Abstract
Myeloid/lymphoid or mixed-lineage leukemia (MLL)-family genes encode histone lysine methyltransferases that play important roles in epigenetic regulation of gene transcription. MLL genes are frequently mutated in human cancers. Unlike MLL1, MLL2 (also known as ALR/MLL4) and its homolog MLL3 are not well-understood. Specifically, little is known regarding the extent of global MLL2 involvement in the regulation of gene expression and the mechanism underlying its alterations in driving tumorigenesis. Here we profile the global loci targeted by MLL2. A combinatorial analysis of the MLL2 binding profile and gene expression in MLL2 wild-type versus MLL2-null isogenic cell lines identified direct transcriptional target genes and revealed the connection of MLL2 to multiple cellular signaling pathways, including the p53 pathway, cAMP-mediated signaling, and cholestasis signaling. In particular, we demonstrate that MLL2 participates in retinoic acid receptor signaling by promoting retinoic acid-responsive gene transcription. Our results present a genome-wide integrative analysis of the MLL2 target loci and suggest potential mechanisms underlying tumorigenesis driven by MLL2 alterations.
Keywords: tumor suppressor, somatic targeting, S100A gene cluster
Modulation of chromatin accessibility through chromatin remodeling and histone modification is a critical step in regulating eukaryotic gene transcription. Histone H3 lysine 4 (H3K4) methylation by histone methyltransferases is an evolutionarily conserved epigenetic mark for active gene transcription (1). In mammalian cells, six complexes with H3K4 methyltransferase activity have been identified, each with one SET domain-containing methyltransferase subunit, including Set1A, Set1B, and four MLL-family proteins. In humans, these MLL-family genes include MLL1, MLL2 (GenBank accession no. NM_003482; also known as ALR/MLL4), MLL3, and MLL4 (GenBank accession no. NM_014727; also known as MLL2 or Wbp7, conflicting with the official symbol for MLL2). Each of these proteins is associated with a complex that modulates histone methylation (2). Among the MLL genes, MLL1 and its homolog MLL4 have been the most extensively studied, as the former is frequently involved in genetic alterations in leukemia (reviewed in ref. 3). In addition, MLL4 has been found to be overexpressed in breast and colorectal cell lines (4). MLL1 and MLL4 play essential roles in regulating the expression of genes that are involved in cell differentiation and early embryonic development, among which the most notable and best-established are the Homeobox (Hox) genes (5, 6).
MLL2 was originally cloned as a human homolog of Drosophila trithorax (7). MLL2 and its homolog, MLL3, function distinctly from MLL1 and MLL4. Each of them associates with nuclear receptor coactivator 6 (also known as activating signaling cointegrator 2) to form a complex that contains other subunits including ASH2L, RbBP5, WDR5, DPY30, PTIP, PA1, and UTX (8–10). Together with the chromatin-remodeling complex SWI/SNF, the MLL2 or MLL3 (hereafter referred to as MLL2/3) complex has been found to play essential roles as a coactivator for transcriptional activation by nuclear hormone receptors (11). Consistent with this notion, previous studies have shown that MLL2/3 complexes regulate Hox gene transcription in an estrogen receptor-dependent manner, and that they play critical roles in PPARγ-dependent adipogenesis (12–14).
Somatically acquired epigenetic changes are prevalent in human cancers. Recent cancer genetics studies have uncovered frequent somatic loss-of-function mutations in the genes encoding MLL2/3 complex subunits in a variety of cancer types. UTX was found to be mutated in numerous human cancer types (15–17). MLL3 was found to be mutated in a subset of colorectal cancers and in transitional cell carcinoma (16, 18–20). Most notably, several recent studies identified frequent MLL2-inactivating mutations in non-Hodgkin B-cell lymphomas (21–23). Finally, genes encoding MLL2/3 complex subunits, particularly MLL2, MLL3, and UTX, were found to be mutated in medulloblastoma, establishing them as critical tumor suppressors (24–26).
In contrast to the compelling genetic evidence, much less is known regarding the mechanism underlying the tumor suppressor roles of the MLL2/3 complex. It was shown that knockdown of MLL2 resulted in reduced cancer cell growth and altered adhesion in HeLa cells (9). An MLL3-knockout mouse model study showed that MLL3 plays a role in DNA damage response in a p53-dependent manner, suggesting that the complex serves as a transcriptional coactivator for p53 (27). The link to p53 is consistent with the tumor suppressor role of the MLL3 complex. However, it is most likely that MLL2/3 epigenetic complexes have a broader role biochemically and that the deficiency of the pathway has a more profound impact on cellular processes. To further elucidate the tumor suppressor role of MLL2, we performed a direct profiling of the global loci that are targeted by MLL2. Moreover, we show that somatic knockout of MLL2 in human cells affects the expression of a variety of genes. We note a set of genes that are retinoic acid-responsive and may be directly regulated by MLL2. Finally, integrative pathway analysis uncovered various signaling pathways that are regulated by the MLL2 complex. Our results reveal the global and pathway-specific roles of MLL2 and suggest that it participates in regulating a wide range of pathways with relevance to its role in oncogenesis.
