Abstract
Processive movements of unconventional myosins on actin filaments generally require motor dimerization. A commonly accepted myosin dimerization mechanism is via formation of a parallel coiled-coil dimer by a stretch of amino acid residues immediately carboxyl-terminal to the motor’s lever-arm domain. Here, we discover that the predicted coiled-coil region of myosin X forms a highly stable, antiparallel coiled-coil dimer (anti-CC). Disruption of the anti-CC either by single-point mutations or by replacement of the anti-CC with a parallel coiled coil with a similar length compromised the filopodial induction activity of myosin X. We further show that the anti-CC and the single α-helical domain of myosin X are connected by a semirigid helical linker. The anti-CC–mediated dimerization may enable myosin X to walk on both single and bundled actin filaments.
Keywords: Myo10, molecular motor, filopodial growth
Myosins are actin-based molecular motors that play essential roles in numerous cellular processes, including cell growth and migration, cell cycle regulation, and mechanotransduction. Many of these processes involve the myosin-mediated transport of proteins, nucleic acids, and cellular organelles along actin filaments. In most cases, processive movement of a myosin along actin filaments requires its dimerization. A number of myosins, including muscle myosin II and unconventional myosins V, VI, VII, X, and XVIII contain predicted coiled-coil domains C-terminal to their lever arms (Fig. S1). It is widely believed that these coiled-coil domains dimerize in parallel, positioning the motor heads of the dimer side-by-side on single actin filaments (1–6). In this prevailing model, the parallel dimerization allows the proper transduction of the mechanic forces generated by ATP hydrolysis and is thus necessary for the “walking” of myosins along actin filaments (1–6). However, the parallel coiled-coil dimerization model has been directly demonstrated only in myosin II and V (7, 8). Instead of coiled-coil–mediated dimerization, certain unconventional myosins have been shown to undergo cargo binding-mediated dimerization (9, 10). It is possible that the coiled-coil region interactions and cargo binding function synergistically in modulating the dimerization of some myosins.
Filopodia, organelles that function as antenna for cells to communicate with their surroundings, are enriched with bundled actin filaments. Myosin X (Myo10) has been found to localize at the tips of filopodia and is required for their formation (11–14). Myo10 contains an N-terminal motor head and three IQ motifs, followed by a predicted coiled-coil domain and a tail region with three PH domains and a MyTH4-FERM tandem (Fig. 1A). Both in vitro single-molecule–based experiments and in vivo near single-molecule imaging assays have shown that Myo10 is capable of processive movements along actin filaments; thus it is expected that Myo10 can dimerize (15, 16). Additionally, the role that Myo10 plays in filopodial induction suggests that it may function as a bundled actin filament-selective motor (17, 18). However, neither Myo10’s supposed coiled-coil dimerization nor its selectivity for bundled actin is well substantiated. A fragment consisting of the first 35 residues of the predicted coiled coil has been shown to form a stable single α-helix (SAH), which may function to extend the lever arm of the motor (19); it is not clear whether the remaining ∼100 residues of the coiled coil can mediate dimer formation (20).
Fig. 1.
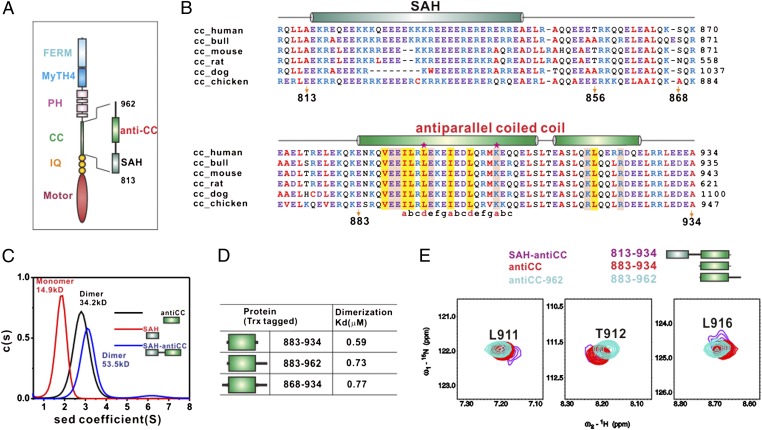
The minimal dimerization domain of Myo10. (A) The domain organizations of Myo10. (B) Sequence alignment of SAH and anti-CC of Myo10 from different species. The secondary structural elements are indicated above the alignment and colored as in A. Residues corresponding to the heptad repeats of the coiled coil are indicated below the alignment. L893 and K904, of which mutations lead to disruption of the Myo10 dimer, are highlighted with stars. The conserved hydrophobic residues that are involved in the dimer interface are highlighted in yellow, and the charged residues in pink. (C) Sedimentation velocity analysis showing that SAH (19.6 kDa) is a monomer, whereas both CC (20.4 kDa) and SAH-CC (29 kDa) form a dimer. (D) Dissociation constants of various dimerization domains of Myo10 derived from sedimentation equilibrium analysis. (E) Nicely overlapping amide peaks of SAH–anti-CC, anti-CC, and anti–CC-962 indicate that anti-CC (aa 883–934) is the minimal and sufficient region for Myo10 dimerization. For clarity, only selected peaks are shown (also refer to Fig. S3 for the full spectra).
