Abstract
Nuclear factor-erythroid 2–related factor 2 (Nrf2) is a key transcriptional regulator for antioxidant and anti-inflammation enzymes that binds to its endogenous inhibitor protein, Kelch-like ECH (erythroid cell-derived protein with CNC homology)-associated protein 1, in the cytoplasm under normal conditions. Various endogenous or environmental oxidative stresses, such as ionizing radiation (IR), can disrupt the Nrf2–Kelch-like ECH-associated protein 1 complex. This allows Nrf2 to translocate from the cytoplasm into the nucleus to induce transcription of heme oxygenase-1 and other cytoprotective enzymes through binding to antioxidant responsive elements. However, how Nrf2 protects cells from IR-induced damage remains unclear. Here, we report that Nrf2 activation by the synthetic triterpenoids, bardoxolone methyl (BARD) and 2-cyano-3,12-dioxooleana-1,9 (11)-dien-28-oic acid–ethyl amide, protects colonic epithelial cells against IR-induced damage, in part, by enhancing signaling of the DNA damage response. Pretreatment with BARD reduced the frequency of both G1 and S/G2 chromosome aberrations and enhanced the disappearance of repairosomes (C-terminal binding protein interacting protein, Rad51, and p53 binding protein-1 foci) after IR. BARD protected cells from IR toxicity in a Nrf2-dependent manner. The p53 binding protein-1 promoter contains three antioxidant responsive elements in which Nrf2 directly binds following BARD treatment. In addition, 2-cyano-3,12-dioxooleana-1,9 (11)-dien-28-oic acid–ethyl amide provided before exposure to a lethal dose of whole-body irradiation protected WT mice from DNA damage and acute gastrointestinal toxicity, which resulted in improved overall survival. These results demonstrate that Nrf2 activation by synthetic triterpenoids is a promising candidate target to protect the gastrointestinal tract against acute IR in vitro and in vivo.
Keywords: radioprotection, enhanced DNA repair, antioxidant anti-inflammatory modulators (AIMs)
Ionizing radiation (IR)-induced DNA damage activates cellular defense mechanisms (1), such as DNA damage sensing and double-strand break (DSB) repair, to protect the cellular genome. DNA DSBs are the most lethal events among IR-induced DNA lesions, and both numerical and structural chromosome aberrations increase if not properly repaired (2). To enhance cell survival of normal tissues postirradiation, identification of novel agents that activate or enhance DNA DSB repair processes and rescue cells from unrepaired DNA damage are needed. Cell killing by IR correlates with the failure to repair DNA DSBs, and normal cells may have less efficient DNA repair capabilities to protect against IR-associated cell death. There are two major DNA DSB repair pathways: nonhomologous end-joining (NHEJ) and homologous recombination (HR). Key NHEJ proteins include Ku70–Ku80 and DNA-dependent protein kinase catalytic subunit (3, 4). Although NHEJ in highly effective, its imprecise nature makes it prone to mutations (4). In contrast, HR is considered to be an error-free pathway and typically uses the intact sister chromatid as a template for synthesis-dependent repair in mitotic cells (5, 6). Shortly after IR, the Rad51-covered filament initiates the homology search and catalyzes strand exchange to allow priming of DNA replication and repair initiation (7). The C-terminal binding protein interacting protein (CtIP), also known as DNA endonuclease RBBBP8, is recruited to sites of DNA damage in human cells and is involved in resection and intrachromosomal associations during DNA DSB repair by HR (8). CtIP also has a critical role in checkpoint maintenance in G2/M-phase checkpoint and in the intra–S-phase checkpoint at later time points (4–8 h) following IR (9). Detailed molecular analysis of the cellular DNA damage detection and repair response has identified specific proteins that form foci (repairosomes) contributing to intrinsic individual radiosensitivity as well as potential molecular targets for reducing IR-related toxicity. Therefore, these DNA repair-associated protein foci are surrogate markers of identifying DNA DSBs, which are commonly used for determining the kinetics of DNA damage response (DDR) and repair.