Results
Generation of Endogenous Flag-Tagged MLL2-Expressing Cells.
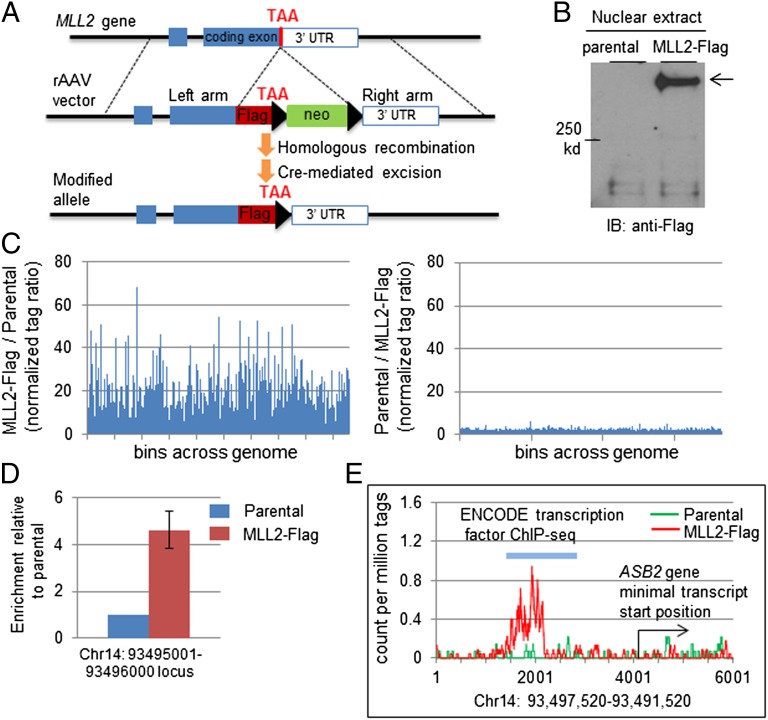
Identifying genomic binding sites is a powerful strategy to reveal genes regulated by transcriptional regulators (5, 28, 29). However, two hurdles confront the direct global profiling of MLL2-targeted loci: (i) It is a massive protein of 5,537 amino acids (∼600 kDa), which complicates biochemical studies that require ectopic expression; and (ii) neither a proven-quality antibody nor a human cell assay is available for chromatin immunoprecipitation-sequencing (ChIP-seq). To overcome these obstacles, we have adopted a somatic knockin-based approach in which an endogenous protein is fused with a short tagging peptide, which facilitates antibody-based ChIP studies (30, 31). We chose the HCT116 carcinoma cell line for our study, as it is a near-diploid cell line with well-defined key cancerous pathways. Furthermore, this cell line has homozygous MLL3-inactivating mutations, thus preventing compensatory effects from obscuring functional analysis (19). An rAAV-based somatic knockin construct (30, 32, 33) enabled the insertion of a DNA fragment encoding Flag tags immediately upstream of the stop codon of the MLL2 gene (Fig. 1A and Fig. S1A). In the parental cells, only the nontagged MLL2 transcript could be detected, whereas in the heterozygous recombinant clones (MLL2-Flag), both the nontagged and Flag-tagged transcripts could be detected (Fig. S1B). Anti-Flag immunoblot detected full-length MLL2-Flag proteins in the nuclear extract (Fig. 1B and Fig. S1C). In addition, the endogenous MLL2-Flag protein was suitable for ChIP, as evidenced by the Flag signal in the ChIP fraction from the somatic knockin clones (Fig. S1D). Collectively and consistent with previous studies using the same strategy for ChIP studies (30), we have established a human cell assay that consists of isogenic cell lines that differ only in the presence or absence of an MLL2-Flag allele, thus permitting the direct profiling of MLL2-targeted loci by a proven-quality antibody-based ChIP-seq.
Fig. 1.
Genome-wide profiling of MLL2 binding sites. (A) rAAV vector and somatic targeting of Flag epitope to the carboxyl terminus of MLL2 protein. (B) Immunoblot detection of the endogenous MLL2-Flag protein. (C) ChIP enrichment analysis identified Flag-tagged MLL2-enriched loci across the genome specifically in MLL2-Flag–expressing cells. (Left) For each qualified locus, the ratio of the normalized distinct tag number in the MLL2-Flag–expressing cells to the distinct tag number in the parental cells is plotted. (Right) For each qualified locus, the reverse ratio of normalized distinct tag number (parental to MLL2-Flag 1) is plotted. (D) ChIP coupled with qPCR to confirm the locus identified by ChIP-seq. Results are from three independent ChIP experiments. Error bar stands for standard deviation from three independent ChIP assays. (E) An MLL2-Flag–enriched locus. The distinct coverage of each base in a 6-kb region is plotted. The gene that is most closely associated with the enriched locus and the corresponding transcription factor binding site identified by ENCODE are indicated.
