Abstract
The hippocampus is a highly plastic brain region particularly susceptible to the effects of environmental stress; it also shows dynamic changes in epigenetic marks in response to stress and learning. We have previously shown that, in the rat, acute (30 min) restraint stress induces a substantial, regionally specific, increase in hippocampal levels of the repressive histone H3 lysine 9 trimethylation (H3K9me3). Because of the large magnitude of this effect and the fact that stress can induce the expression of endogenous retroviruses and transposable elements in many systems, we hypothesized that the H3K9me3 response was targeted to these elements as a means of containing potential genomic instability. We used ChIP coupled with next generation sequencing (ChIP-Seq) to determine the genomic localization of the H3K9me3 response. Although there was a general increase in this response across the genome, our results validated this hypothesis by demonstrating that stress increases H3K9me3 enrichment at transposable element loci and, using RT-PCR, we demonstrate that this effect represses expression of intracisternal-A particle endogenous retrovirus elements and B2 short interspersed elements, but it does not appear to have a repressive effect on long interspersed element RNA. In addition, we present data showing that the histone H3K9-specific methyltransferases Suv39h2 is up-regulated by acute stress in the hippocampus, and that this may explain the hippocampal specificity we observe. These results are a unique demonstration of the regulatory effect of environmental stress, via an epigenetic mark, on the vast genomic terra incognita represented by transposable elements.
Keywords: heterochromatin, ncRNA, posttraumatic stress disorder, transposon
Epigenetic activity in the brain has become a significant focus of neuroscience research (1–4), and a number of epigenetic marks have been identified in association with models of neuropsychiatric disease (5). Environmental stress, particularly via its effects on limbic structures, such as the hippocampus, is among the most important contributors to susceptibility to depression and posttraumatic stress disorder (PTSD) (6–8). Notably, the hippocampus is both structurally and functionally responsive to stress. Indeed, the hippocampus was one of the first brain regions to show an epigenetic response to an exogenous stimulus (9). Evidence has begun to accumulate that the hippocampus responds to stress at the epigenetic and even genomic levels (with regard to transposon activity) (10, 11). Our own recent work has demonstrated a pronounced hippocampus-specific increase in histone H3 lysine 9 trimethylation (H3K9me3) after acute stress, on a time scale similar to those observed for the adjacent H3S10phos mark in the aforementioned article (9). We found that a single traumatic stressor, acute restraint stress, produced a substantial elevation of H3K9me3 levels coupled with an equally substantial reduction in H3K9me1 levels in a regionally specific fashion. H3K9me3 is associated with heterochromatin formation and the large changes observed suggested a rapid (less than 2 h) and global chromatin reorganization (12). Similar effects have been observed as a result of cocaine treatment in the nucleus accumbens, where Maze et al. showed that the H3K9 mark was associated with repetitive and transposable elements in the genome (13). Given, this finding and the significant metabolic expense of such an event, the addition of the third methyl group being, energetically, a very expensive epigenetic modification, we hypothesized that H3K9me3 suppresses the stress-induced activation of repetitive and transposable elements. These elements comprise a much larger proportion of the mammalian genome (∼50%) than do genes themselves (less than 5%) (14) and contribute to genomic instability (15), a significant problem for postmitotic neurons that must remain phenotypically stable for years or even decades. Here we show that stress-induced increases in H3K9me3 in the hippocampus are strongly targeted to retrotransposable elements and rapidly silence their expression.
Results
Acute Stress-Induced Increase in H3K9me3 Is Hippocampus-Specific.
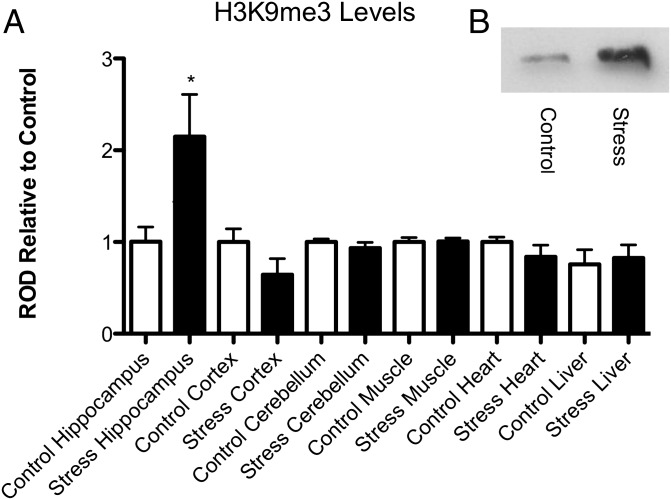
We used acute restraint stress to extend our examination of the tissue specificity of the stress-induced rise in H3K9me3. Rats were immobilized for 30 min in a plastic bag restrainer, with holes for ventilation, and allowed to recover for 60 min before killing and hippocampal tissue collection. Using Western blotting of H3K9me3 in hippocampus, as well as cerebellum, frontal cortex, heart, liver, and skeletal muscle, we show acute immobilization stress selectively up-regulates H3K9me3 levels in the hippocampus by 114% (±2%, n = 10, P < 0.03), but produced no significant changes elsewhere (Fig. 1).