The synthetic triterpenoid 2-cyano-3,12-dioxooleana-1,9 (11)-dien-28-oic acid (CDDO), along with chemically modified derivatives CDDO-methyl ester [also known as bardoxolone methyl (BARD)] and CDDO-EA (ethyl amide), are noncytotoxic, highly multifunctional, and orally administered available drugs that have been studied as anti-inflammatory, antiangiogenic, and anticancer agents in vivo and in vitro (10–12). Nanomolar concentrations of synthetic triterpenoids have been shown to induce the expression of proteins associated with the nuclear factor-erythroid 2–related factor 2 (Nrf2) antioxidant response element (ARE) pathway. Synthetic triterpenoids stabilize Nrf2 by interacting with Kelch-like ECH (erythroid cell-derived protein with CNC homology)-associated protein 1. Stabilized Nrf2 is phosphorylated by PKC, inducing nuclear Nrf2 translocation and the activation of AREs (13, 14). Therefore, the increase of both Nrf2 and phospho-Nrf2 is important in its activity. Treatment of nontumorigenic “normal” immortalized human colonic epithelial cells (HCECs) with BARD inhibits Nrf2 degradation and stabilizes Nrf2, which leads to ARE activation and IR protection (15). Here, we demonstrate that pretreatment with this class of synthetic triterpenoids protects HCECs in vitro and in mice in vivo from IR-induced toxicity by enhancing DNA damage repair efficiency through a unique mechanism of action involving Nrf2.
Results
Nrf2 Is a Key Factor for BARD-Mediated Radioprotection Activity.
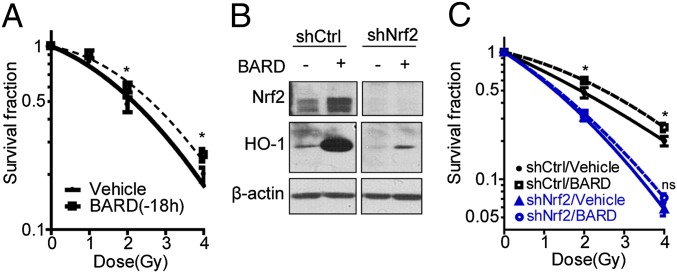
To explore the potential radioprotective effects of synthetic triterpenoids in HCECs containing a nonrandom premalignant chromosome alteration, trisomy 7 (CT7), we assessed clonogenic survival post-IR. When BARD was administered in the culture medium 18 h prior to exposure to graded IR doses, an increase in clonogenic survival was observed compared with sham-treated cells (Fig. 1A). The dose-modifying factor (DMF), the ratio of absorbed radiation doses with and without a particular agent that produce the same biological effect (16), for BARD as a radioprotector was calculated to be 1.61 (Table S1). To demonstrate the importance of Nrf2 in BARD-mediated radioprotection, Nrf2 in HCEC CT7s (HCEC CT7/short hairpin Nrf2) was depleted by specific shRNA knockdown and Nrf2 depletion failed to increase heme oxygenase-1 (HO-1) expression with BARD treatment, a downstream Nrf2 effector (Fig. 1B). This resulted in the loss of radioprotection activity (DMF = 1.05) when administered 18 h before irradiation (Fig. 1C).
Fig. 1.
BARD protects HCECs from irradiation through Nrf2. (A) Clonogenic survival for countermeasure effect. HCECs were treated with BARD for 18 h, and cells were then irradiated with the indicated radiation doses. (B) Establishment of Nrf2 knockdown cell lines. HCEC CT7s were infected by a lentiviral vectors expressing a shRNA against Nrf2 and were treated with or without BARD. Cell extracts were separated by SDS/PAGE and blotted using anti-Nrf2 or anti–HO-1 antibody. (C) Clonogenic survival of control or Nrf2 knockdown cell line. HCEC CT7/short hairpin Nrf2 (shNrf2) cells were treated with BARD for 18 h and were then irradiated with the indicated radiation doses. *P < 0.05 compared with vehicle control; ns, not significant differences in the unpaired Student t test; shCtrl, short hairpin control.
BARD Reduces IR-Induced Chromosome Aberrations.