Global Identification of MLL2-Targeted Loci.
The parental cell line and MLL2-Flag–expressing cells (clone MLL2-Flag1 and 2) were subjected to anti-Flag antibody-based ChIP-seq procedures (Fig. S2A). We modified a previously described differential tag-density analysis (34, 35) to obtain a global view of MLL2-bound loci. We generated bins (regions) across the human genome and compared the tag density for each bin between the parental and MLL2-Flag–expressing cell lines. The relative tag density of the MLL2-Flag–expressing cell line to the parental line showed frequent spikes (Fig. 1C), indicating an MLL2-Flag–specific enrichment. In contrast, such enrichment was absent in the parental line, as indicated by the reverse tag-density ratio (Fig. 1C). The same set of MLL2-Flag–specific enrichments was also apparent in MLL2-Flag–expressing cell line 2 (Fig. S2B).
To identify MLL2-targeted regions, we applied a criterion by which at least fivefold enrichment was required to identify differential tag density (35). A total of 2,060 MLL2 binding events were identified. The enrichment was validated by independent quantitative PCR (qPCR) of the originally sequenced libraries and independent ChIP-qPCR experiments (Fig. 1D and Fig. S2 C and D). Analysis of the global distribution of the binding loci showed that about 59% of them were intragenic (Fig. S3A). The remaining 41% of them were located outside the defined boundary, or intergenic region (Fig. S3A), demonstrating a concentration in intragenic regions of the genome (P < 0.0001). Among those intragenic loci, about 15% were within a distal promoter region, whereas most were within a transcript boundary (Fig. S3B). These results are in agreement with previous studies showing the broad distribution of MLL1 across the genome, including many localized within the gene boundary (36).
We then focused on those binding events that surrounded presumed transcript start sites, as they were most likely linked to direct transcriptional regulation (Dataset S1). Close examination of these loci found that all were previously identified as transcription factor binding sites by the ENCODE Project (28), further supporting that they could be directly targeted by transcription regulators (examples in Fig. 1E, Fig. S2D, and Dataset S1). Genes linked to the MLL2-associated loci encode products of diverse functions, including those of signaling transducers, cellular structure proteins, and microRNAs (Dataset S1). It should be noted that the ChIP assay was performed when cells were under standard culture conditions. It is likely that the extent of the MLL2 complex’s binding to any locus and its specific function are exogenous signaling- and/or cellular context-dependent and thus will be underestimated by a single study. In this regard, the global map of MLL2 binding sites provides valuable information and opportunities for further studies of this essential epigenetic regulator.
Generation of MLL2-Knockout Colorectal Cancer Cells.
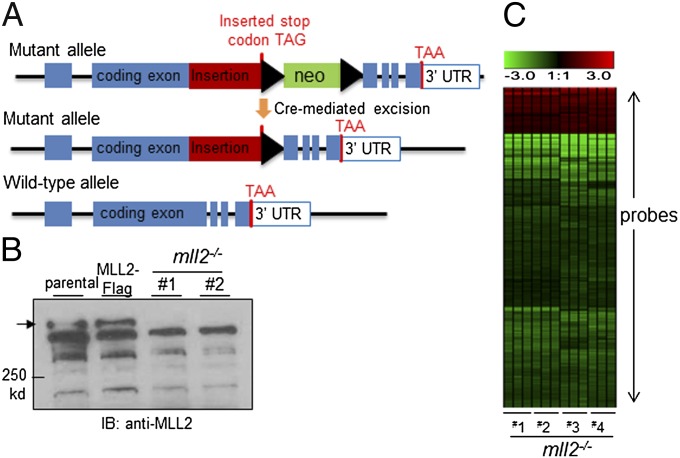
To further investigate the function of MLL2, we generated MLL2-null HCT116 cell lines by modifying the somatic recombination such that a fragment of DNA was inserted into the MLL2 gene, effectively generating inactivating mutations similar to those that are frequently found in cancers (Fig. 2A). As expected, cell lines with insertions in both MLL2 alleles (MLL2−/−) had complete loss of the wild-type MLL2 genes and transcript, and had no detectable MLL2 protein due to the nonsense mutation and/or altered splicing (Fig. 2B and Fig. S1 E and F).
Fig. 2.
Expression profiling of parental and MLL2-knockout cells. (A) Somatic MLL2 knockout by insertion. A DNA fragment containing a stop codon and a neomycin (neo) selection marker was inserted at the position right before the SET domain-coding sequence. The insertion disrupts MLL2 with or without the presence of the neo selection marker. (B) Anti-MLL2 immunoblot confirms the absence of the expected full-length MLL2 protein in MLL2−/− cells. (C) Unsupervised hierarchical clustering for probes with average linkage was performed to reveal differential expression genes in MLL2−/− clones (1–4) in comparison with parental cells. Triplicates for each clone were included and compared with the average expression value of parental cells. Red denotes relative overexpression, and green denotes relative reduced expression level. The exact levels of change for these probes are shown in Dataset S2.
