Abstract
O-GlcNAcylation is an abundant posttranslational modification in the brain implicated in human neurodegenerative diseases. We have exploited viable null alleles of the enzymes of O-GlcNAc cycling to examine the role of O-GlcNAcylation in well-characterized Caenorhabditis elegans models of neurodegenerative proteotoxicity. O-GlcNAc cycling dramatically modulated the severity of the phenotype in transgenic models of tauopathy, amyloid β-peptide, and polyglutamine expansion. Intriguingly, loss of function of O-GlcNAc transferase alleviated, whereas loss of O-GlcNAcase enhanced, the phenotype of multiple neurodegenerative disease models. The O-GlcNAc cycling mutants act in part by altering DAF-16–dependent transcription and modulating the protein degradation machinery. These findings suggest that O-GlcNAc levels may directly influence neurodegenerative disease progression, thus making the enzymes of O-GlcNAc cycling attractive targets for neurodegenerative disease therapies.
Keywords: neurodegeneration, Alzheimer's disease, Huntington disease, insulin
Proteotoxicity is a common pathological feature of human neurodegenerative diseases. The disease-associated protein aggregates include senile plaques from amyloid β-peptide, neurofibrillary tangles from hyperphosphorylated tau in Alzheimer’s disease (AD) (1, 2), and huntingtin aggregates from mutant huntingtin protein containing polyglutamine repeats in Huntington disease (HD) (3). The precise mechanism involved in triggering protein aggregation in neurodegeneration is less clear. Recent work has suggested that changes in insulin-like signaling, cell cycle progression, or protein clearance mechanisms may all be involved in the progression of neurodegenerative diseases (4–6).
O-GlcNAcylation is a dynamic posttranslational modification similar to phosphorylation, in which the monosaccharide β-N-acetyl-d-glucosamine (O-GlcNAc) is attached to serine or threonine residues of target proteins (7). In the nucleoplasm and the cytoplasm, O-GlcNAc is actively added and removed by two evolutionarily conserved enzymes: O-GlcNAc transferase (OGT) and β-N-acetylglucosaminidase (OGA), respectively. This process is also referred to as O-GlcNAc cycling. The deregulation of O-GlcNAc modification has been linked to several human diseases, including diabetes mellitus, cardiovascular disease, and cancer (8–10). The O-GlcNAc modification is also implicated in neurodegenerative diseases (11, 12). OGT and OGA are highly expressed in the brain (13), and many neuronal proteins, including tau and amyloid precursor protein, are modified by O-GlcNAc directly (14–17). The human OGA gene, MGEA5, is not only linked to a late-onset form of AD (18), but also shows significant alternative splicing in AD (19). More importantly, O-GlcNAcylation plays a key role in neural growth, learning, and long-term memory (20).
To facilitate genetic analysis of this group of diseases, many well-characterized Caenorhabditis elegans transgenic models have been developed, and have been used to investigate the roles of insulin and protein degradation pathways in the regulation of proteotoxicity (21–23). C. elegans also provides unique genetic advantages to study the effects of altered O-GlcNAc cycling; loss of function of Ogt in vertebrate systems results in embryonic lethality (24, 25). However, C. elegans animals bearing null mutations of ogt-1 or oga-1 are alive and fertile and appear phenotypically wild type (WT) with respect to development, growth, and movement (26, 27). In the present study, we examined the phenotype of neurodegenerative proteotoxicity models in O-GlcNAcylation null mutants, and demonstrated that the loss of OGT function alleviated, whereas loss of OGA function exacerbated, the proteotoxicity associated with aggregate-prone proteins. The mechanism of this protective effect of decreased O-GlcNAcylation on proteotoxicity may be linked to deregulation of the protein homeostasis machinery, signaling pathways and DAF-16, a key regulator of the stress response and proteotoxicity in C. elegans.
Results
The O-GlcNAc Cycling Mutants Affect Toxicity of Two HD Models.
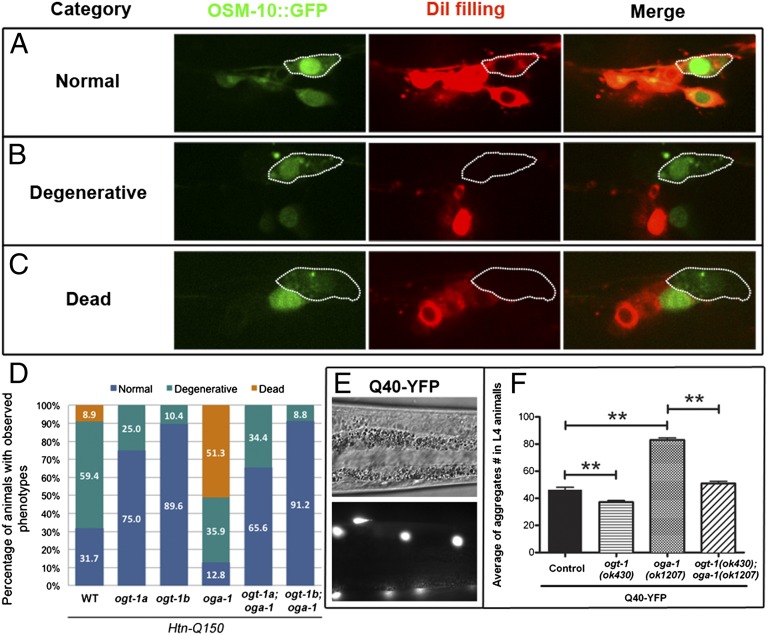
We first examined the influence of O-GlcNAc cycling on proteotoxicity with Htn-Q150, an HD model with a fusion protein of the 171 N-terminal amino acids of the human huntingtin protein and a tract of 150 glutamine residues (Htn-Q150) expressed in a small subset of sensory neurons (ASH, ASI, PHA, and PHB) (28). Htn-Q150 expression causes age-dependent degenerative changes and even death of the ASH neurons, and the phenotype can be scored by observing the expression of OSM-10::GFP coexpressed with Htn-Q150 and 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) dye filling (28). Normal ASH neurons were positive for OSM-10::GFP expression and DiI filling (Fig. 1A). Degenerative ASH neurons showed normal OSM-10::GFP expression, but had a DiI-filling defect caused by degenerative changes of the ASH dendrites in response to the toxic Htn-Q150 fusion protein (Fig. 1B). Dead ASH neurons had neither OSM-10::GFP expression nor DiI filling (Fig. 1C). We found that the oga-1(ok1207) mutation, which causes accumulation of O-GlcNAcylation, dramatically enhanced the toxicity of Htn-Q150 in the ASH neurons in animals at day 8 of adulthood, with 51.3% dead ASH neurons compared with 8.9% in the control strain expressing Htn-Q150 alone (Fig. 1D). In contrast, two ogt-1 null alleles (ok430 and ok1474) that are devoid of O-GlcNAcylation alleviated the toxicity of Htn-Q150 in ASH neurons. No dead, and fewer degenerative, ASH neurons were observed by day 8 in the ogt-1 mutant adults compared with the control strain (Fig. 1D). In addition to enhancing the severity of the Htn-Q150 toxicity, oga-1(ok1207) also enhanced the onset of the dye-filling defect. In animals at day 3 of adulthood, more degenerative ASH neurons (40.6%) were observed in the oga-1(ok1207) mutant compared with the control strain (17.7%). In contrast, an alleviated dye-filling defect was observed in the ogt-1 mutants (Fig. S1).