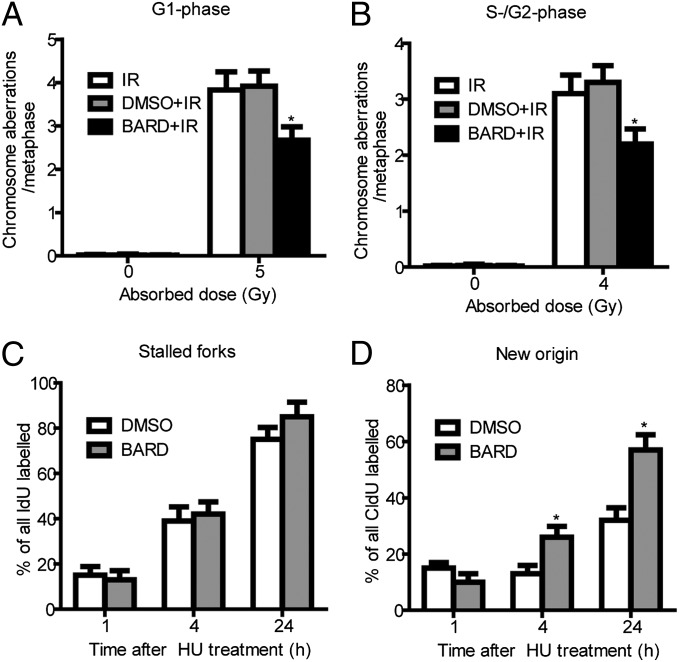
To understand if radioprotective activity correlates with DNA DSB repair, we determined the residual frequencies of IR-induced chromosomal aberrations in G1-phase and S/G2-phase cells. Chromosome aberrations observed at metaphase represent unrepaired DNA DSBs, which are repaired by NHEJ as well as HR pathways. Cell cycle phase-specific chromosome aberrations were ascertained based on the frequency of chromosomal and chromatid-type aberrations observed at metaphase. G1-specific aberrations detected at metaphase are mostly of the chromosomal type and include a high frequency of dicentric chromosomes (17) (Fig. S1A). The S- and G2-phase types of aberration detected at metaphase are chromosomal, as well as chromatids (Fig. S1B). To determine chromosome damage, exponentially growing cells were irradiated and collected either after 14 h to study G1-type aberrations or after 4 h to study S/G2 aberrations at metaphase. Aberrations were scored at metaphase as described previously (18, 19). A significant (P < 0.05, Student t test) reduction in residual IR-induced G1 chromosomal aberrations (Fig. 2A) as well as S/G2 chromosomal aberrations (Fig. 2B) was observed in metaphase cells pretreated with BARD. Furthermore, cells pretreated with BARD have reduced tri- and quadriradials (Fig. 2B). These results suggest that treatment with BARD before IR exposure enhances repair capability and HR activation.
Fig. 2.
BARD reduces chromosome aberrations and releases DNA replication block after IR. Chromosome aberrations were studied at metaphase postirradiation. (A) G1-type aberrations were analyzed at metaphase 14 h after exposure to 5 Gy of IR (Fig. S1A). The histogram shows the G1-chromosome aberrations. (B) S- and G2-type chromosomal aberrations were analyzed at metaphases after 4 Gy of IR. Cells were exposed to 4 Gy of IR and collected at different times points to analyze S- or G2-type aberrations (Fig. S1B). The histogram shows S- and G2-type chromosomal aberrations per metaphase. For each phase, 50 metaphases were analyzed from two independent experiments for each phase of the cell cycle. The mean of four experiments is presented in the histogram. (C and D) The restart of replication forks was analyzed using DNA fiber assay. The percentages of stalled forks (C) and new origins of replication (D) were measured at various time points posttreatment with HU. For each experiment, 100 fibers were analyzed in different sections of the slides. The results are the mean of three experiments. *P < 0.05 (compared with vehicle control) in the unpaired Student t test.