In this work, we show that a 50-residue fragment C-terminal to the SAH of Myo10 forms a stable, antiparallel coiled-coil dimer. This antiparallel coiled coil (anti-CC) is connected to SAH by a semirigid α-helix. Replacing the antiparallel coiled-coil dimer of Myo10 with a stable parallel coiled-coil dimer of similar length compromises filopodia formation, showing that the antiparallel coiled-coil dimer is absolutely required for Myo10’s role in filopodial induction. We propose that the antiparallel coiled-coil dimerization of Myo10 allows it to walk in a straddled, duck-like manner with relatively small step size on bundled actin filaments, in addition to the canonical hand-over-hand manner for single filaments.
Results
Identification of a Minimal Region Responsible for Myo10 Dimer Formation.
The predicted coiled-coil region of Myo10 contains ∼150 residues (aa 813–962) immediately C-terminal to its IQ motifs (Fig. 1 A and B). The first 35-residues (aa 813–847) of this predicted coiled coil have been shown to form a monomeric SAH (19), and we confirmed this observation using sedimentation velocity analysis (Fig. 1C). Interestingly, the entire predicted coiled coil (aa 813–934) was found to form a dimer, and a smaller 52-residue fragment (aa 883–934) lacking the SAH region also formed a dimer (Fig. 1C). Using sedimentation equilibrium analysis, we showed that this 52-residue fragment (aa 883–934) is the minimal dimer formation region of the predicted coiled coil, as extensions of this fragment at both termini did not significantly alter dimerization strength (Fig. 1D). The dissociation constant (Kd) of this fragment is ∼0.6 μM (Fig. 1D and Fig. S2), indicating that concentrated Myo10 in filopodia is capable of forming dimers.
To provide direct evidence that the 52-residue fragment (aa 883–934) represents the entire dimerization region of the Myo10 coiled coil, we compared the NMR spectra of three overlapping fragments of Myo10 (i.e., aa 813–934/SAH/anti-CC, 883–934/anti-CC, and 883–962/anti–CC-962). We found that the signals arising from residues 883–934 in the three samples can be nicely overlapped with each other (Fig. 1E), indicating that this 52-residue fragment adopts the same conformation in all three constructs (i.e., the 52-residue fragment is necessary and sufficient for the dimer formation). NMR-based analysis further shows that a stretch of conserved residues C-terminal to 934 in the 883–962 construct are unstructured (Fig. S3).
Myo10 Dimerization Domain Forms a Stable Antiparallel Coiled Coil in Solution.
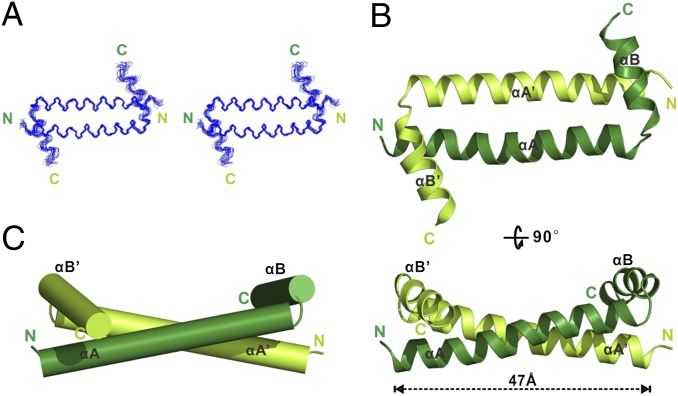
The 1H-15N hetero-nuclear single quantum coherence (HSQC) spectrum of the minimal dimerization domain of Myo10 (aa 883–934) is well dispersed and contains only one set of peaks (Fig. S3), indicating that the domain forms a well-folded, symmetric dimer. The solution structure of the dimer was determined to a high resolution using NMR spectroscopy (Fig. 2 and Table 1; also see SI Materials and Methods for the strategies used for the structure determination). Unexpectedly, the first 28 residues of the dimerization domain (aa 883–910) formed a long α-helix (αA) that interacts with the corresponding α-helix from another molecule to form an anti-CC that is 47 Å in length. The C-terminal 14 residues of the domain form another α-helix (αB), and the two α-helices form a perpendicular α-helical corner connected by a two-residue linker (Leu911 and Thr912, Fig. 2). The αB helix makes extensive contact with the N-terminal of αA on the other molecule, and thus αB is expected to be critical for the stability of the Myo10 dimer.