Fig. 1.
The O-GlcNAc cycling mutants regulate HD models. (A–C) In 8-d-old adult worms, ASH neurons (outlined) exposed to Htn-Q150 toxicity were categorized as normal (A), degenerative (B), and dead (C). (D) The percentage of the three categories of ASH neurons from O-GlcNAc single and double mutants: ogt-1a for ogt-1(ok430), and ogt-1b for ogt-1(ok1474). (E) Differential interference contrast and fluorescent images of Q40-YFP aggregates (white dots) in body wall muscles. (F) Quantification of Q40-YFP aggregates in L4 animals. Data are shown as mean ± SEM (**P < 0.001).
OGT and OGA are multidomain proteins capable of mediating other potential functions in addition to catalyzing addition and removal of O-GlcNAc (26, 27). To confirm that alterations in the proteotoxicity observed in the ogt-1 and oga-1 null mutant backgrounds depended upon deregulation of the O-GlcNAc modification per se, we compared the proteotoxic phenotype of Htn-Q150 in the ogt-1;oga-1 double mutants to those in the ogt-1 or oga-1 single mutants. In O-GlcNAc cycling, the need for OGA activity is wholly dependent on the presence of functional OGT to generate the O-GlcNAcylated protein substrate. Thus, similar phenotypes should be observed in the ogt-1 single mutant and the ogt-1;oga-1 double mutants. Indeed, the ogt-1;oga-1 double mutants completely rescued the ASH cell death phenotype associated with Htn-Q150 proteotoxicity in the oga-1(ok1207) single mutant (Fig. 1D and Fig. S1).
To test whether the effect of O-GlcNAcylation on PolyQ proteotoxicity is specific for Htn-Q150, we used a second HD model. In this model, a fusion protein containing 40 glutamine residues fused to YFP (Q40-YFP) is expressed in somatic muscles by using the promoter from the unc-54 gene (29). The Q40-YFP transgenic animals, in an otherwise WT background, show an age-dependent accumulation of fluorescent aggregates in the body wall muscles (Fig. 1E) (29). The oga-1(ok1207) mutation increased the number of Q40-YFP aggregates twofold in the fourth larval stage (L4) animals (Fig. 1F). In contrast, the ogt-1(ok430) mutation decreased the Q40-YFP aggregates by 23% (Fig. 1F). As with the Htn-Q150 model, we also investigated the Q40-YFP phenotype in the ogt-1;oga-1 double mutant; as before, the enhanced phenotypes seen in the oga-1 mutant were suppressed in the double mutant background (Fig. 1F). Thus, the failure to remove O-GlcNAc by loss of OGA activity increased the severity and decreased the age of onset for the Htn-Q150 and Q40-YFP proteotoxicity models. Conversely, the absence of O-GlcNAc addition by the loss of OGT activity suppressed the toxic effects of polyQ in both HD models, demonstrating that alterations in proteotoxicity were dependent on O-GlcNAcylation.
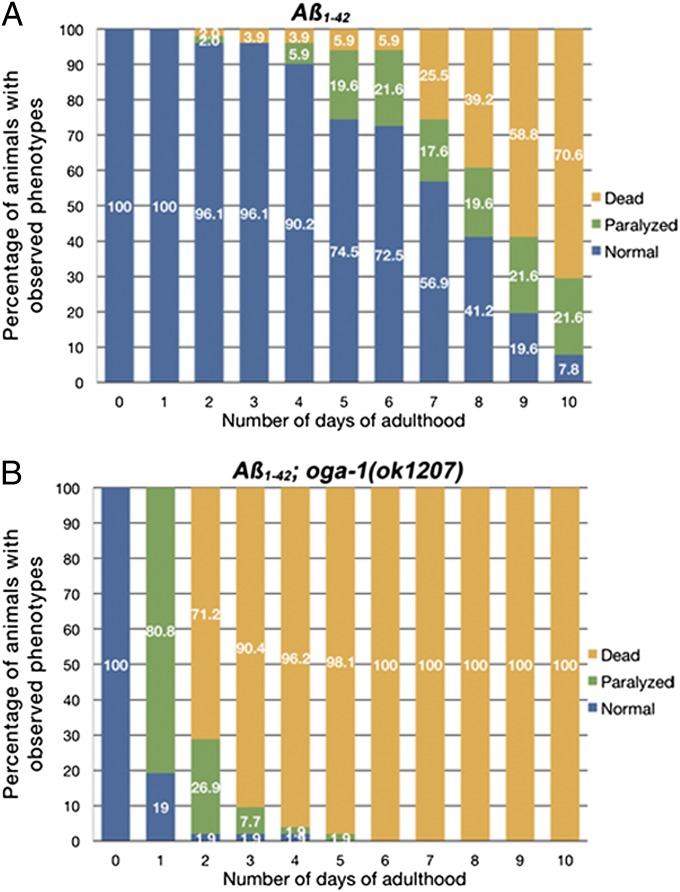
Age-Dependent Paralysis of Aβ1–42 Animals Is Exacerbated in an OGA Mutant Background.