We next compared the restart of replication forks after different periods of hydroxyurea (HU) treatment in cells with and without BARD treatment using the DNA fiber technique (20). Replication forks are immediately stalled by HU treatment due to deoxyribonucleotide pool depletion (21). Cells with and without BARD treatment were pulse-labeled with 5-iododeoxyuridine (IdU) for 20 min, washed and treated with HU for 1 h, and then washed and pulse-labeled with 5-chlorodeoxyuridine (CIdU) for 1 or 4 or 24 h (Fig. S2) to analyze stalled replication forks as previously described (22, 23). No major difference in the frequency of stalled forks was observed in cells treated with and without BARD (Fig. 2C). We quantified the replication fork restart by determining the total number of replication tracks labeled with CIdU. We observed a higher frequency of forks (the signal of CIdU) in BARD-treated cells from HU block compared with control cells (Fig. 2D). These results indicate that the higher frequency of new initiation of replication restarts in BARD treated cells may be due to enhanced protection of DNA strands at replication forks.
BARD Increases DNA Damage Signaling After Irradiation Through Nrf2 Activation.
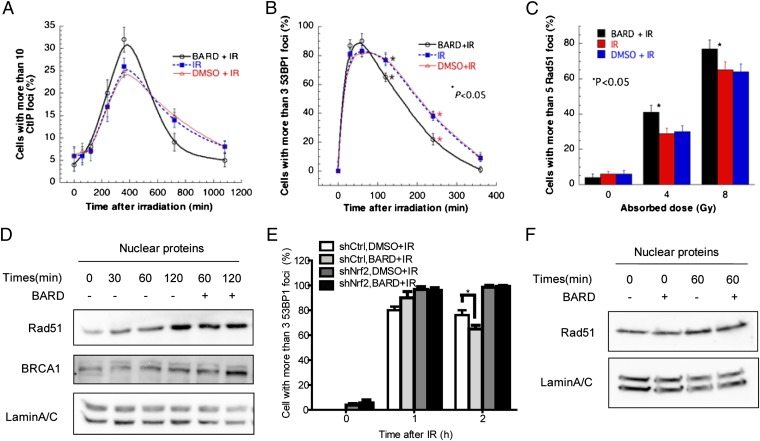
We next examined the appearance and disappearance of repairosomes, which are involved in DNA damage recognition and repair. CtIP, p53 binding protein-1 (53BP1), and Rad51 foci were detected by immunostaining, and positive cells were counted at the indicated time point after exposure to IR. The cutoffs were determined by the number of spontaneous foci in 200 untreated control cells. Consistent with the chromosome data, we observed that BARD treatment before irradiation results in the rapid disappearance of CtIP-positive (Fig. 3A) and 53BP1-positive (Fig. 3B) cells. We also observed that BARD treatment results in increased Rad51 foci formation (Fig. 3C) and nuclear translocation of both Rad51 and BRCA1 (Fig. 3D). Rapid nuclear translocation of Rad51 was observed 60 min following IR in BARD-treated cells, whereas no significant differences in localization were detected after 30 min (Fig. 3D). Total Rad51 protein expression was not changed by BARD treatment (Fig. S3). In contrast, Nrf2 knockdown cells failed to increase DNA damage signaling (Fig. 3E) and nuclear Rad51 import (Fig. 3F) after exposure to IR with BARD pretreatment. This suggests that pretreatment with BARD increases DNA damage signaling, which includes rapid reduction of repairosome foci at later time points after IR in an Nrf2-dependent manner.
Fig. 3.
BARD increases DNA damage signaling. Effect of BARD on appearance and disappearance of DNA damage-associated repairosome foci of CtIP (A) and 53BP1 (B) after 5-Gy doses of IR. (C) Rad51-positive cells with vehicle or BARD 4 h after 5-Gy doses of IR. For each point, 200 cells were counted, and the mean of three experiments is given in the figures. (D) Western blot of Rad51 and BRCA1 levels in the nuclear fraction of HCECs 30, 60, and 120 min after 5-Gy doses of IR with or without BARD pretreatment. 53BP1 foci-positive cells (E) and nuclear localization of Rad51 (F) in Nrf2 knockdown cell line 60 min after 5-Gy doses of IR. *P < 0.05 (compared with vehicle control) in the unpaired Student t test. shCtrl, short hairpin control; shNrf2, short hairpin Nrf2.
BARD Increases 53BP1 Expression Through Nrf2 Activation.