Fig. 2.
Solution structure of the Myo10 anti-CC dimer. (A) Stereoview showing the backbones of 20 superimposed NMR structures of the Myo10 anti-CC dimer. (B) Ribbon diagram of a representative NMR structure of the Myo10 anti-CC dimer at two different view angles. The two monomers are shown in lime and dark green, respectively. (C) Cylinder representation of anti-CC.
Table 1.
NMR structural statistics for the family of 20 structures of Myo10 antiparallel coiled coil
| Protein | |
| NMR distance and dihedral constraints | |
| Total NOE | 2,591 |
| Intraresidue | 312 |
| Interresidue | |
| Sequential (|i – j| = 1) | 573 |
| Medium range (|i – j| < 4) | 561 |
| Long range (|i – j| > 5) | 27 |
| Intermolecular | 659 |
| Ambiguous NOEs restraints (i.e., either intra- or intermolecular couplings) | 382 |
| Hydrogen bonds | 104 |
| NMR total dihedral angle restraints | 164 |
| ϕ | 82 |
| ψ | 82 |
| Structure statistics | |
| Violations (mean and SD) | |
| Distance constraints (Å) | 0.004 ± 0.000 |
| Dihedral angle constraints (°) | 0.099 ± 0.017 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.001 ± 0.000 |
| Bond angles (°) | 0.331 ± 0.004 |
| Impropers (°) | 0.154 ± 0.010 |
| Average pairwise rmsd* (Å) | |
| Well-ordered residues (Myo 10 anti-CC883–925) | |
| Heavy | 1.272 |
| Backbone | 0.67 |
*Pairwise rmsd was calculated among 20 refined structures.
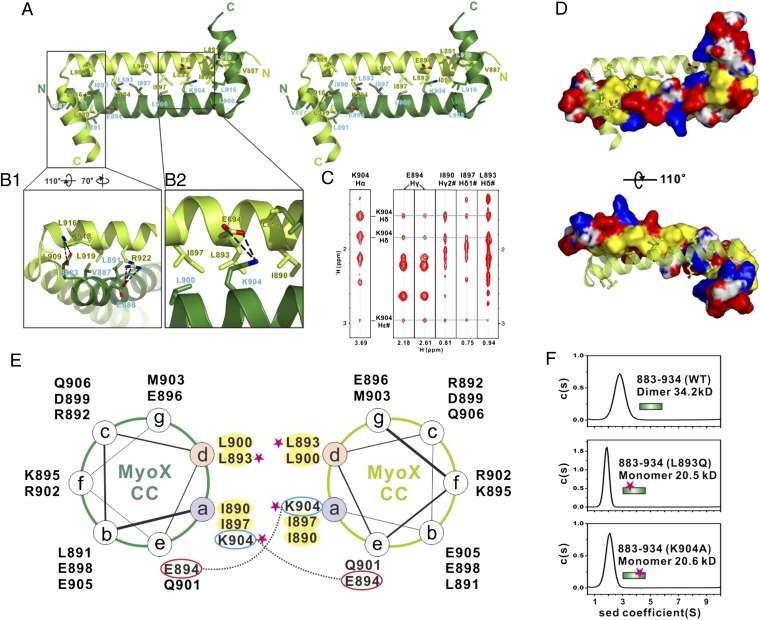
The dimerization of the Myo10 CC is mediated by both hydrophobic and charge–charge interactions. The amino acids at the center of αA show a characteristic heptad repeat pattern with hydrophobic residues located in the a and d positions and charged residues in the e and g positions (Figs. 1B and 3A). The highly conserved hydrophobic residues, including Ile890, Leu893, Ile897, and Leu900 at the a/d positions of the two αA helices, form an extensive hydrophobic interface in which their aliphatic side chains interact with each other in a “knobs-into-holes” manner (Fig. 3 D and E). Numerous long-distance NOEs are detected between the side chain of Lys904 and those of Glu894, Ile890, Leu893, and Ile897 (Fig. 3C).These residues are located more than 10 Å apart within the same molecule of this 52-residue fragment and thus are too far apart to generate intramolecular NOEs (Fig. S4). These definitive, long-distance intermolecular NOEs reveal that the dimer must be antiparallel. A pair of interhelical salt bridges formed between Glu894 and Lys904 at the two ends of the anti-CC further stabilizes dimer formation. Additionally, the aliphatic portion of the Lys904 side chain intimately interacts with Ile890, Leu893, and Ile897 from the neighboring αA and Leu900 on the same helix (Fig. 3B). Thus, Lys904 is expected to be critical for the stability of the Myo10 anti-CC dimer. Consistent with this analysis, single substitution of either Leu893 with Gln or Lys904 with Ala completely abolished Myo10 dimerization (Fig. 3F).