In the HD models described earlier, the phenotypic onset was age-dependent and enhanced in oga-1(ok1207) (Fig. 1). We then turned to a C. elegans model expressing amyloid β-peptide, which is implicated in AD and inclusion body myositis (2, 30). In this model, the amyloid β1–42 peptide is expressed in the muscles (i.e., Aβ1–42) (31). Aβ1–42 animals show a variety of phenotypes, including age-dependent paralysis in adults (31) (Movie S1), constipation caused by disruption of the defecation muscle functions, and egg-laying defects (Egl) in hermaphrodites caused by defects in sex muscle functions (Fig. S2). Severe Egl phenotypes can lead to early adult death as a result of progeny hatching inside the hermaphrodite uterus (i.e., bagging). Approximately 50 Aβ1–42 or Aβ1–42;oga-1(ok1207) animals were scored daily for the phenotypes from days 1 to 10 of adulthood. The paralysis phenotype caused by the Aβ1–42 peptide was more severe with earlier onset in oga-1(ok1207). Most Aβ1–42;oga-1(ok1207) adults (>80%) were paralyzed at day 1 of adulthood, compared with less than 20% at day 5 of adulthood in the Aβ1–42 animals (Fig. 2 A and B). In addition, many animals showed a strong Egl phenotype, and quickly died from bagging, with all animals dead by day 6 of adulthood (Fig. 2B). WT N2 or oga-1(ok1207) single mutant animals lacking Aβ1–42 did not show a paralyzing phenotype, and most animals were still alive at day 5 of adulthood under the same conditions (Fig. S3), suggesting that the toxic Aβ1–42 peptide was the cause of the paralysis. Several efforts to cross the Aβ1–42 strain into the ogt-1 mutant background were unsuccessful. As an alternative, we carried out ogt-1 feeding RNAi experiments on the Aβ1–42 strain, but no obvious effects were seen, possibly as a result of incomplete knockdown of OGT activity.
Fig. 2.
The oga-1(ok1207) mutation significantly enhanced the toxicity of Aβ1–42 peptide. (A) Aβ1–42 transgenic animals. (B) Aβ1–42 animals in the oga-1(ok1207) mutant background.
Null Alleles of ogt-1 Are Neuroprotective in C. elegans Frontotemporal Dementia with Parkinsonism Chromosome 17 Tauopathy Model.
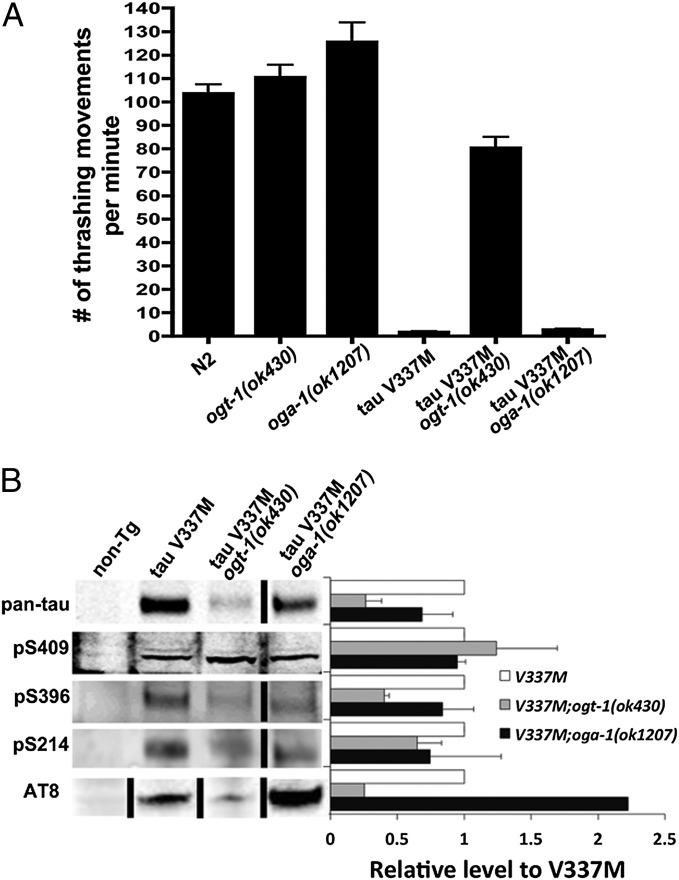
As a final test of the influence of O-GlcNAc cycling on proteotoxicity, we selected a neuronal tauopathy model. A mutation (V337M) in the microtubule-associated protein tau has been identified as the defect in human frontotemporal dementia with parkinsonism chromosome 17 (FTDP-17) (32). FTDP-17 is one of many human tauopathies in which characteristic neurofibrillary tangles are formed from hyperphosphorylated tau. The C. elegans transgenic model expressing the tau V337M throughout the nervous system shows phenotypes of uncoordinated locomotion, axon degeneration, and neuronal death (33). A simple, yet effective, method to assay neuronal function in C. elegans is by assessing “thrashing” behavior. When placed in liquid, WT animals thrash actively by repeatedly moving the head and tail to the same side of body (Movie S2), the rate of which can be determined by counting the number of body bends per minute. WT and ogt-1 or oga-1 single mutant animals exhibit a similar thrashing rate of 100 to 120 bends per minute (Fig. 3A). In contrast, tau V337M transgenic animals showed a significant decrease in the number of thrashing movements (Fig. 3A), with a kinked tail unable to move coordinately with the head (Movie S3) as a result of the severe neurotoxicity of this mutant protein, as previously described (33).