Gene Expression Analysis of Parental and MLL2-Knockout Cells.
To identify downstream events that were regulated by the MLL2 complex, we used microarrays to analyze gene expression in the parental HCT116 cells and MLL2−/− clones. This analysis revealed that a subset of genes that were associated with MLL2-enriched loci displayed reduced expression in MLL2−/− cell lines (Dataset S2). The decreased expression was accompanied by reduced H3K4 trimethylation (Fig. S4). Compared with the overall frequency of genes that were down-regulated by MLL2 deficiency, the ChIP-identified subset of genes was significantly enriched for those whose expression was attenuated in the MLL2−/− cell lines [5.54% (24/433) versus 1.10% (228/20,722) overall when the same cutoff was applied; P < 0.0001]. Gene expression changes were further verified by qPCR of transcripts from the original MLL2−/− clones and by microarray analysis in an additional pair of MLL2−/− lines (Fig. 2C and Dataset S2). Interestingly, there were also genes that displayed higher expression levels in MLL2−/− cells than in the parental cells (Fig. 2C and Dataset S2), consistent with an indirect effect. Overall, most of the down-regulation of gene expression was moderate, in agreement with the observation that deletion of a single MLL-family gene has only minimal effects on global H3K4 methylation (37, 38). The moderate expression changes might also reflect the relatively low expression level of some genes in the parental cells without an exogenous stimulus, suggesting that MLL2 can be poised at various loci independent of transcriptional activation. Nevertheless, these results identified a subset of genes as candidate targets regulated by MLL2 in the specified cellular context.
MLL2 Binds S100A-Family Genes and Regulates Their Expression.
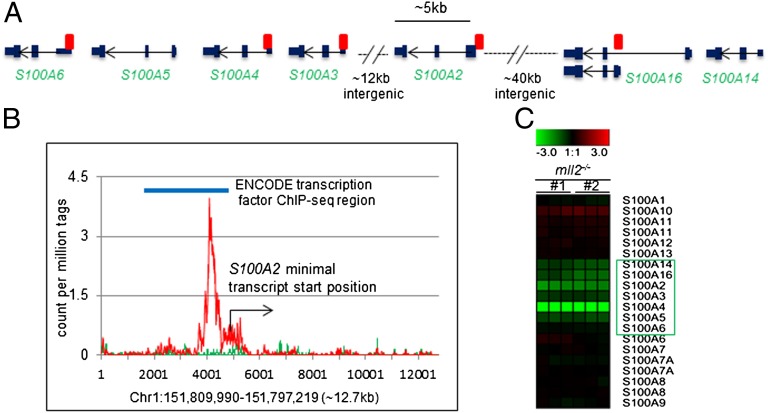
Further examination of the expression profile revealed genes that were potentially direct targets of MLL2 (Dataset S2). Unlike MLL1, MLL2 was not associated with the Hox-family gene cluster in the present study, probably due to the difference in gene functions and/or distinct cellular contexts (5). Instead, we noticed that MLL2 was associated with and likely directly regulated another family gene cluster, the S100 alpha (S100A) genes (Fig. 3 A and B and Fig. S5). Among the 16 S100A genes, a phylogenetically related subgroup of five (S100A6, 5, 4, 3, and 2) cluster within an ∼30-kb region in chromosome 1q21, a region that is frequently altered in cancers (39, 40). MLL2 binding loci were found to localize around the transcriptional start sites of S100A2–6, and at ∼40 kb upstream of this cluster, where S10016, 14, 13, and 1 are located (Fig. 3A). Microarray analysis revealed the apparent down-regulation of the S100A2, 3, 4, 5, 14, and 16 genes and a slight reduction of the S100A6 gene (Fig. 3C). Independent microarray analysis and qPCR confirmed the down-regulation of these genes in another pair of MLL2−/− cell lines derived from a different progenitor MLL2+/− line (Fig. S6).
Fig. 3.
MLL2 binding to the S100A gene cluster. (A) Schematics of the S100A gene cluster region on chromosome 1q21.3. MLL2 bindings within this region (as identified in the present study) are marked (red rectangles). Among these binding events, the S100A2 site was the best-enriched and identified in the original ChIP-seq. The other binding sites have less than fivefold enrichment in the ChIP-seq analysis but were confirmed by an independent ChIP-qPCR. (B) The MLL2 binding site near the transcript start of the S100A2 gene is depicted at single–base-pair resolution based on the normalized distinct tag coverage from the ChIP-seq experiment. (C) Microarray profiling shows reduced expression of the clustered S100A genes in MLL2−/− cells, in comparison with parental cells.
MLL2 Regulates Gene Expression of Multiple Signaling Pathways.