Fig. 3.
Molecular details of the Myo10 anti-CC dimer. (A) Stereoview showing the dimerization interface of the Myo10 anti-CC dimer. The side chains of the residues involved in the dimer interface are drawn in the stick model. (B) Detailed interactions showing the interface of αB with the N-terminal end of αA (B1) and the interface of K904 with residues from the neighboring subunit (B2). (C) Selected strips from the 3D 13C-NOESY spectrum of Myo10 (aa 883–934) showing long-distance NOEs between the side chain of K904 with the side chains of E894, I890, I897, and L893. (D) Combined-surface and ribbon representation of the Myo10 anti-CC dimer. In the surface diagram, the positively charged amino acids are drawn in blue, the negatively charged residues in red, the hydrophobic residues in yellow, and the others in white. (E) Helical wheel presentation showing the heptad repeats at the center of αA. Interhelical salt bridges between the residues at a and e positions are depicted by dashed lines. Residues at “a” and “d” positions forming the hydrophobic core of the coiled coil are highlighted in yellow. Single substitution mutations that disrupt the Myo10 dimer formation are highlighted with pink stars. (F) Sedimentation velocity analysis showing that the L893Q and the K904A mutants of anti-CC are monomer in solution.
Numerous NOEs were detected between the side chain of Leu919, Arg922 in αB, and those of Val887 at the N-terminal end of αA (Fig. S5); once again, these NOEs are consistent only with an anti-CC conformation. Also, two pairs of intermolecular salt bridges formed between Lys918 and Glu883 and between Arg922 and Glu888 further stabilize the intermolecular interaction between αB and αA as well as the orientations of the α-helices in the dimer. Removal of even part of the αB helix can seriously alter the anti-CC conformation of the Myo10 dimer and thus is expected to disrupt motor function (see Discussion for more details).
The anti-CC arrangement of Myo10 is highly unexpected and distinctly different from the parallel coiled coils observed in myosin II and myosin V (7, 8). A parallel coiled-coil dimer naturally positions the two heads of a myosin dimer side-by-side, allowing them to walk in a hand-over-hand fashion on actin filaments. In contrast, the formation of an anti-CC dimer would position the two motor heads at opposite ends of the coiled coil, a unique arrangement among known motor proteins.
Anti-CC of Myo10 Is Highly Stable.
An NMR-based amide exchange experiment showed that the backbone amides of the anti-CC (aa 883–934) are highly resistant to hydrogen/deuterium (H/D) exchange (Fig. S6A), indicating the Myo10 anti-CC is highly stable and lacks even very slow time-scale conformational exchange, such as the exchange between antiparallel and parallel CC. A steady-state 1H-15N NOE experiment also showed that the entire backbone of the anti-CC is highly rigid (Fig. S6B).
We further note that the residues connecting the SAH and the anti-CC (aa 856–882) adopt a semirigid, helix-like structure. Although we were not able to obtain sequence-specific assignment of NMR signals for residues 856–882 due to extensive signal overlap, we could measure heteronuclear NOE values for more than 15 residues from this connection sequence (Fig. S6C), which clearly indicates that the linker is partially structured. Their narrow chemical shift dispersion (Fig. S3) as well as many NOEs between amide protons (Fig. S6D) indicate that this linker likely adopts an α-helix like structure. The fast H/D-exchange rates of these amides suggest that this linker helix-like structure is semirigid (Fig. S6A). Taken together, the above NMR-based experiments indicate that the predicted coiled-coil region of Myo10 consists of a rigid SAH and a highly stable anti-CC connected by a semirigid α-helix.
Anti-CC Is Essential for Myo10’s Role in Filopodial Induction.