Fig. 3.
Thrashing-defective phenotype of tau V337M animals was rescued by the ogt-1(ok430) mutation. (A) Mean number of thrashing movements ± SEM. (B) Immunoblot of tau protein in WT and O-GlcNAc cycling mutant animals. Protein extract was analyzed by immunoblotting with the following antibodies: phosphorylation-independent tau (pan-tau), pS409, pS396, and pS214 for specific phosphorylated serine residues; and AT-8 for pS202/pT205. Data are shown as mean ± SD.
We crossed the transgenic tau V337M mutant strain into the ogt-1 or oga-1 null mutant background. The uncoordinated locomotion phenotype was completely rescued and the thrashing rate was significantly improved when the tau V337M was crossed into the ogt-1(ok430) mutant background (Fig. 3A and Movie S4). The oga-1(ok1207) did not worsen the thrashing defect of the tau V337M transgenic animals, as observed in other neurodegenerative models tested (Fig. 3A and Movie S5). The tau protein is modified by O-GlcNAc and phosphorylation (14, 34), and hyperphosphorylated tau forms the characteristic neurofibrillary tangles (1). To examine whether the phenotypes seen in the O-GlcNAc cycling mutants result from changes in the phosphorylation state of tau V337M protein, we examined the accumulation of the total and phosphorylated tau by using specific antisera by Western blotting. In general, the levels of the phosphorylation-independent tau were decreased in ogt-1 and oga-1 null mutants. Interestingly, there was less pS396 and pS202/pT205 in ogt-1(ok430) and increased pS202/pT205 in oga-1(ok1207) (Fig. 3B). We concluded that the ogt-1 mutation decreased the overall level of the tau protein and multiple phosphorylated forms thus alleviated tau V337M proteotoxicity. The oga-1 mutation did not further worsen the existing severe phenotype even though it increased tau phosphorylation as detected by certain tau phospho-specific antibodies.
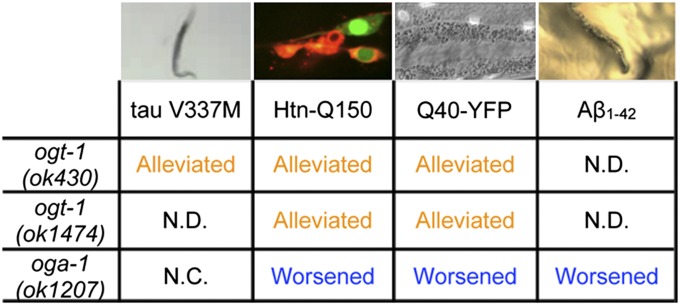
For multiple C. elegans neurodegenerative disease models, loss of O-GlcNAcylation (ogt-1 mutants) alleviated the proteotoxicity of the exogenous, aggregate-prone proteins whereas an excess of O-GlcNAcylation (oga-1 mutant) enhanced it (except for the tau V337M model; Fig. 4). Interestingly, O-GlcNAcylation modulated the proteotoxicity when the toxic protein was expressed in neurons (Htn-Q150 and tau V337M) or muscles (Q40-YFP and Aβ1–42). Additionally, the effect of O-GlcNAcylation was not universal to all types of proteotoxicity. O-GlcNAcylation had no effects on a generic proteotoxicity model (GFP-degron; Fig. S4) (35). Quantitative measurements (i.e., with quantitative RT-PCR) of the expression of different transgenes suggested that differential expression of the toxic transgene was not responsible for the phenotypes observed in the O-GlcNAc cycling mutants (Fig. S5). We next tested whether the effects of disrupted O-GlcNAc cycling on these proteotoxicity models were linked to protein stability and turnover.
Fig. 4.
CeOGT and CeOGA mutations regulate proteotoxicity in multiple C. elegans models of neurodegenerative diseases differently. N.C., no change in the phenotype; N.D., not determined.
O-GlcNAc Cycling Influences Pathways of Protein Degradation: Proteasome and Autophagy.