Because coactivators such as MLL2 function together with other transcription factors in regulating gene transcription, we investigated which transcription factors were associated with the MLL2 pathway. Genes that were found to be associated with MLL2-bound loci (Dataset S1; among them, 476 were annotated with official gene symbols) were subjected to Ingenuity Pathway Analysis (IPA) (www.ingenuity.com) for the identification of transcription factors potentially cooperating with MLL2. Interestingly, among the enriched transcription factors were NR3C1, a nuclear receptor, and p53 (Dataset S3). In support of this finding, we also found that many MLL2 binding sites overlapped with previously identified p53-targeted regions (Dataset S1) (29). Although this analysis did not point to a direct interaction between MLL2 and the identified transcription factors, these findings provided promising candidate pathways for further studies. To further gain an integrative view of the functional consequence of MLL2 in the context of cellular signaling pathways, we selected the genes whose expression was down-regulated by MLL2 loss of function and subjected them to IPA canonical pathway analysis (Dataset S2). Analysis of pathways that may be regulated by MLL2 revealed the potential effects of MLL2 on a broad range of cellular processes. Among those signaling pathways that were found to be significantly related were cAMP-mediated signaling (Table 1). Most intriguingly, this canonical pathway and the p53 pathway are among the unique signature pathways that define a particular subtype of medulloblastoma (41).
Table 1.
Ingenuity Pathway Analysis of genes down-regulated by MLL2 loss of function
| Pathway | P value | Gene list |
| cAMP-mediated signaling | 0.0023 | PDE12, PKIA, PKIB, RPS6KA1, PPP3CA |
| LPS/IL-1–mediated inhibition of RXR function | 0.0024 | FABP6, ABCC2, ABCC3, ACOX2, IL-18 |
| Hepatic cholestasis | 0.0032 | ABCC2, ABCC3, FABP6, IL-18 |
| FXR/RAR activation | 0.0062 | ABCC2, FABP6, IL-18 |
| B-cell development | 0.01 | IL7, IL7R |
| Cardiac β-adrenergic signaling | 0.0199 | PDE12, PKIA, PKIB |
| Role of JAK1 and JAK3 in Yc cytokine signaling | 0.0299 | IL7, IL7R |
| PXR/RXR activation | 0.0354 | ABCC2, ABCC3 |
| WNT/β-catenin signaling | 0.0368 | GJA1, SOX4, SOX9 |
| Embryonic stem cell differentiation into cardiac lineages | 0.0427 | ISL1 |
| HER-2 signaling in breast cancer | 0.0433 | ITGB4, NRG1 |
cAMP, cyclic adenosine monophosphate; FXR, farnesoid X receptor; LPS, lipopolysaccharide; RXR, retinoid X receptor; PXR, pregnane X receptor; RAR, retinoic acid receptor.
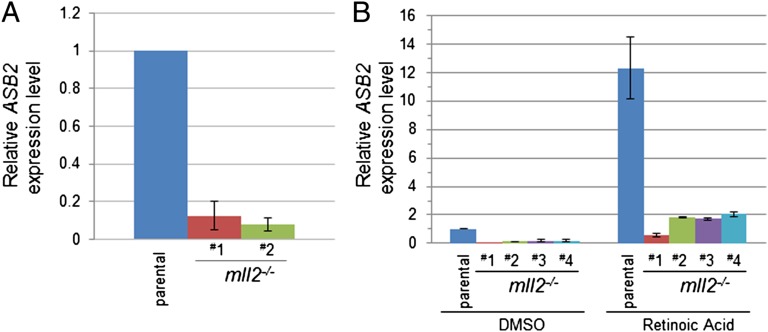
MLL2 Regulates the Expression of the Retinoic Acid-Responsive Gene ASB2.
We next sought to validate the functional role of MLL2 in regulating the transcription of the genes we identified. The pathway analysis revealed that one of the interesting categories of genes was those that respond to nuclear hormones. Interestingly, one of the pathways overrepresented in both MLL2-bound and MLL2-responsive gene sets was retinoic acid signaling. A more careful examination of MLL2-enriched loci revealed that many of them indeed overlapped with retinoic X receptor α ChIP-seq loci (28) (Dataset S1). One of the genes that was linked to the MLL2-enriched loci was ASB2 (Fig. 1E), whose expression was previously found to be induced by retinoic acid in leukemia cells (42, 43). This gene was expressed at a relatively low level in parental HCT116 cells, and quantitative analysis of the ASB2 transcript showed that its expression was further reduced in the MLL2−/− cell lines (Fig. 4A). To verify the role of the MLL2 complex in mediating the retinoic acid response, we tested whether HCT116 cells responded to retinoic acid transcriptionally using ASB2 as a marker gene. Treatment of parental HCT116 cells with retinoic acid induced robust ASB2 expression (Fig. 4B). Furthermore, whereas retinoic acid treatment also led to an increased ASB2 transcript level over a very low basal level in the MLL2−/− cells, the expression level of ASB2 was significantly lower than the level seen in the MLL2-intact parental cells upon retinoic acid treatment (Fig. 4B), indicating that MLL2 involvement accounted for the majority of increased ASB2 expression in response to retinoic acid. Collectively, these results validated the roles of MLL2 uncovered by ChIP-seq and gene expression studies, and suggest an extensive involvement of MLL2 in a variety of cellular signaling pathways.