Fig. 1.
(A) quantification of H3K9me3 Western blot immunoreactivity relative to control tissue in rat hippocampus, frontal cortex, cerebellum, skeletal muscle, cardiac muscle, and liver (*P < 0.03, n = 10). (B, Inset) Representative blot with control hippocampal H3K9me3 (Left) and stress (Right).
Acute Stress Globally and Selective Increases H3K9me3 Levels at Transposable Element Loci.
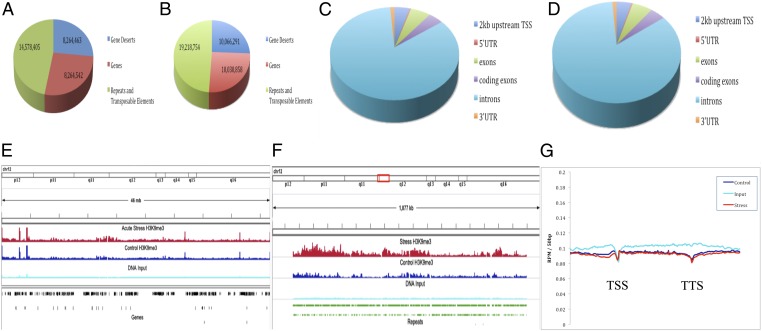
To test the hypothesis that the stress-induced increase in H3K9me3 was targeted toward repetitive and transposable elements, we performed ChIP sequencing (ChIP-Seq) against the H3K9me3 mark in the hippocampi of acutely stressed and control male Sprague–Dawley rats. This process produced over 40 million raw 75-bp sequence reads for both stress and control samples (see SI Methods for more detail). Using both the RN4 version of the rat genome and a database of rat repetitive and transposable elements derived from Repbase (16), we aligned these reads to genomic features (Fig. 2). We found that stress did increase alignment of H3K9me3 reads to transposable and repeat elements by 32%, or 4.6 million unique alignments (Fig. 2 A, B, and F). Intergenic regions showed a more moderate increase but alignment within gene bodies (promoters, introns, exons, and untranslated regions) was flat between stress and control groups (Fig. 2 A, B, and E). Within genes the majority of alignments was to intronic regions (Fig. 2 C and D) in both groups. Fig. 2G shows reads mapped to subregions of genes, including the transcription start site (TSS) and transcription termination site (TTS), showing that, within genes, H3K9me3-enriched reads are at lower levels than unimmunoprecipitated input DNA.
Fig. 2.
Genomic regions enriched for H3K9me3 showed little change with stress within genes proper; regions containing repetitive and transposable elements showed increased H3K9me3 enrichment with stress: (A) the breakdown of H3K9me3 reads in control rat hippocampus by genes, gene deserts, and repetitive sequences; (B) the breakdown in stressed hippocampus. (C and D) The regional breakdown of reads aligned within genes in control and stressed animals; no significant changes in alignments within genes were observed. (E) A representative 46-Mb segment of chromosome 12 comparing H3K9me3 and DNA input reads with known genes; little change is observable in H3K9me3 at annotated gene loci. (F) A representative 1.9-MB region of chromosome 12 with low gene density, aligned with known repetitive elements (green), demonstrating the high level of stress induced enrichment in H3K9me3 binding (red) compared with control (dark blue) and DNA input (light blue) reads. (G) Read alignments within gene bodies (RPM, reads per million mapped) showing a drop in H3K9me3 enrichment around TSS and TTS.