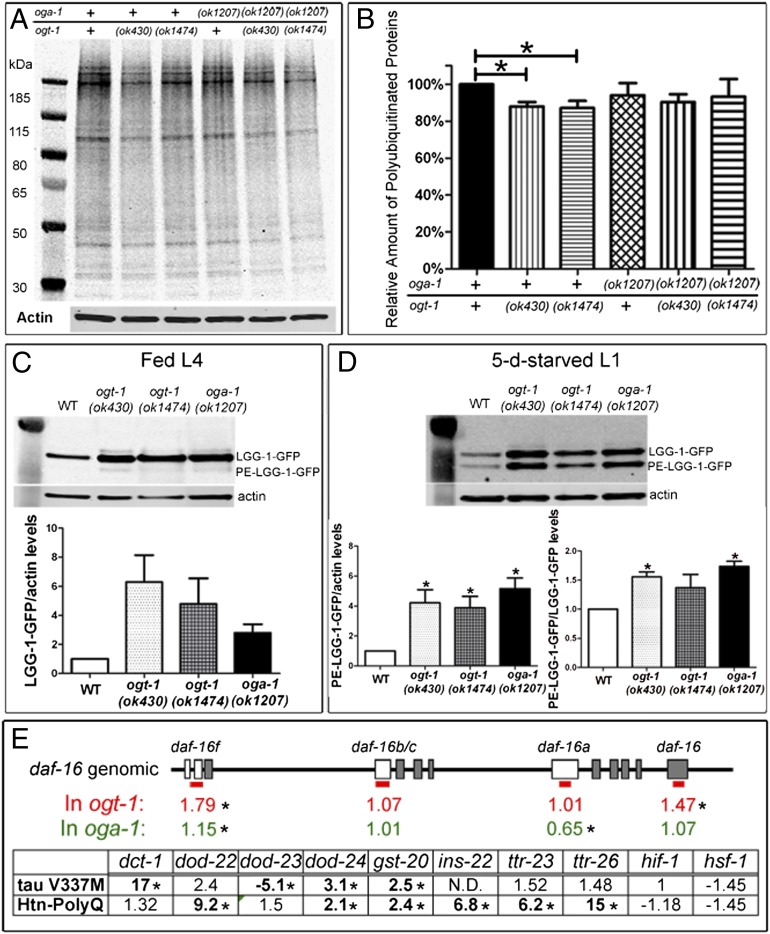
Many cellular protein aggregates are ubiquitinated and proteins involved in several neurodegenerative diseases are known substrates of the ubiquitin proteasome system (36, 37). To explore the possibility that O-GlcNAc cycling modifies proteotoxicity by regulating proteasome activity, we examined the accumulation of polyubiquitinated proteins in WT and the O-GlcNAc cycling mutants. As shown in Fig. 5 A and B, there was a slight (13%) decrease in the level of polyubiquitinated proteins observed in the ogt-1 mutants compared with WT animals, consistent with a modest activation of proteasome activity and the reduction in proteotoxicity observed in these mutants. However, we did not see significant changes in the level of polyubiquitinated proteins in the oga-1(ok1207) and ogt-1;oga-1 double mutants (Fig. 5 A and B), suggesting regulation of proteasome activity is not the only mechanism responsible for the effects of O-GlcNAcylation on proteotoxicity.
Fig. 5.
The O-GlcNAc cycling mutants modulate proteotoxicity through multiple pathways in C. elegans. (A and B) Proteasome activity was activated in the ogt-1 mutants. (A) Polyubiquitinated proteins. (B) Relative level of polyubiquitinated proteins to WT animals. Data are shown as mean ± SEM. WT alleles are indicated with plus signs (*P < 0.05). (C and D) Autophagy was activated in the ogt-1 and oga-1 mutants. (C) LGG-1-GFP was induced in the ogt-1 and oga-1 mutants in fed L4 larvae, with little PE-conjugated LGG-1-GFP. (D) PE-LGG-1-GFP was induced in 5-d starved stage 1 larvae (Upper) and normalized to actin (Lower, Left) or unmodified LGG-1-GFP (Lower, Right). Data are shown as mean ± SEM (*P < 0.05). (E) Quantitative PCR analysis of daf-16 isoforms in the O-GlcNAc cycling mutants and DAF-16 downstream targets and insulin pathway-related genes in two neuronal proteotoxicity models in the ogt-1 mutant background. Blank boxes indicate exons unique for daf-16 isoforms, which are detected with specific primer sets labeled by a red bar (*Significant at P < 0.05). N.D., not determined because transcript was undetectable.
Autophagy has also been suggested to play a role in the clearance of protein aggregates in neurodegenerative diseases (38). The formation of autophagosomes is triggered under conditions of starvation and stress. The O-GlcNAc cycling machinery acts as a nutrient sensor and has been suggested to regulate autophagy (39, 40). To test this hypothesis, we used a marker of autophagy in C. elegans, LGG-1-GFP, to investigate the effects of O-GlcNAcylation on autophagy (41, 42). In fed animals, we observed increased expression of LGG-1-GFP in ogt-1 and oga-1 mutants, but with little phosphatidylethanolamine (PE)-conjugated isoform (PE-LGG-1-GFP; Fig. 5C), which is the marker of autophagy induction (41). Under starvation, ogt-1 and oga-1 mutants showed increased LGG-1-GFP expression as in the fed state. Moreover, there was an increase in the active form (PE-LGG-1-GFP) when normalized to actin or LGG-1-GFP (Fig. 5D). These findings suggest that O-GlcNAc cycling plays a significant role in regulating the process of autophagy in C. elegans. However, it is unclear to what extent O-GlcNAcylation regulates proteotoxicity through autophagy because OGA loss of function mutants enhanced proteotoxicity despite the induction of autophagy.
The O-GlcNAc Cycling Mutants Deregulate daf-16 Isoforms and Downstream Targets.
DAF-16 has been shown to be a key regulator of longevity, stress response, and proteotoxicity in C. elegans (21, 43). Multiple daf-16 splicing isoforms have been identified, with different expression patterns (44) and distinctive O-GlcNAc marks at their promoters (45). To ask whether the O-GlcNAc cycling mutants regulate proteotoxicity through a DAF-16–dependent pathway, the expression level of daf-16 isoforms and DAF-16 targets was measured with quantitative RT-PCR. The O-GlcNAc cycling mutants significantly altered the transcript levels of isoforms daf-16f and daf-16a. Whereas daf-16f and total daf-16 transcripts were significantly up-regulated in the ogt-1 mutant, daf-16a was uniquely down-regulated in the oga-1 mutant (Fig. 5E). This deregulation of daf-16 was further confirmed by looking at known DAF-16 targets in the two neuronal proteotoxicity models (Fig. 5E). In the ogt-1 mutant background, multiple DAF-16 targets, including two transthyretin-like genes, were significantly deregulated in the proteotoxic models, whereas hif-1 and hsf-1 were not significantly changed (Fig. 5E and Datasets S1 and S2). HSF-1 has been demonstrated to have a joint action in regulating proteotoxicity in C. elegans with DAF-16 (21, 43). Datasets S1 and S2 provide expression data in the oga-1 mutant, muscular proteotoxicity models, and a complete list of genes tested. These findings, and our previously published work (45), suggest that O-GlcNAc cycling may alter DAF-16–dependent gene expression, thus providing a mechanistic link between the hexosamine signaling pathway and proteotoxicity.