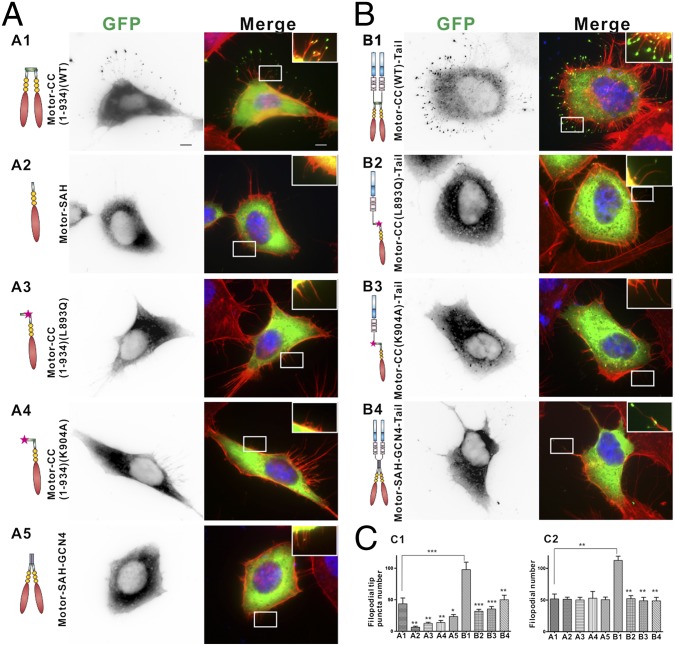
We next tested the role of the anti-CC in Myo10-induced filopodial formation. Myo10 is well known for its processive movement within filopodia and filopodial induction activity. First, we quantified GFP-Myo10–positive filopodial tip puncta number in HeLa cells as an indication of the motor’s processive movements along filopodia. We overexpressed the wild-type GFP-tagged tailless Myo10 [“Motor-CC(1-934)WT”], which terminates at the end of the dimerization domain. As shown in Fig. 4A1, Motor-CC(1-934)WT is capable of forming puncta at the tips of filopodia. In sharp contrast, both the removal of the anti-CC and the single-point substitution mutations that disrupt anti-CC formation (the L893Q mutant and the K904A mutant shown in Fig. 3F) lead to significant decreases in the filopodial tip puncta localization of Motor-CC(1-934) (Fig. 4 A2 and C1). To further test the importance of the anti-CC for Myo10’s function, we substituted the anti-CC part of Motor-CC(1-934) with GCN4, which can form a stable parallel coiled coil similar in length to the Myo10 anti-CC. This GCN4-substituted construct (Motor-SAH-GCN4) also decreased filopodial tip puncta localization (Fig. 4 A5 and C1), indicating that anti-CC–mediated dimerization is critical for Myo10’s processive movement and localization to the filopodial tip. This GCN4-substituted Myo10 head still showed significantly more filopodial tip puncta than anti–CC-disruption mutants, consistent with previous reports showing that the forced parallel coiled-coil dimerization of the Myo10 head can trigger filopodial tip localization (15, 21). To further assay the anti-CC’s function in Myo10-induced filopodial induction, we counted elongated filopodial numbers in both HeLa and COS7 cells using the GFP-tagged full-length Myo10. In agreement with previously reported findings (22), inclusion of the tail domains of the motor tremendously increases filopodia tip puncta number and induces the formation of numerous elongated filopodia in transfected cells (Fig. 4 B1 and C2; Fig. S7 A3 and B2). Fully consistent with the observations made using the tailless motor constructs, the two single-point substitution mutants of the full-length Myo10 (the L893Q mutant and the K904A mutant) displayed diminished filopodial induction activity in both HeLa (Fig. 4 B2, B3, and C2) and COS7 (Fig. S7 A4, A5, and B2) cells. Finally, replacing the anti-CC with the GCN4 parallel CC also compromised the filopodial induction activity of Myo10 (Fig. 4 B4 and C2; Fig. S7 A6 and B2). The tail cargo binding domains may enhance the filopodial induction activity of Myo10 by interacting with membranes or membrane-localized cargoes (23, 24). Alternatively, the binding of cargoes may further stabilize the dimer conformation of Myo10 (25). Regardless of the precise mechanism, our results shown in Fig. 4 and Fig. S7 demonstrate that the anti-CC–mediated dimerization is required for Myo10’s filopodial induction activity.
Fig. 4.
Anti-CC is essential for Myo10’s function. (A and B) Representative images of HeLa cells transfected with various GFP-tagged Myo10 constructs, actin filament with phalloidin (red), and nuclei with DAPI (blue): (A1), Motor-CC(1-934)(WT); (A2), Motor-SAH; (A3), Motor-CC(1-934)(L893Q); (A4), Motor-CC(1-934)(K904A); (A5), Motor-SAH-GCN4; (B1), Motor-CC(WT)-Tail; (B2), Motor-CC(L893Q)-Tail; (B3), Motor-CC(K904A)-Tail; (B4), Motor-SAH-GCN4-Tail. (Scale bar, 5 μm.) (C) Quantitative analysis of GFP puncta number per cell at the tip of filopodia (C1) and filopodial number (C2) (with length >0.5 μm). Values are means ± SD from three independent experiments with 10 cells per experiment, using unpaired t test, *P < 0.05, *P < 0.01, ***P < 0.001. Also refer to Fig. S7 for data in COS7 cells.