Fig. 4.
MLL2 regulates the expression of retinoic acid target genes. (A) Reduced expression of ASB2 (a retinoic acid response gene) in MLL2−/− cells, as determined by RT-qPCR. (B) Attenuated ASB2 gene expression in response to retinoic acid treatment in MLL2−/− cells. Four MLL2−/− lines derived from two different MLL2+/− progenitor lines were treated with DMSO or with 1 μM retinoic acid for 48 h. Quantitative RT-PCR was performed to determine the relative expression level of ASB2 genes. Error bars stand for standard deviation from three independent experiments.
Discussion
In this study, we profiled the global binding loci of the MLL2 complex to reveal the scope of its gene regulation. In a parallel effort, we generated MLL2-null cell lines for the identification of genes whose expression was affected by MLL2 loss of function. A combinatorial analysis of these results provided a list of genes that are regulated by the MLL2 complex. These genes encode proteins of distinct functions, which support the notion that MLL2 plays roles in multiple signaling pathways and cellular processes. Among those that are direct targets of the MLL2 complex are the S100A-family genes. The S100A-family genes encode proteins containing EF-hand calcium-binding motifs that regulate intracellular calcium signaling and participate in a number of cellular processes (44). In particular, S100A2 was identified as a putative tumor suppressor (45, 46), possibly through regulating the p53 pathway (47). The regulation of the S100A gene cluster by the MLL2 complex reveals an additional gene cluster that is transcriptionally coregulated by an MLL-family epigenetic regulator, reminiscent of the Hox gene cluster’s regulation by MLL1 (5).
Our findings provide evidence to expand previously established links between H3K4 methyltransferases and other pathways, and suggest that the roles of MLL2 may be extensive. p53 and nuclear hormone receptors are among those in the top of the list in the transcription factor and pathway analysis from our unbiased binding site and gene expression profiling for MLL2. The link of MLL2 to the p53 pathway is in good agreement with the previous MLL3-p53 findings (27), and supports the role of the MLL2 complex as a tumor suppressor in a variety of cancer types. Another example of interest was the potential role of the MLL2 complex in cholestasis signaling, as previously the MLL3 complex was found to play a role in adipogenesis and cholestasis (13, 48). This is supported by the discovery that HNF4A, a nuclear receptor involved in hepatic lipid metabolism and pancreatic islet function (49, 50), is among the transcription factors linked to the MLL2 complex. Finally, among the most interesting discoveries are the signaling pathways that were not previously known to be associated with MLL2 or MLL3 complexes, such as cAMP signaling. These findings, and those from previous work, provide insight that may inform the future search for mechanisms underlying the role of MLL2 in pathogenesis: (i) It is possible that one or more of these currently described pathways contributes to MLL2 mutation-driven tumorigenesis in a cell type-specific manner, whereas the p53 pathway acts independently of cancer type; (ii) comparing gene expression profiles (MLL2 wild-type versus mutant) in different cellular contexts may suggest the primacy of these pathways in tumors bearing MLL2 pathway alterations; and (iii) gene expression signatures characteristic of MLL2 mutations may be exploited for tumor classification, a possibility raised by the findings that several MLL2-affected pathways overlap with the pathways that distinguish particular subgroups of medulloblastoma (41), a cancer that has frequent MLL2 mutations (24–26).
Our profiling study uncovered global distribution characteristics of MLL2 that mirror those of MLL1. Studies of MLL1 have shed light on its global effect on gene expression, including the broad genomic distribution and its specific roles in Hox gene transcription (5). These results have illuminated mechanisms underlying the role of MLL1 in normal hematopoiesis and in leukemogenesis. Previous studies of MLL2 and MLL3 identified roles in nuclear hormone signaling, particularly estrogen receptor signaling, and revealed their similarity to MLL1 in regulating inducible gene expression (5, 12, 51). Further studies will be necessary to test the association of the genes/pathways described herein with MLL-family members. It is important to point out that although the MLL2 binding events we identified were highly specific, we could not rule out binding of loci that were not positively identified. Just like any antibodies used in similar studies, nonspecific antibody binding would have prevented real binding events from being discovered if they colocalize with the former. Secondarily, the cellular context limitation was also notable, because certain MLL2 binding events may be detectable only upon specific exogenous signals or only in certain types of cells.