Discussion
Viable mutants in C. elegans provide a unique opportunity to examine the effects of loss of O-GlcNAc cycling in well-defined neurodegenerative models. Our findings clearly demonstrate that O-GlcNAc cycling can influence the severity of proteotoxic phenotypes in C. elegans. Direct O-GlcNAc modification on amyloid precursor protein regulates its proteolytic processing (15, 46), which was not the focus in the present study because Aβ1–42 is directly expressed in muscles intracellularly. Tau can be modified by O-GlcNAc and O-phosphorylation (14, 34). Hyperphosphorylated tau forms the characteristic neurofibrillary tangles found in AD and other tauopathies, including FTDP-17 as modeled in our study with tau V337M (1, 33, 47). Based on the extensive crosstalk between these two modifications on tau (11, 34), one working model is that increased O-GlcNAcylation (when OGA is mutated or inhibited) will decrease tau phosphorylation and inhibit aggregate formation. A recent study with Thiamet-G (an O-GlcNAcase inhibitor) in transgenic mice overexpressing P301L tau, another mutation identified in FTDP-17, decreases tau aggregate formation and neuronal loss without altering tau phosphorylation (12). In contrast, we observe that increased O-GlcNAcylation (in oga-1 mutant) has no effect on the already severe tauopathy phenotype, whereas loss of the OGT (in ogt-1 mutant) rescued the tauopathy (Fig. 3A). We also observe unchanged tau phosphorylation at S409, S396, and S214, and increased phosphorylation at S202/T205 in the O-GlcNAcase null mutant (Fig. 3B). Thus, the relationship between tau O-GlcNAcylation and phosphorylation is likely to be more complex than a simple competition model might suggest. The discrepancy observed in FTDP-17 models in mouse and C. elegans could be a result of differences between the model organisms used in these studies. In addition, one must also be cautious in comparing the effects of treating animals with a pharmacological agent blocking O-GlcNAc cycling and the consequences of mutant alleles that have a chronic loss of activity. Adaptive changes in protein homeostasis and gene expression may occur (and even be selected for) in mutant animals, and these may mask physiological effects seen upon relatively acute pharmacological treatments.
O-GlcNAcylation can also modulate protein homeostasis indirectly through the stress response, insulin signaling, and protein degradation pathways (45, 48). Excessive O-GlcNAcylation has been shown to block proteasome activity and proteasome-dependent degradation of Sp1, p53, and delta-lactoferrin (49–52). Moreover, knocking down OGT activity leads to proteasome activation (51). Consistent with this finding, we observe reduced proteotoxicity and overall lower levels of polyubiquitinated proteins in ogt-1 mutants (Figs. 4 and 5A). O-GlcNAcylation has also been proposed to regulate autophagy (39, 40). RNAi knockdown of autophagy components in C. elegans enhances the proteotoxicity of Htn-Q150 and Q40-YFP (23). We show that both O-GlcNAc cycling mutants substantially up-regulate the expression of the autophagy marker LGG-1-GFP and enhance autophagy induction during starvation, whereas ogt-1 and oga-1 mutants regulated proteotoxicity in opposite directions. Thus, modulation of the autophagy and proteasome activity may not provide a full explanation for the altered proteotoxic phenotype in the O-GlcNAc cycling mutants.
Insulin/insulin-like growth factor–like signaling (IIS) has been linked to neurodegenerative diseases and associated proteotoxicity (21, 53, 54). O-GlcNAc cycling is a key regulator of the insulin pathway in C. elegans, impacting dauer formation and longevity (26, 27, 45). Decreased IIS signaling, and the resulting release of inhibition of the downstream transcription factor, DAF-16, ameliorate the phenotype of C. elegans proteotoxicity models; conversely, RNAi knockdown of DAF-16 enhances it (21, 22, 43). Here, we have shown that daf-16 isoforms are differentially regulated in the O-GlcNAc cycling mutants, resulting in deregulation of multiple downstream targets of DAF-16 in the proteotoxicity models. The deregulated targets include the transthyretin-like genes, whose products are generally believed to be neuroprotective proteins in AD (55). Thus, modulation of the IIS pathway is likely to be one of the key mechanisms by which changes in O-GlcNAcylation influences proteotoxicity. Our findings extend the growing body of evidence linking insulin signaling to neurodegeneration (4, 56). The striking effects of O-GlcNAcylation on multiple neurodegenerative disease models open the possibility of exploiting the O-GlcNAc cycling pathway therapeutically for these types of disorders.
Materials and Methods
Strains.
O-GlcNAc cycling mutant strains used in this paper include ogt-1(ok430) (26), ogt-1(ok1474) (45), and oga-1(ok1207) (27). Details of C. elegans proteotoxic models are provided in SI Materials and Methods.
Scoring Degeneration and Death of ASH Neurons.
The proteotoxicity of Htn-Q150 in ASH neurons was assessed as described previously with minor modification (28). Day-3 or day-8 adult animals were incubated in the fluorescent dye DiI (20 μg/mL) for 1 h at room temperature. Then, after a 1-h recovery on NGM plates to remove extra dye attached to the body surface, the animals were mounted on 2% (wt/vol) agarose pads. The DiI dye staining and OSM-10::GFP expression in ASH neurons were scored with a confocal spin disk microscope.