Discussion
The canonical walking model for myosins on actin filaments is based on the study of Myo5 (4–6). In this model, small conformational changes in the ATP-binding pocket of the head domain are amplified to induce a large swing of the lever arm, which allows the motor to take ∼36-nm hand-over-hand steps along actin filaments. The formation of the stable parallel coiled-coil dimer by the stretch of residues right after the lever arm of Myo5 ensures that the two heads of the motor dimer are juxtaposed to the actin filament in parallel, with at least one head attached to the filament at any given time, and prevents the dissipation of the torque generated by the ATP hydrolysis. Thus, the formation of the parallel coiled-coil dimer is pivotal for the processive movement of Myo5. It is widely believed that similar parallel coiled-coil dimers are also formed in other myosins that contain predicted coiled coils after their lever arm domains.
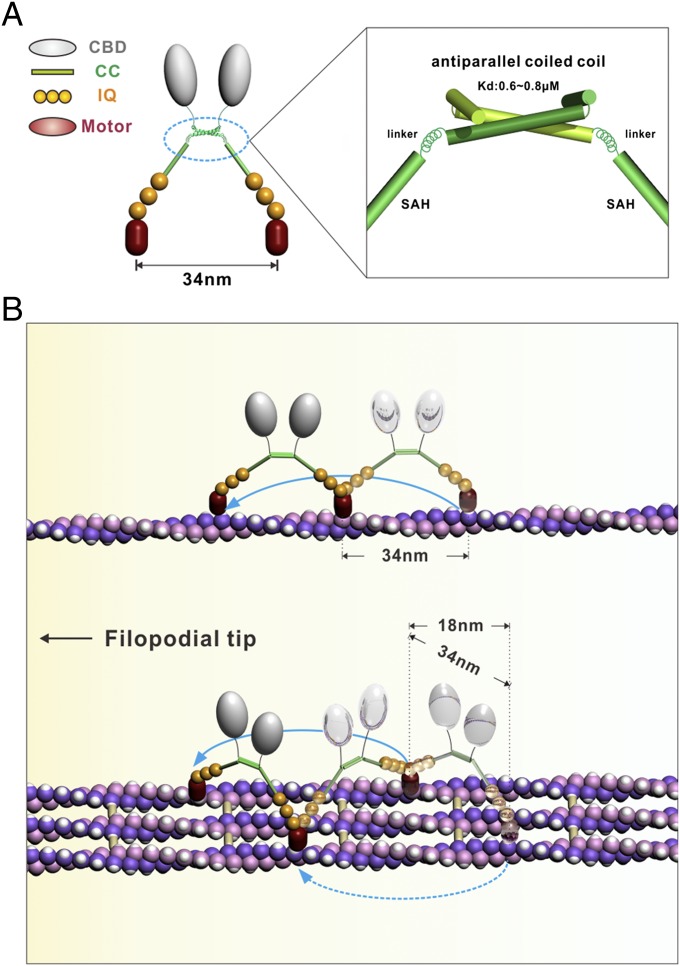
The discovery of the anti-CC dimer in Myo10 suggests a different walking model (Fig. 5). In this model, the anti-CC dimer forms a “shoulder” with a width of ∼5 nm (the width of this “shoulder” would be considerably smaller if the anti-CC fragment forms a parallel coiled-coil dimer). Two extended lever arms, each composed of a SAH and three IQ motifs, emanate from opposite sides of this “shoulder.” Each lever arm is connected to the “shoulder” by a semirigid helix much like the human arm/shoulder joint (Fig. 5A). This allows the two motor heads in the Myo10 dimer to bind to two parallel actin filaments simultaneously. The most salient implication of this anti-CC–mediated Myo10 dimerization model is that the motor should be capable of walking on both single and bundled actin filaments (Fig. 5B), which has been directly demonstrated by in vitro single-molecule–based stepping assays (16). When walking on single actin filaments, Myo10 walked in the well-characterized hand-over-hand mode with a step size of ∼34 nm. On bundled actin filaments, the Myo10 dimer can walk either with two motor heads attached to different actin filaments in a straddled, duck-like manner or on a single filament of the bundle in the canonical hand-over-hand fashion (Fig. 5B). As a result, when walking on bundled actin filaments, the average step size of Myo10 should be considerably smaller. Supporting our model, Goldman and colleagues have recently shown that the stepping behavior of Myo10 on bundled actin is consistent with two different walking modes, with step sizes of 34 and 18 nm (16), presumably corresponding to the hand-over-hand and straddled walking modes, respectively.
Fig. 5.