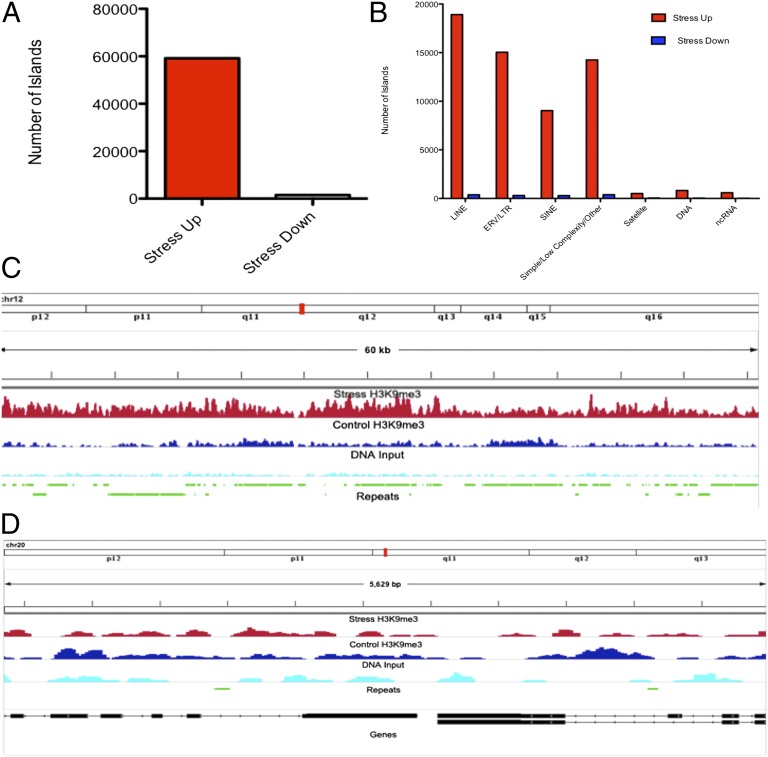
Analysis of significantly altered peaks in H3K9me3 after stress further demonstrated the dramatic effect of acute stress on this mark at repetitive and transposable elements in the genome. Stress caused a significant increase (stress up) in H3K9me3 alignments at 59,147 islands associated with repetitive elements, but it was associated with reductions (stress down) at only 1,438 loci (Fig. 3A) [false-discovery rate (FDR) <0.0001, P < 0.05]. When categorized by repetitive-element class, differences emerge in the enrichment of stress-up versus stress-down peaks: the autonomous long interspersed element (LINE) retrotransposons and endogenous retrovirus/long terminal repeat (LTR) classes showed 50-fold differences in the number up and down peaks, but short interspersed elements (SINEs) showed 31-fold differentials, low complexity, and simple repeats showed 37-fold differentials; DNA transposons showed 25-fold, satellite repeats showed 10-fold, and noncoding RNA loci showed 39-fold differentials, respectively (Fig. 3B). Representative stress-up and -down regions are displayed in Fig. 3 C and D, respectively.
Fig. 3.
Analysis of genomic regions showing statistically significant differences in H3K9me3 enrichment between stress and control conditions revealed profound increases in H3K9me3 enrichment after stress. Total number of genomic islands aligned to repetitive elements showing significant increases in H3K9me3 enrichment after stress (stress up, red) or decreased enrichment (stress down, blue) are shown (A). (B) These reads are categorized by repeat type. (C) A representative stress up 60-Kb region of chromosome 12, along with known repetitive elements. (D) A stress-down region along with gene and repeat alignments, demonstrating the pronounced bias toward stress induced increases in H3K9me3 levels in regions of high repetitive-element density.