Aggregate Quantification of Q40-YFP Animals.
Synchronized first larval stage animals were kept at 20 °C for 2 d, and the resulting L4 animals were observed under a stereomicroscope with an epifluorescence illuminator. Aggregates of Q40-YFP protein were counted as described previously (29).
Paralysis Assay of Animals Expressing Amyloid β-Peptide or GFP-Degron in Body Wall Muscles.
Fifty synchronized adult animals were scored daily for their movement phenotype from days 1 to 10 of adulthood (35). The phenotypes were scored in three categories: normal, paralyzed, and dead. Animals that moved freely on the plate or responded well to light touch (i.e., moving in the opposite direction of the touch) were scored as normal; those that did not respond promptly or had a “halo” formed around their head (as a result of limited body movement and feeding on the bacterial lawn) were scored as paralyzed; and those that could not move at all and did not have pharyngeal pumping were scored as dead.
Worm Thrashing Assay.
L4 larvae were picked and placed in 15 μL of M9 buffer on a glass slide. The animals were allowed to acclimate for 1 to 2 min, after which their movements in the liquid were recorded with a camera attached to a stereomicroscope. The number of thrashes per minute was counted in a slow-motion analysis of the recording for each animal.
Western Blot Analysis.
Ten 1-d-old adult animals were lysed at 95 °C in the presence of LDS sample buffer and β-mercaptoethanol. After brief centrifugation, the proteins in the lysates were separated on a 4% to 12% NuPAGE Bis-Tris gel. The procedures of blocking and blotting followed the standard protocol for the Odyssey infrared imaging system (Li-COR Biosciences). Primary antibodies used in the study are listed in SI Materials and Methods.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (NIH). We thank Drs. A. Hart, C. Link, and B. Kraemer for transgenic strains and the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.W.H. is a guest editor invited by the Editorial Board.
See Commentary on page 17319.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205748109/-/DCSupplemental.
References
- 1.Grundke-Iqbal I, et al. Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J Biol Chem. 1986;261:6084–6089. [PubMed] [Google Scholar]
- 2.Masters CL, et al. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci USA. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 4.Cohen E, Dillin A. The insulin paradox: Aging, proteotoxicity and neurodegeneration. Nat Rev Neurosci. 2008;9:759–767. doi: 10.1038/nrn2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat Rev Neurosci. 2007;8:368–378. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- 6.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 7.Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259:3308–3317. [PubMed] [Google Scholar]
- 8.Caldwell SA, et al. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene. 2010;29:2831–2842. doi: 10.1038/onc.2010.41. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, et al. Increased hexosamine biosynthesis and protein O-GlcNAc levels associated with myocardial protection against calcium paradox and ischemia. J Mol Cell Cardiol. 2006;40:303–312. doi: 10.1016/j.yjmcc.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 10.McClain DA, et al. Altered glycan-dependent signaling induces insulin resistance and hyperleptinemia. Proc Natl Acad Sci USA. 2002;99:10695–10699. doi: 10.1073/pnas.152346899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuzwa SA, et al. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat Chem Biol. 2008;4:483–490. doi: 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
- 12.Yuzwa SA, et al. Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat Chem Biol. 2012;8:393–399. doi: 10.1038/nchembio.797. [DOI] [PubMed] [Google Scholar]
- 13.Liu K, et al. Accumulation of protein O-GlcNAc modification inhibits proteasomes in the brain and coincides with neuronal apoptosis in brain areas with high O-GlcNAc metabolism. J Neurochem. 2004;89:1044–1055. doi: 10.1111/j.1471-4159.2004.02389.x. [DOI] [PubMed] [Google Scholar]
- 14.Arnold CS, et al. The microtubule-associated protein tau is extensively modified with O-linked N-acetylglucosamine. J Biol Chem. 1996;271:28741–28744. doi: 10.1074/jbc.271.46.28741. [DOI] [PubMed] [Google Scholar]
- 15.Griffith LS, Mathes M, Schmitz B. Beta-amyloid precursor protein is modified with O-linked N-acetylglucosamine. J Neurosci Res. 1995;41:270–278. doi: 10.1002/jnr.490410214. [DOI] [PubMed] [Google Scholar]
- 16.Dong DL, et al. Glycosylation of mammalian neurofilaments. Localization of multiple O-linked N-acetylglucosamine moieties on neurofilament polypeptides L and M. J Biol Chem. 1993;268:16679–16687. [PubMed] [Google Scholar]
- 17.Cole RN, Hart GW. Cytosolic O-glycosylation is abundant in nerve terminals. J Neurochem. 2001;79:1080–1089. doi: 10.1046/j.1471-4159.2001.00655.x. [DOI] [PubMed] [Google Scholar]
- 18.Bertram L, et al. Evidence for genetic linkage of Alzheimer’s disease to chromosome 10q. Science. 2000;290:2302–2303. doi: 10.1126/science.290.5500.2302. [DOI] [PubMed] [Google Scholar]
- 19.Twine NA, Janitz K, Wilkins MR, Janitz M. Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer’s disease. PLoS ONE. 2011;6:e16266. doi: 10.1371/journal.pone.0016266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rexach JE, et al. Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat Chem Biol. 2012;8:253–261. doi: 10.1038/nchembio.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 22.Florez-McClure ML, Hohsfield LA, Fonte G, Bealor MT, Link CD. Decreased insulin-receptor signaling promotes the autophagic degradation of beta-amyloid peptide in C. elegans. Autophagy. 2007;3:569–580. doi: 10.4161/auto.4776. [DOI] [PubMed] [Google Scholar]
- 23.Jia K, Hart AC, Levine B. Autophagy genes protect against disease caused by polyglutamine expansion proteins in Caenorhabditis elegans. Autophagy. 2007;3:21–25. doi: 10.4161/auto.3528. [DOI] [PubMed] [Google Scholar]