In summary, the present study advances our understanding of MLL2 in the following ways: (i) It revealed the global binding profile of the MLL2 protein and the broad extent and characterization of MLL2 distribution across the genome, and thus provides insight into the potential roles of this important epigenetic regulator; (ii) it identified various downstream pathways that are regulated by MLL2, establishing a basis for studying the mechanism underlying the role of MLL2 in development and disease; (iii) it uncovered the regulatory role of MLL2 in the transcription of clustered S100A-family genes that encode for a group of broadly distributed yet understudied proteins; (iv) it further established the extensive role of MLL2 in nuclear hormone signaling and provided a rationale for future studies on the link between nuclear hormone signaling and diseases associated with MLL2 alterations; and finally, (v) our work further validated the somatic recombination-based approach for studying massive proteins, such as MLL2, for which overexpression is a challenging task. The findings and approaches undertaken here have laid the foundation for further understanding the function of MLL2 and have implications for future research on the rapidly accumulating list of epigenetic regulator genes that have been found to be altered in cancers and other diseases.
Materials and Methods
Somatic Targeting.
The vector and protocol for epitope tag targeting and gene knockout have been described previously (28, 49, 50). Briefly, homology arms surrounding the DNA insertion site were amplified by and cloned into an rAAV vector. Following rAAV infection, cells were screened for recombinant clones. Positive clones were infected with an adenovirus expressing Cre recombinase (Vector Biolabs) to remove the drug-resistance cassette. For homozygous gene disruption, the second allele of the MLL2 gene was then targeted following the same procedure, except that the drug-resistance cassette was not removed. Two independent MLL2+/− clones were used for the second MLL2 allele knockout to generate four homozygous lines (MLL2−/−) for experiments.
Nuclear Extraction, Immunoprecipitation, and Western Blot.
Nuclear extraction and IP were performed following a previously described protocol (8). Immunoblot detection of MLL2 was performed by following the standard Western blot procedure, except that a TE22 tank transfer unit (Hoefer) was used for the protein transfer.
ChIP-Seq and Data Analysis.
The ChIP procedure is described in detail in SI Materials and Methods. Briefly, cells were cross-linked and cell lysate was then prepared and subjected to sonication to shear the chromatin. Magnetic beads (New England BioLabs) conjugated with anti-Flag antibody (F3165; Sigma) were used for IP. ChIP-derived DNA was recovered for Illumina GAII-compatible library preparation. Libraries were used for sequencing or qPCR analysis. High-throughput sequencing was performed on the Illumina GAII system. Tags were mapped to the human genome (release hg18) by using the Illumina alignment software Eland. Detailed tag assignment and differential density analysis are described in SI Materials and Methods.
Quantitative PCR.
SYBR label-based qPCR was performed on a 7900HT fast real-time PCR system (Applied Biosystems) and analyzed with the machine software. For qPCR evaluation of gene expression, reverse transcription was performed to convert total RNA into single-strand cDNA using the SuperScript II system (Invitrogen) before qPCR.
Supplementary Material
Acknowledgments
We thank Zhenghe Wang (Case Western Reserve University) for the somatic knockin construct and advice on generating somatic knockin cell lines and the Duke Microarray Core Facility (a Duke National Cancer Institute and Duke Institute for Genome Sciences and Policy shared-resource facility) for technical support, microarray data management, and feedback on the generation of the microarray data reported in this manuscript. This work was supported by the Pediatric Brain Tumor Foundation and The Preston Robert Tisch Brain Tumor Center at Duke (Y.H.) and the Duke Cancer Institute (Y.H.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208807109/-/DCSupplemental.
References
- 1.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25(1):15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 2.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13(8):713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 3.Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annu Rev Pathol. 2012;7:283–301. doi: 10.1146/annurev-pathol-011811-132434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Natarajan TG, et al. Epigenetic regulator MLL2 shows altered expression in cancer cell lines and tumors from human breast and colon. Cancer Cell Int. 2010;10:13. doi: 10.1186/1475-2867-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guenther MG, et al. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci USA. 2005;102(24):8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glaser S, et al. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133(8):1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- 7.Prasad R, et al. Structure and expression pattern of human ALR, a novel gene with strong homology to ALL-1 involved in acute leukemia and to Drosophila trithorax. Oncogene. 1997;15(5):549–560. doi: 10.1038/sj.onc.1201211. [DOI] [PubMed] [Google Scholar]