A stepping model for Myo10. (A) A schematic model showing anti-CC–mediated dimerization of Myo10. The anti-CC and SAH are shown with ribbon and cylinders, respectively. The semirigid α-helix connecting anti-CC and SAH is shown by a spring. (B) The stepping model for Myo10. In this model, Myo10 can processively walk both on a single actin filament and on bundle filaments. Myo10 walks following the well-characterized hand-over-hand fashion with a step size of ∼34 nm on single actin filaments (Upper). On bundled actin filaments, Myo10 can walk like a duck with two motor heads attached to different actin filaments or walk on the same filament via the hand-over-hand fashion (Lower).
The straddled walking model of Myo10 proposed here has a number of implications. One possible advantage of Myo10’s capability to walk on bundled actin filaments is that this would lower its chances of being obstructed by actin filament-decorating proteins during walking, allowing it to walk for long distances. Indeed, Myo10 has been shown to be able to walk for very long distances on bundled actin filaments (15), presumably due to the abundance of landing sites on actin filament bundles (i.e., by shortening the searching time of the motor head for binding to actin filaments; Fig. 5B). Additionally, the straddled model implies that Myo10 is expected to be able to walk comfortably on actin bundles cross-linked by bundling proteins with different sizes (e.g., espin, fimbrin, and plastin). Finally, the straddled actin filament binding of Myo10 indicates that the motor itself may function as an actin filament bundling protein at the tips of filopodia and actively promote elongation of filopodia (21).
Several recent papers by Rock and colleagues have suggested that Myo10 displays strong selectivity for bundled actin (17, 18). This finding contradicts the findings of a recent study by Sun et al. (16). Careful analysis of the Myo10 constructs used in these studies revealed that the two groups have used Myo10 constructs of slightly different lengths (Fig. S8A). In single-molecule–based stepping assays, Myo10 dimerization must be enhanced by fusion of an additional dimerization domain to the end of the protein. Sun et al. (16) fused the dimer-enhancing coiled-coil domain of Myo5 at Ala939 of Myo10, well beyond the anti-CC dimerization domain of Myo10. Thus, the Myo5 coiled coil should enhance the dimerization of Myo10 without altering the dimer’s anti-CC conformation. In the Nagy et al. studies (17, 18), the parallel GCN4 coiled coil was fused to Leu920 of Myo10. Leu920 is in the middle of αB of the Myo10 anti-CC dimer, and the truncation of Myo10 and fusion of GCN4 at this residue is likely to alter the structure of the Myo10 anti-CC dimer. To test this possibility, we investigated the structure of the Myo10 anti-CC/GCN4 chimera used in the Rock et al. studies (residue 883–920 of Myo10 fused to GCN4) using NMR spectroscopy. We found that, although the chimera still dimerizes, the structure of the Myo10 part of the chimera is obviously different from that of the wild-type Myo10 (Fig. S8 B–D). The observed chemical shift changes suggest some structural changes of the GCN4-substituted Myo10 anti-CC. Thus, we believe that the bundled actin selectivity of the Myo10 chimera shown in the Rock et al. studies is likely due to the conformational change of the anti-CC dimer induced by the GCN4 fusion. Nonetheless, the detailed single-molecule stepping assays performed in the Rock et al. studies have provided additional evidence, albeit from a different angle, supporting the critical role that the anti-CC plays in Myo10’s processive movement along actin filaments (17, 18).
In summary, we discovered that the predicted coiled-coil domain C-terminal to the lever arm domain of Myo10 forms a stable, anti-CC dimer. This anti-CC–mediated dimerization provides a mechanistic basis for the capability of Myo10 to walk on both single and bundled actin filaments for long distances. The unexpected dimerization mechanism discovered in Myo10 may also be relevant to other coiled-coil–containing myosins. Finally, the anti-CC–mediated dimerization mode of Myo10 provides hints for designing nano-scale molecular devices with desired functions.
Materials and Methods
Recombinant Myo10 anti-CC protein was expressed in Escherichia coli and purified by Ni2+-nitrilotriacetic acid affinity chromatography. The anti-CC structure was determined by NMR spectroscopy. The structure determination of Myo10 anti-CC was completed in a stepwise manner by first using definitive intermolecular NOEs. HeLa and COS7 cells were transiently transfected with a lipofectamine PLUS kit (Invitrogen) and imaged with a Nikon TE2000E inverted fluorescent microscope. An extended description of the materials and methods used can be found in SI Materials and Methods.
The atomic coordinates of Myo10 anti-CC have been deposited in the Protein Data Bank under PDB ID code 2LW9.