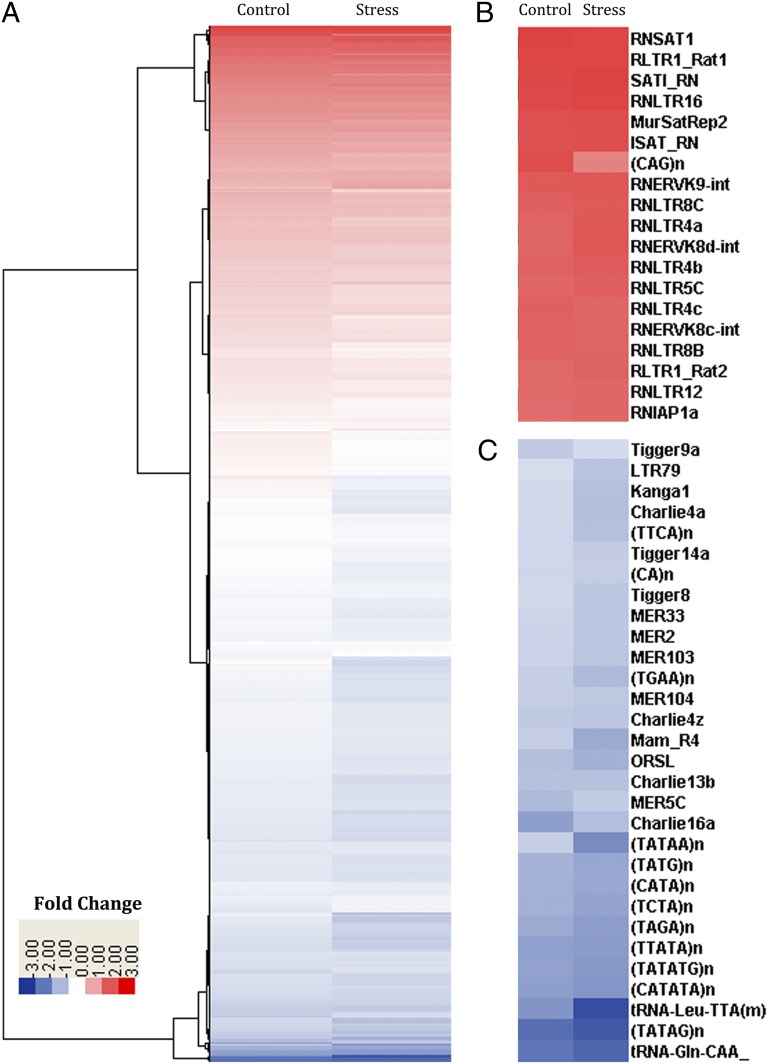
To further assess if the stress-induced increase in H3K9me3 showed a selective bias toward a particular group of repetitive elements, read alignments using the phylogenetic clustering and instance approaches used in ref. 17 were performed to compare the H3K9me3 enrichment at repeat and transposon loci in the stress and control samples compared with input DNA. Using an equal number of reads from input, stress, and control samples (15 M) allowed us to see if the increase in H3K9me3 was generalized or specific to particular families of repetitive or transposable elements. As can be seen in Fig. 4A, stress produces increased enrichment in the families at the top of the distribution and reduced enrichment in the classes, which already showed reduced enrichment in the control. The highly enriched classes were largely endogenous retrovirus (ERV)/LTR retrotransposons and some satellites (Fig. 4B), whereas the low H3K9me3 regions were more likely to contain DNA transposons or simple repeats (Fig. 4C).
Fig. 4.
Heat map comparison and phylogenetic clustering of 15-M reads from both control and stress H3K9me3 ChIP-Seq compared with an equal number of reads from input DNA (A). Enlargement of the top end of this enrichment range (B) demonstrates selective enrichment of ERV/LTR class retrotransposons associated with the H3K9me3 mark and reduced enrichment associated with the mark at simple repeats and DNA transposons (C).
Acute Stress Down-Regulates Transposable Element RNA Expression.
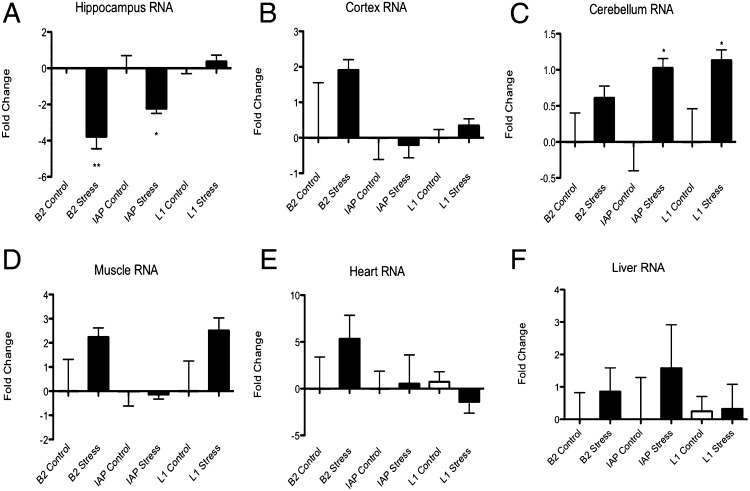
To determine if any of the transposable elements that showed large increases in H3K9me3 or potential heterochromatinization, as measured by H3K9me3 reads, we performed RT-PCR against specific elements within the LINE, SINE, and ERV/LTR classes: an ERV intracisternal-A particle (IAP) element (RNIAP1a) the B2_RN SINE element and L1_RN. We found that, in the hippocampus, RNAIP RNA was reduced 3.2-fold (±0.27, n = 12, P < 0.04), B2_RN RNA was reduced 4.8-fold (±0.67, n = 12, P < 0.005) (Fig. 5A), demonstrating that the increase in H3K9me3 induced by acute stress is correlated with silencing of transposable elements in the hippocampal genome. L1 RNA showed no significant change as a consequence of stress in the hippocampus, given previous findings showing that H3K9me3 is consistently associated with ERV/LTR but not LINE elements; it is possible that there are other chromatin elements, which prevent silencing at these loci (17, 18). No significant changes were observed in the other tissues or brain regions examined (Fig. 5 B–F), save in the cerebellum, where both IAP (twofold, 0.13, n = 6, P < 0.02) and L1_RN RNA (2.1-fold, ±0.15, n = 6, P < 0.03) were up-regulated by stress. The latter result suggests that the lack of a stress-induced H3K9me3 response is permissive for increased transposable element RNA expression.