- 8.Cho YW, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282(28):20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Issaeva I, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27(5):1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goo YH, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23(1):140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S, et al. Crucial roles for interactions between MLL3/4 and INI1 in nuclear receptor transactivation. Mol Endocrinol. 2009;23(5):610–619. doi: 10.1210/me.2008-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ansari KI, Hussain I, Shrestha B, Kasiri S, Mandal SS. HOXC6 is transcriptionally regulated via coordination of MLL histone methylase and estrogen receptor in an estrogen environment. J Mol Biol. 2011;411(2):334–349. doi: 10.1016/j.jmb.2011.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, et al. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci USA. 2008;105(49):19229–19234. doi: 10.1073/pnas.0810100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho YW, et al. Histone methylation regulator PTIP is required for PPARγ and C/EBPα expression and adipogenesis. Cell Metab. 2009;10(1):27–39. doi: 10.1016/j.cmet.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gui Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43(9):875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41(5):521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sjöblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe Y, et al. Frequent alteration of MLL3 frameshift mutations in microsatellite deficient colorectal cancer. PLoS One. 2011;6(8):e23320. doi: 10.1371/journal.pone.0023320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vakoc CR, et al. Low frequency of MLL3 mutations in colorectal carcinoma. Cancer Genet Cytogenet. 2009;189(2):140–141. doi: 10.1016/j.cancergencyto.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012;109(10):3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331(6016):435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pugh TJ, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488(7409):106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488(7409):100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, et al. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc Natl Acad Sci USA. 2009;106(21):8513–8518. doi: 10.1073/pnas.0902873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers RM, et al. ENCODE Project Consortium A user’s guide to the Encyclopedia of DNA Elements (ENCODE) PLoS Biol. 2011;9(4):e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124(1):207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, et al. Epitope tagging of endogenous proteins for genome-wide ChIP-chip studies. Nat Methods. 2008;5(2):163–165. doi: 10.1038/nmeth1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z. Epitope tagging of endogenous proteins for genome-wide chromatin immunoprecipitation analysis. Methods Mol Biol. 2009;567:87–98. doi: 10.1007/978-1-60327-414-2_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rago C, Vogelstein B, Bunz F. Genetic knockouts and knockins in human somatic cells. Nat Protoc. 2007;2(11):2734–2746. doi: 10.1038/nprot.2007.408. [DOI] [PubMed] [Google Scholar]
- 33.Kohli M, Rago C, Lengauer C, Kinzler KW, Vogelstein B. Facile methods for generating human somatic cell gene knockouts using recombinant adeno-associated viruses. Nucleic Acids Res. 2004;32(1):e3. doi: 10.1093/nar/gnh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4(8):651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 35.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 36.Milne TA, et al. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci USA. 2005;102(41):14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lubitz S, Glaser S, Schaft J, Stewart AF, Anastassiadis K. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol Biol Cell. 2007;18(6):2356–2366. doi: 10.1091/mbc.E06-11-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang P, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29(22):6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Craig RW, et al. Human and mouse chromosomal mapping of the myeloid cell leukemia-1 gene: MCL1 maps to human chromosome 1q21, a region that is frequently altered in preneoplastic and neoplastic disease. Genomics. 1994;23(2):457–463. doi: 10.1006/geno.1994.1523. [DOI] [PubMed] [Google Scholar]
- 40.Engelkamp D, Schäfer BW, Mattei MG, Erne P, Heizmann CW. Six S100 genes are clustered on human chromosome 1q21: Identification of two genes coding for the two previously unreported calcium-binding proteins S100D and S100E. Proc Natl Acad Sci USA. 1993;90(14):6547–6551. doi: 10.1073/pnas.90.14.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Northcott PA, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29(11):1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guibal FC, et al. ASB-2 inhibits growth and promotes commitment in myeloid leukemia cells. J Biol Chem. 2002;277(1):218–224. doi: 10.1074/jbc.M108476200. [DOI] [PubMed] [Google Scholar]
- 43.Kohroki J, et al. ATRA-regulated Asb-2 gene induced in differentiation of HL-60 leukemia cells. FEBS Lett. 2001;505(2):223–228. doi: 10.1016/s0014-5793(01)02829-0. [DOI] [PubMed] [Google Scholar]
- 44.Wolf S, Haase-Kohn C, Pietzsch J. S100A2 in cancerogenesis: A friend or a foe? Amino Acids. 2011;41(4):849–861. doi: 10.1007/s00726-010-0623-2. [DOI] [PubMed] [Google Scholar]
- 45.Feng G, Xu X, Youssef EM, Lotan R. Diminished expression of S100A2, a putative tumor suppressor, at early stage of human lung carcinogenesis. Cancer Res. 2001;61(21):7999–8004. [PubMed] [Google Scholar]
- 46.Lee SW, Tomasetto C, Swisshelm K, Keyomarsi K, Sager R. Down-regulation of a member of the S100 gene family in mammary carcinoma cells and reexpression by azadeoxycytidine treatment. Proc Natl Acad Sci USA. 1992;89(6):2504–2508. doi: 10.1073/pnas.89.6.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mueller A, et al. The calcium-binding protein S100A2 interacts with p53 and modulates its transcriptional activity. J Biol Chem. 2005;280(32):29186–29193. doi: 10.1074/jbc.M505000200. [DOI] [PubMed] [Google Scholar]
- 48.Ananthanarayanan M, et al. Histone H3K4 trimethylation by MLL3 as part of ASCOM complex is critical for NR activation of bile acid transporter genes and is downregulated in cholestasis. Am J Physiol Gastrointest Liver Physiol. 2011;300(5):G771–G781. doi: 10.1152/ajpgi.00499.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Odom DT, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303(5662):1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21(4):1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dreijerink KM, et al. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res. 2006;66(9):4929–4935. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.