs were recovered from WT (pSD01-08) or mutant (pSD01-08-KO1 or pSD01-08-KO2) full-length cDNA clones as described previously (18). Growth kinetics was examined by infecting MARC-145 cells with passage 2 WT or mutant virus at a multiplicity of infection of 0.1. Supernatants from infected cells were collected at 12, 24, 36, 48, 60, and 72 h p.i., and virus titers were determined by fluorescent focus or plaque assay as described previously (18).
Supplementary Material
Acknowledgments
We thank Glenn Björk for his input and Linda Boomaars for technical assistance. This work was supported in part by Wellcome Trust Grant 088789 (to A.E.F.) and Science Foundation Ireland Grant 08/IN.1/B1889 (to J.F.A.).
Footnotes
Conflict of interest statement: The authors have filed a provisional patent application that relates to some aspects of this work.
This article is a PNAS Direct Submission. I.M. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JX258842 and JX258843).
See Author Summary on page 17321 (volume 109, number 43).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211145109/-/DCSupplemental.
References
- 1.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Firth AE, Brierley I. Non-canonical translation in RNA viruses. J Gen Virol. 2012;93:1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Groot RJ, et al. Order Nidovirales. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Waltham MA: Academic Press; 2012. pp. 785–795. [Google Scholar]
- 4.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Nidovirales: Evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snijder EJ, Meulenberg JJM. The molecular biology of arteriviruses. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 6.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.den Boon JA, et al. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang Y, Snijder EJ. The PRRSV replicase: Exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res. 2010;154:61–76. doi: 10.1016/j.virusres.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziebuhr J, Snijder EJ, Gorbalenya AE. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 10.Pasternak AO, Spaan WJ, Snijder EJ. Nidovirus transcription: How to make sense...? J Gen Virol. 2006;87:1403–1421. doi: 10.1099/vir.0.81611-0. [DOI] [PubMed] [Google Scholar]
- 11.Brierley I, Gilbert RJC, Pennell S. Pseudoknot-dependent programmed -1 ribosomal frameshifting: Structures, mechanisms and models. In: Atkins JF, Gesteland RF, editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Heidelberg: Springer; 2010. pp. 149–174. [Google Scholar]
- 12.Meulenberg JJM, et al. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993;192:62–72. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller WA, Giedroc DP. Ribosomal frameshifting in decoding plant viral RNAs. In: Atkins JF, Gesteland RF, editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Heidelberg: Springer; 2010. pp. 193–220. [Google Scholar]
- 14.Firth AE, Wills NM, Gesteland RF, Atkins JF. Stimulation of stop codon readthrough: Frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res. 2011;39:6679–6691. doi: 10.1093/nar/gkr224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loughran G, Firth AE, Atkins JF. Ribosomal frameshifting into an overlapping gene in the 2B-encoding region of the cardiovirus genome. Proc Natl Acad Sci USA. 2011;108:E1111–E1119. doi: 10.1073/pnas.1102932108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jagger BW, et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauck M, et al. Novel, divergent simian hemorrhagic fever viruses in a wild Ugandan red colobus monkey discovered using direct pyrosequencing. PLoS ONE. 2011;6:e19056. doi: 10.1371/journal.pone.0019056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang Y, et al. A full-length cDNA infectious clone of North American type 1 porcine reproductive and respiratory syndrome virus: Expression of green fluorescent protein in the Nsp2 region. J Virol. 2006;80:11447–11455. doi: 10.1128/JVI.01032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Tas A, Snijder EJ, Fang Y. Identification of porcine reproductive and respiratory syndrome virus ORF1a-encoded non-structural proteins in virus-infected cells. J Gen Virol. 2012;93:829–839. doi: 10.1099/vir.0.039289-0. [DOI] [PubMed] [Google Scholar]