- 24.Webster DM, et al. O-GlcNAc modifications regulate cell survival and epiboly during zebrafish development. BMC Dev Biol. 2009;9:28. doi: 10.1186/1471-213X-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shafi R, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci USA. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanover JA, et al. A Caenorhabditis elegans model of insulin resistance: Altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc Natl Acad Sci USA. 2005;102:11266–11271. doi: 10.1073/pnas.0408771102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forsythe ME, et al. Caenorhabditis elegans ortholog of a diabetes susceptibility locus: Oga-1 (O-GlcNAcase) knockout impacts O-GlcNAc cycling, metabolism, and dauer. Proc Natl Acad Sci USA. 2006;103:11952–11957. doi: 10.1073/pnas.0601931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faber PW, Alter JR, MacDonald ME, Hart AC. Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc Natl Acad Sci USA. 1999;96:179–184. doi: 10.1073/pnas.96.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Askanas V, Engel WK, Alvarez RB. Light and electron microscopic localization of beta-amyloid protein in muscle biopsies of patients with inclusion-body myositis. Am J Pathol. 1992;141:31–36. [PMC free article] [PubMed] [Google Scholar]
- 31.Link CD. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc Natl Acad Sci USA. 1995;92:9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spillantini MG, Bird TD, Ghetti B. Frontotemporal dementia and Parkinsonism linked to chromosome 17: A new group of tauopathies. Brain Pathol. 1998;8:387–402. doi: 10.1111/j.1750-3639.1998.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraemer BC, et al. Neurodegeneration and defective neurotransmission in a Caenorhabditis elegans model of tauopathy. Proc Natl Acad Sci USA. 2003;100:9980–9985. doi: 10.1073/pnas.1533448100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu F, Iqbal K, Grundke-Iqbal I, Hart GW, Gong CX. O-GlcNAcylation regulates phosphorylation of tau: A mechanism involved in Alzheimer’s disease. Proc Natl Acad Sci USA. 2004;101:10804–10809. doi: 10.1073/pnas.0400348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Link CD, et al. Conversion of green fluorescent protein into a toxic, aggregation-prone protein by C-terminal addition of a short peptide. J Biol Chem. 2006;281:1808–1816. doi: 10.1074/jbc.M505581200. [DOI] [PubMed] [Google Scholar]
- 36.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 37.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 38.Komatsu M, Kominami E, Tanaka K. Autophagy and neurodegeneration. Autophagy. 2006;2:315–317. doi: 10.4161/auto.2974. [DOI] [PubMed] [Google Scholar]
- 39.Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta. 2010;1800:80–95. doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marsh SA, Powell PC, Dell’italia LJ, Chatham JC. Cardiac O-GlcNAcylation blunts autophagic signaling in the diabetic heart. Life Sci. 2012 doi: 10.1016/j.lfs.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang C, You YJ, Avery L. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev. 2007;21:2161–2171. doi: 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meléndez A, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 43.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 44.Kwon ES, Narasimhan SD, Yen K, Tissenbaum HA. A new DAF-16 isoform regulates longevity. Nature. 2010;466:498–502. doi: 10.1038/nature09184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love DC, et al. Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc Natl Acad Sci USA. 2010;107:7413–7418. doi: 10.1073/pnas.0911857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacobsen KT, Iverfeldt K. O-GlcNAcylation increases non-amyloidogenic processing of the amyloid-β precursor protein (APP) Biochem Biophys Res Commun. 2011;404:882–886. doi: 10.1016/j.bbrc.2010.12.080. [DOI] [PubMed] [Google Scholar]
- 47.Bancher C, et al. Accumulation of abnormally phosphorylated tau precedes the formation of neurofibrillary tangles in Alzheimer’s disease. Brain Res. 1989;477:90–99. doi: 10.1016/0006-8993(89)91396-6. [DOI] [PubMed] [Google Scholar]
- 48.Keembiyehetty CN, Krzeslak A, Love DC, Hanover JA. A lipid-droplet-targeted O-GlcNAcase isoform is a key regulator of the proteasome. J Cell Sci. 2011;124:2851–2860. doi: 10.1242/jcs.083287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang WH, et al. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- 50.Hardivillé S, Hoedt E, Mariller C, Benaïssa M, Pierce A. O-GlcNAcylation/phosphorylation cycling at Ser10 controls both transcriptional activity and stability of delta-lactoferrin. J Biol Chem. 2010;285:19205–19218. doi: 10.1074/jbc.M109.080572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang F, et al. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell. 2003;115:715–725. doi: 10.1016/s0092-8674(03)00974-7. [DOI] [PubMed] [Google Scholar]
- 52.Wang K, et al. Increased O-GlcNAc causes disrupted lens fiber cell differentiation and cataracts. Biochem Biophys Res Commun. 2009;387:70–76. doi: 10.1016/j.bbrc.2009.06.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen E, et al. Temporal requirements of insulin/IGF-1 signaling for proteotoxicity protection. Aging Cell. 2010;9:126–134. doi: 10.1111/j.1474-9726.2009.00541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steen E, et al. Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer’s disease—is this type 3 diabetes? J Alzheimers Dis. 2005;7:63–80. doi: 10.3233/jad-2005-7107. [DOI] [PubMed] [Google Scholar]
- 55.Schwarzman AL, et al. Transthyretin sequesters amyloid beta protein and prevents amyloid formation. Proc Natl Acad Sci USA. 1994;91:8368–8372. doi: 10.1073/pnas.91.18.8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pilcher H. Alzheimer’s disease could be “type 3 diabetes”. Lancet Neurol. 2006;5:388–389. doi: 10.1016/s1474-4422(06)70434-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.