Fig. 5.
RT-PCR reveals that stress-induced increases in H3K9me3 are correlated with rapid (1 h poststress) reductions in transposable element RNA in the hippocampus (*P < 0.05, **P < 0.005, n = 12 for hippocampus, 6 for cerebellum) (A), but not in tissues where the stress induced increase in H3K9me3 was absent, such as the frontal cortex (B), cerebellum (C), skeletal muscle (D), cardiac muscle (E), or liver (F).
Acute Restraint Selectively Increases Hippocampal Suv39h2 Expression.
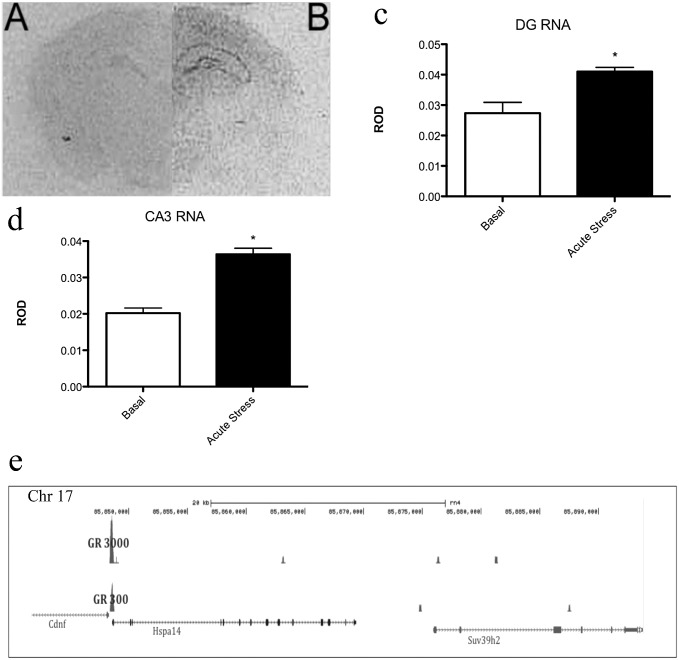
To determine if the hippocampal specificity we observed may be in part because of selective regulation of H3K9-specific histone methyltransferases, we examined the expression of Suv29h1 and Suv39h2 after acute stress using in situ hybridization. We found that, although hippocampal expression of Suv39h1 and h2 (Fig. 6A) was low under basal conditions, acute stress up-regulated the expression of the latter by 50% (±3.2%, n = 4, P < 0.025) (Fig. 6 B and C) in the dentate gyrus, 81% (±4.6%, n = 4, P < 0.002) (Fig. 6 B and D) in the CA3, and showed no significant change in expression in the CA1. No significant change was observed in Suv39h1 expression after stress in any region.
Fig. 6.
In situ hybridization shows that acute stress increases the expression of Suv39h2 in the rat hippocampus. (A) A representative section from an animal under basal; (B) a section from a rat stressed acutely for 30 min. (C) Quantification of the stress induced increase in Suv39h2 (*P < 0.03, n = 4) in the dentate gyrus. (D) Relative levels of Suv39h2 in basal and stressed CA3 (P < 0.002, n = 4) no significant changes in CA1 Suv39h2 expression were observed. (F) Binding of GR to hippocampal DNA in the region of the suv39h2 gene, as measured by ChIP-Seq. Rats were systemically injected with either 300 or 3,000 μg/kg of corticosterone; GR300, 300 μg/kg corticosterone; GR3000, 3,000 μg/kg corticosterone.