- 20.Pedersen KW, van der Meer Y, Roos N, Snijder EJ. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J Virol. 1999;73:2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knoops K, et al. Ultrastructural characterization of arterivirus replication structures: Reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J Virol. 2012;86:2474–2487. doi: 10.1128/JVI.06677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov IP, Atkins JF. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: Close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 2007;35:1842–1858. doi: 10.1093/nar/gkm035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsufuji S, Matsufuji T, Wills NM, Gesteland RF, Atkins JF. Reading two bases twice: Mammalian antizyme frameshifting in yeast. EMBO J. 1996;15:1360–1370. [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Z, Gilbert RJ, Brierley I. Spacer-length dependence of programmed -1 or -2 ribosomal frameshifting on a U6A heptamer supports a role for messenger RNA (mRNA) tension in frameshifting. Nucleic Acids Res. 2012;40:8674–8689. doi: 10.1093/nar/gks629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J, Hendrix RW, Duda RL. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol Cell. 2004;16:11–21. doi: 10.1016/j.molcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Liu HW, Chu YD, Tai JH. Characterization of Trichomonas vaginalis virus proteins in the pathogenic protozoan T. vaginalis. Arch Virol. 1998;143:963–970. doi: 10.1007/s007050050345. [DOI] [PubMed] [Google Scholar]
- 27.Goodman RP, et al. Clinical isolates of Trichomonas vaginalis concurrently infected by strains of up to four Trichomonasvirus species (Family Totiviridae) J Virol. 2011;85:4258–4270. doi: 10.1128/JVI.00220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–259. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 29.Guarraia C, Norris L, Raman A, Farabaugh PJ. Saturation mutagenesis of a +1 programmed frameshift-inducing mRNA sequence derived from a yeast retrotransposon. RNA. 2007;13:1940–1947. doi: 10.1261/rna.735107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung BY, Firth AE, Atkins JF. Frameshifting in alphaviruses: A diversity of 3′ stimulatory structures. J Mol Biol. 2010;397:448–456. doi: 10.1016/j.jmb.2010.01.044. [DOI] [PubMed] [Google Scholar]
- 31.Han J, Rutherford MS, Faaberg KS. Proteolytic products of the porcine reproductive and respiratory syndrome virus nsp2 replicase protein. J Virol. 2010;84:10102–10112. doi: 10.1128/JVI.01208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacks T, Townsley K, Varmus HE, Majors J. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc Natl Acad Sci USA. 1987;84:4298–4302. doi: 10.1073/pnas.84.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.den Boon JA, et al. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: Identification of two papainlike cysteine proteases. J Virol. 1995;69:4500–4505. doi: 10.1128/jvi.69.7.4500-4505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snijder EJ, Wassenaar ALM, Spaan WJM, Gorbalenya AE. The arterivirus Nsp2 protease. An unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J Biol Chem. 1995;270:16671–16676. doi: 10.1074/jbc.270.28.16671. [DOI] [PubMed] [Google Scholar]
- 35.Wassenaar AL, Spaan WJ, Gorbalenya AE, Snijder EJ. Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: Evidence that NSP2 acts as a cofactor for the NSP4 serine protease. J Virol. 1997;71:9313–9322. doi: 10.1128/jvi.71.12.9313-9322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snijder EJ, Wassenaar AL, Spaan WJ. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J Virol. 1994;68:5755–5764. doi: 10.1128/jvi.68.9.5755-5764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frias-Staheli N, et al. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Z, Chen Z, Lawson SR, Fang Y. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J Virol. 2010;84:7832–7846. doi: 10.1128/JVI.00217-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Z, Li Y, Ransburgh R, Snijder EJ, Fang Y. Nonstructural protein 2 of porcine reproductive and respiratory syndrome virus inhibits the antiviral function of interferon-stimulated gene 15. J Virol. 2012;86:3839–3850. doi: 10.1128/JVI.06466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Kasteren PB, et al. Arterivirus and nairovirus ovarian tumor domain-containing Deubiquitinases target activated RIG-I to control innate immune signaling. J Virol. 2012;86:773–785. doi: 10.1128/JVI.06277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z, et al. Immunodominant epitopes in nsp2 of porcine reproductive and respiratory syndrome virus are dispensable for replication, but play an important role in modulation of the host immune response. J Gen Virol. 2010;91:1047–1057. doi: 10.1099/vir.0.016212-0. [DOI] [PubMed] [Google Scholar]
- 42.Brown E, et al. Antibody response to porcine reproductive and respiratory syndrome virus (PRRSV) nonstructural proteins and implications for diagnostic detection and differentiation of PRRSV types I and II. Clin Vaccine Immunol. 2009;16:628–635. doi: 10.1128/CVI.00483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 44.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 45.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 46.Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu WH, et al. The 2b protein as a minor structural component of PRRSV. Virus Res. 2005;114:177–181. doi: 10.1016/j.virusres.2005.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Akker J, et al. The redox state of transglutaminase 2 controls arterial remodeling. PLoS ONE. 2011;6:e23067. doi: 10.1371/journal.pone.0023067. [DOI] [PMC free article] [PubMed] [Google Scholar]