Supplementary Material
Acknowledgments
We thank Anthony Zhang for critical editing of the manuscript. The NMR facility used in this work was supported by funds from the Hong Kong Jockey Club and by special Grant SEG_HKUST06 from the Research Grants Council of Hong Kong. This work was also supported by RGC of Hong Kong Grants 663808, 664009, 660709, 663610, 663811, HKUST6/CRF/10, and SEG_HKUST06 (to M.Z.); HKUST6/CRF/09 (to Z. Wen), and 662710 (to Z. Wei). Z. Wei is a Postdoctoral Fellow of the Tin Ka Ping Foundation of the Institute for Advanced Study (IAS) at Hong Kong University of Science and Technology (HKUST), and M.Z. is a Senior Fellow of IAS at HKUST.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.E.C. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID code 2LW9 (Myo10 anti-CC)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208642109/-/DCSupplemental.
See Commentary on page 17313.
References
- 1.Sweeney HL, Houdusse A. Structural and functional insights into the Myosin motor mechanism. Annu Rev Biophys. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- 2.Hartman MA, Finan D, Sivaramakrishnan S, Spudich JA. Principles of unconventional myosin function and targeting. Annu Rev Cell Dev Biol. 2011;27:133–155. doi: 10.1146/annurev-cellbio-100809-151502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howard J. Molecular motors: Structural adaptations to cellular functions. Nature. 1997;389:561–567. doi: 10.1038/39247. [DOI] [PubMed] [Google Scholar]
- 4.Mehta AD, et al. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 5.Purcell TJ, Morris C, Spudich JA, Sweeney HL. Role of the lever arm in the processive stepping of myosin V. Proc Natl Acad Sci USA. 2002;99:14159–14164. doi: 10.1073/pnas.182539599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yildiz A, et al. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, et al. Visualization of an unstable coiled coil from the scallop myosin rod. Nature. 2003;424:341–345. doi: 10.1038/nature01801. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Taylor DW, Krementsova EB, Trybus KM, Taylor KA. Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography. Nature. 2006;442:208–211. doi: 10.1038/nature04719. [DOI] [PubMed] [Google Scholar]
- 9.Yu C, et al. Myosin VI undergoes cargo-mediated dimerization. Cell. 2009;138:537–548. doi: 10.1016/j.cell.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 10.Mukherjea M, et al. Myosin VI dimerization triggers an unfolding of a three-helix bundle in order to extend its reach. Mol Cell. 2009;35:305–315. doi: 10.1016/j.molcel.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berg JS, Derfler BH, Pennisi CM, Corey DP, Cheney RE. Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J Cell Sci. 2000;113:3439–3451. doi: 10.1242/jcs.113.19.3439. [DOI] [PubMed] [Google Scholar]
- 12.Berg JS, Cheney RE. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat Cell Biol. 2002;4:246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- 13.Tokuo H, Ikebe M. Myosin X transports Mena/VASP to the tip of filopodia. Biochem Biophys Res Commun. 2004;319:214–220. doi: 10.1016/j.bbrc.2004.04.167. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, et al. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat Cell Biol. 2004;6:523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- 15.Kerber ML, et al. A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Curr Biol. 2009;19:967–973. doi: 10.1016/j.cub.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y, et al. Single-molecule stepping and structural dynamics of myosin X. Nat Struct Mol Biol. 2010;17:485–491. doi: 10.1038/nsmb.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy S, et al. A myosin motor that selects bundled actin for motility. Proc Natl Acad Sci USA. 2008;105:9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagy S, Rock RS. Structured post-IQ domain governs selectivity of myosin X for fascin-actin bundles. J Biol Chem. 2010;285:26608–26617. doi: 10.1074/jbc.M110.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knight PJ, et al. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J Biol Chem. 2005;280:34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 20.Kerber ML, Cheney RE. Myosin-X: a MyTH-FERM myosin at the tips of filopodia. J Cell Sci. 2011;124:3733–3741. doi: 10.1242/jcs.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tokuo H, Mabuchi K, Ikebe M. The motor activity of myosin-X promotes actin fiber convergence at the cell periphery to initiate filopodia formation. J Cell Biol. 2007;179:229–238. doi: 10.1083/jcb.200703178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohil AB, Robertson BW, Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci USA. 2006;103:12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Q, Yu J, Yan J, Wei Z, Zhang M. Structural basis of the myosin X PH1(N)-PH2-PH1(C) tandem as a specific and acute cellular PI(3,4,5)P(3) sensor. Mol Biol Cell. 2011;22:4268–4278. doi: 10.1091/mbc.E11-04-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Z, Yan J, Lu Q, Pan L, Zhang M. Cargo recognition mechanism of myosin X revealed by the structure of its tail MyTH4-FERM tandem in complex with the DCC P3 domain. Proc Natl Acad Sci USA. 2011;108:3572–3577. doi: 10.1073/pnas.1016567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Umeki N, et al. Phospholipid-dependent regulation of the motor activity of myosin X. Nat Struct Mol Biol. 2011;18:783–788. doi: 10.1038/nsmb.2065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.