Acute Corticosterone Increases Glucocorticoid Receptor-DNA Binding in the Vicinity of the Suv39h2 Gene.
ChIP-Seq analysis of hippocampal glucocorticoid receptor (GR) DNA binding in the hippocampus of rats treated with either 300 μg/kg or 3,000 μg/kg corticosterone demonstrated increased levels of GR in the vicinity of the Suv39h2 gene. The 300-μg/kg dose increased the major peak in GR binding 84-fold (FDR 1.17%) and the larger dose increased GR binding by 268-fold (FDR 8.47%) (Fig. 6E).
Discussion
PTSD, a disorder that shows substantial evidence of transgenerational epigenetic influences in humans (19, 20), is often the proximal result of a single severe stressor. As such, understanding the mechanisms by which an acute stressor in an animal model contributes to functional and molecular changes in the brain is of significance to our understanding of this and other stress-related mental disorders. Here, we have demonstrated that an exogenous stressor can have a rapid and pronounced effect on the regulation of the vast and, to date, relatively unexamined “other genome” represented by transposable elements. Variations in L1 LINE activity are found in Rett syndrome patients, where they can be directly related to MECP2 dysfunction and altered DNA methylation. LINE1 activity is also induced in the hippocampus in exercised rats (11, 21), suggesting a linkage to the machinery in control of neurogenesis and plasticity in this region. The present study extends these findings with regard to environmental stress and transposon class. Maze et al. (13) observed similar alterations in H3K9me3 at various genomic loci, including a number of repetitive elements, in the nucleus accumbens after repeated cocaine. However, the direction of the changes they reported were of mixed directionality in contrast to the pronounced bias toward stress up-regulation of H3k9me3 shown here. Surprisingly, it appears that H3K9me3 may be targeted in a selective fashion toward certain varieties of repetitive or transposable elements; as shown in Fig. 4, H3K9me3 is more enriched at ERV/LTR class retrotransposons and less likely to be found at simple repeats or DNA transposons. This observation is in agreement with the pattern of histone methylation found at repetitive elements in stem cells (17), although the stress-induced increase in the bias toward these elements has not been observed elsewhere to our knowledge.
Our findings raise the question of why the hippocampus shows such a selective H3K9me3 response to stress. Indeed, the absence of an H3K9me3 stress response in the cerebellum is associated with a significant increase in retrotransposon expression (Fig. 5C); interestingly, this brain region is the only brain region where the intact endogenous retrovirus Emv2 is expressed in the mouse (22), suggesting this brain region may be privileged (or perhaps underprivileged) for endogenous retrovirus expression. Part of the answer to this question has to do with the regional expression of the histone H3 lysine 9 selective methyltransferases SUV39h1 and Suv39h2 (KMT1a and -1b, respectively). Using in situ hybridization, we found that both were expressed at low levels in the hippocampus and largely undetectable elsewhere in the brain (Fig. 6 A and B). Suv39h2 was up-regulated within 30 min of the onset of stress in the dentate gyrus and CA3 (Fig. 6 C and D). ChIP-Seq using GR antibodies showed that GR binding in the region of the Suv39h2 gene was increased by corticosterone treatment in a dose-responsive fashion (Fig. 6E). Less is known about the expression of these genes in the peripheral tissues we examined, but binding of GR to GR-elements is highly tissue-specific (23), and although our data do not conclusively demonstrate that GR actively regulates Suv39h2 expression in the hippocampus, they do suggest that the changes in H3K9 methylation we have observed in the hippocampus may use a different mechanism than has been observed elsewhere in the brain (e.g., the accumbens, where G9a appears to be the dominant H3K9 methyltransferase) (24). It should also be added that our examination of stress-responsive brain regions and tissues was not comprehensive, so it is possible that other stress-responsive regions, such as the amygdala or paraventricular hypothalamus, could show similar changes in H3K9 methylation or transposon expression as a consequence of stress.
Our results demonstrating silencing of repetitive elements via the H3K9me3 mark are congruent with previous observations in embryonic stem cells, showing that disruption of H3K9me3, via KAP-1 deletion, results in a profound increase in IAP expression (25). Of interest is the observation that mice bearing a forebrain deletion of KAP-1 show increased anxiety and deficits in hippocampus-dependent memory (26). Those data, taken together with our present findings showing remarkable hippocampal specificity, suggest that dysregulation of heterochromatin-mediated transposon silencing may have an impact on spatial memory, anxiety, and hippocampal function.
Stress has been shown to alter the expression of a number of other epigenetic marks in addition to the H3K9me3 mark, as we have recently discussed elsewhere (4). Our previous work showed that acute stress decreased global levels of the H3K27me3 mark and had no effect on the levels of the H3K4me3 mark (12). However, the later mark has been show to change in a gene-specific manner with social defeat stress (27), suggesting that regulation of H3K4 methylation by stress is much more restricted to specific genomic localities in the hippocampus than H3K9me3. We also observed a decrease in global H3K9me1 levels, and others have observed a stress-induced decrease in H3K9ac, which are consistent with our observations of globally increased H3K9 trimethylation (12, 28). Crosio et al. (9) observed an increase in the H3S10phos mark with a similar regional and temporal specificity to our own observations; whether this change is coupled to the changes in H3K9me3 we see is presently unknown, but of great interest given the role of the H3K9me3-S10phos dual mark as a molecular switch controlling HP1 association with chromatin (29). In contrast, Reul and colleagues have observed increases in phospho acetylation of H3 at serine 10 and lysine 14 following acute stressors; however, these changes were observed in a small subpopulation of dentate gyrus neurons, distinct from the population-wide changes reported here (30).
Finally, it is important to note, that only a few common brain disorders have been shown to result from variations in protein-coding genes and that transposable elements constitute an order of magnitude larger fraction of the genome than genes. Evidence is emerging not only of the capacity of those transposable elements for inducing genomic instability and neuro-inflamation (15, 31), but also of the capacity of some of those elements to regulate gene transcription under normal or stressed conditions (32). Furthermore, it is estimated that each human genome may contain at least on “private” transposition (33) and that as much as 25% of human genetic variation may be because of transposon activity (34, 35). Intriguingly, it has recently been observed that LINE1 shows variations in DNA methylation between veterans with PTSD and combat-deployed controls (36). In view of these considerations, we suggest that these elements may represent a novel basis for understanding susceptibility to a variety of brain disorders, particularly depression and anxiety disorders, such as PTSD or hippocampus-dependent cognitive disorders.
Methods
Animals were handled in accordance with National Institutes of Health and Rockefeller University Institutional Animal Care and Use Committee guidelines. Western blots, RT-PCR, and in situ hybridization were performed using standard methods. Detailed methods can be found in SI Methods.
Supplementary Material
Acknowledgments
We thank C. David Allis of The Rockefeller University for assistance. Support was provided by National Institutes of Health Grants MH MH58911 and MH41256 (to B.S.M.), HD-05751 (to D.W.P.), and The National Alliance for Research on Schizophrenia and Depression Betz Family Young Investigator Award (to R.G.H.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE41217).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215810109/-/DCSupplemental.
References
- 1.Jiang Y, et al. Epigenetics in the nervous system. J Neurosci. 2008;28(46):11753–11759. doi: 10.1523/JNEUROSCI.3797-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60(6):961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zocchi L, Sassone-Corsi P. Joining the dots: from chromatin remodeling to neuronal plasticity. Curr Opin Neurobiol. 2010;20(4):432–440. doi: 10.1016/j.conb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunter RG. Epigenetic effects of stress and corticosteroids in the brain. Front Cell Neurosci. 2012;6:18. doi: 10.3389/fncel.2012.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudley KJ, Li X, Kobor MS, Kippin TE, Bredy TW. Epigenetic mechanisms mediating vulnerability and resilience to psychiatric disorders. Neurosci Biobehav Rev. 2011;35(7):1544–1551. doi: 10.1016/j.neubiorev.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 6.McEwen BS. Protection and damage from acute and chronic stress: Allostasis and allostatic overload and relevance to the pathophysiology of psychiatric disorders. Ann N Y Acad Sci. 2004;1032:1–7. doi: 10.1196/annals.1314.001. [DOI] [PubMed] [Google Scholar]
- 7.Sapolsky RM. Why stress is bad for your brain. Science. 1996;273(5276):749–750. doi: 10.1126/science.273.5276.749. [DOI] [PubMed] [Google Scholar]
- 8.Gilbertson MW, et al. Smaller hippocampal volume predicts pathologic vulnerability to psychological trauma. Nat Neurosci. 2002;5(11):1242–1247. doi: 10.1038/nn958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crosio C, Heitz E, Allis CD, Borrelli E, Sassone-Corsi P. Chromatin remodeling and neuronal response: Multiple signaling pathways induce specific histone H3 modifications and early gene expression in hippocampal neurons. J Cell Sci. 2003;116(Pt 24):4905–4914. doi: 10.1242/jcs.00804. [DOI] [PubMed] [Google Scholar]
- 10.McEwen BS, Eiland L, Hunter RG, Miller MM. Stress and anxiety: Structural plasticity and epigenetic regulation as a consequence of stress. Neuropharmacology. 2012;62(1):3–12. doi: 10.1016/j.neuropharm.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muotri AR, Zhao C, Marchetto MC, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus. 2009;19(10):1002–1007. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci USA. 2009;106(49):20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maze I, et al. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc Natl Acad Sci USA. 2011;108(7):3035–3040. doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumann GG, et al. Unique functions of repetitive transcriptomes. Int Rev Cell Mol Biol. 2010;285:115–188. doi: 10.1016/B978-0-12-381047-2.00003-7. [DOI] [PubMed] [Google Scholar]
- 15.Kazazian HH, Jr, Goodier JL. LINE drive. Retrotransposition and genome instability. Cell. 2002;110(3):277–280. doi: 10.1016/s0092-8674(02)00868-1. [DOI] [PubMed] [Google Scholar]
- 16.Jurka J, et al. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110(1-4):462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 17.Day DS, Luquette LJ, Park PJ, Kharchenko PV. Estimating enrichment of repetitive elements from high-throughput sequence data. Genome Biol. 2010;11(6):R69. doi: 10.1186/gb-2010-11-6-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yehuda R, Bierer LM. Transgenerational transmission of cortisol and PTSD risk. Prog Brain Res. 2008;167:121–135. doi: 10.1016/S0079-6123(07)67009-5. [DOI] [PubMed] [Google Scholar]
- 20.Miller MM, McEwen BS. Establishing an agenda for translational research on PTSD. Ann N Y Acad Sci. 2006;1071:294–312. doi: 10.1196/annals.1364.023. [DOI] [PubMed] [Google Scholar]
- 21.Muotri AR, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468(7322):443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee KH, Horiuchi M, Itoh T, Greenhalgh DG, Cho K. Cerebellum-specific and age-dependent expression of an endogenous retrovirus with intact coding potential. Retrovirology. 2011;8:82. doi: 10.1186/1742-4690-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datson NA, et al. Specific regulatory motifs predict glucocorticoid responsiveness of hippocampal gene expression. Endocrinology. 2011;152(10):3749–3757. doi: 10.1210/en.2011-0287. [DOI] [PubMed] [Google Scholar]
- 24.Maze I, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327(5962):213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowe HM, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463(7278):237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 26.Jakobsson J, et al. KAP1-mediated epigenetic repression in the forebrain modulates behavioral vulnerability to stress. Neuron. 2008;60(5):818–831. doi: 10.1016/j.neuron.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 27.Tsankova NM, et al. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9(4):519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 28.Ferland CL, Schrader LA. Regulation of histone acetylation in the hippocampus of chronically stressed rats: A potential role of sirtuins. Neuroscience. 2011;174:104–114. doi: 10.1016/j.neuroscience.2010.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dormann HL, Tseng BS, Allis CD, Funabiki H, Fischle W. Dynamic regulation of effector protein binding to histone modifications: The biology of HP1 switching. Cell Cycle. 2006;5(24):2842–2851. doi: 10.4161/cc.5.24.3540. [DOI] [PubMed] [Google Scholar]
- 30.Mifsud KR, et al. Epigenetic mechanisms in stress and adaptation. Brain Behav Immun. 2011;25(7):1305–1315. doi: 10.1016/j.bbi.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Perron H, Lang A. The human endogenous retrovirus link between genes and environment in multiple sclerosis and in multifactorial diseases associating neuroinflammation. Clin Rev Allergy Immunol. 2010;39(1):51–61. doi: 10.1007/s12016-009-8170-x. [DOI] [PubMed] [Google Scholar]
- 32.Ponicsan SL, Kugel JF, Goodrich JA. Genomic gems: SINE RNAs regulate mRNA production. Curr Opin Genet Dev. 2010;20(2):149–155. doi: 10.1016/j.gde.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iskow RC, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141(7):1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kidd JM, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143(5):837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318(5849):420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rusiecki JA, et al. DNA methylation in repetitive elements and post-traumatic stress disorder: A case-control study of US military service members. Epigenomics. 2012;4(1):29–40. doi: 10.2217/epi.11.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.