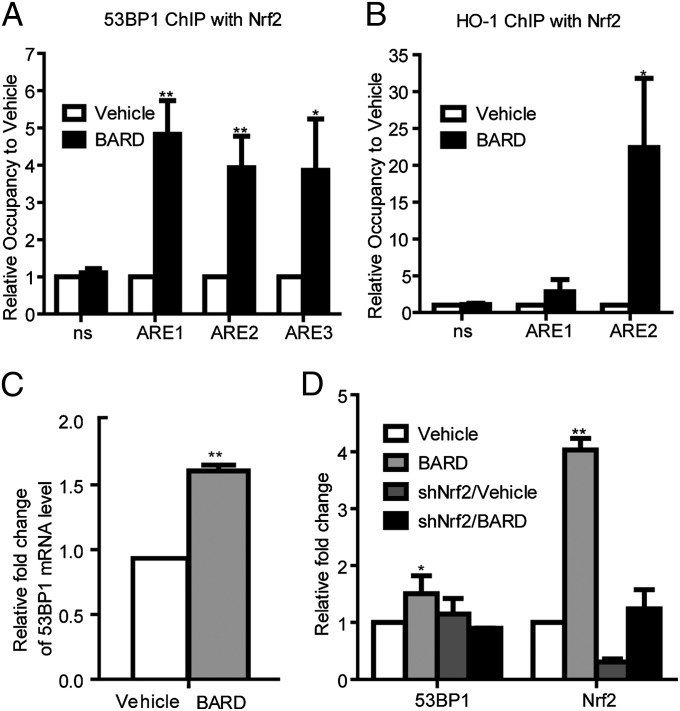
To explore the role of Nrf2 in the BARD-mediated DDR activity, we examined the promoter region of 53BP1, CtIP, and Rad51 for Nrf2 binding sites. Nrf2 binds to the AREs, which share a common TGACnnnGC motif (“core sequence”) (24). The HO-1 promoter has five core sequences (25) (Fig. S4A), and the 53BP1 promoter contains three ARE core sequences (Fig. S4B). However, these sequences were not found in CtIP or Rad51 promoters. Next, we examined if Nrf2 binds to the 53BP1 promoter region harboring AREs with BARD treatment using ChIP-quantitative PCR (qPCR). ChIP from unirradiated cells was performed using an anti-Nrf2 antibody 18 h after BARD treatment, followed by qPCR using specific primers for AREs of 53BP1 (Table S2), and revealed that Nrf2 binds to all AREs of 53BP1 after BARD treatment (Fig. 4A and Fig. S5). Nrf2 strongly (∼20-fold) binds to the HO-1 promoter region harboring ARE2 (Fig. 4B) with BARD treatment. The significant induction of 53BP1 expression by BARD treatment was confirmed by quantitation of mRNA (Fig. 4C) and protein levels (Fig. 4D).
Fig. 4.
Nrf2 increases 53BP1 expression by BARD treatment. ChIP-qPCR analysis of Nrf2 occupancy on the 53BP1 (A) and HO-1 (B) promoters using unirradiated cells following treatment with vehicle or BARD for 18 h. Data shown are from two separate triplicate experiments. (C) Quantitation of 53BP1 mRNA levels by RT-qPCR analysis showing increase of 53BP1 expression with BARD treatment for 18 h. Data shown are from two separate experiments and are normalized to GAPDH. (D) Quantitation of Western blot analysis showing increase of 53BP1 with BARD treatment for 18 h. Data shown are from two separate experiments and are normalized to β-actin. *P < 0.05 and **P < 0.005 (compared with vehicle control) in the unpaired Student t test; ns, not significant differences in the unpaired Student t test.
CDDO-EA Is an Effective Radioprotector in Mice.
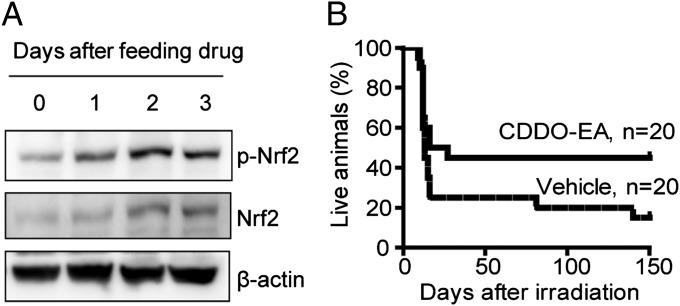
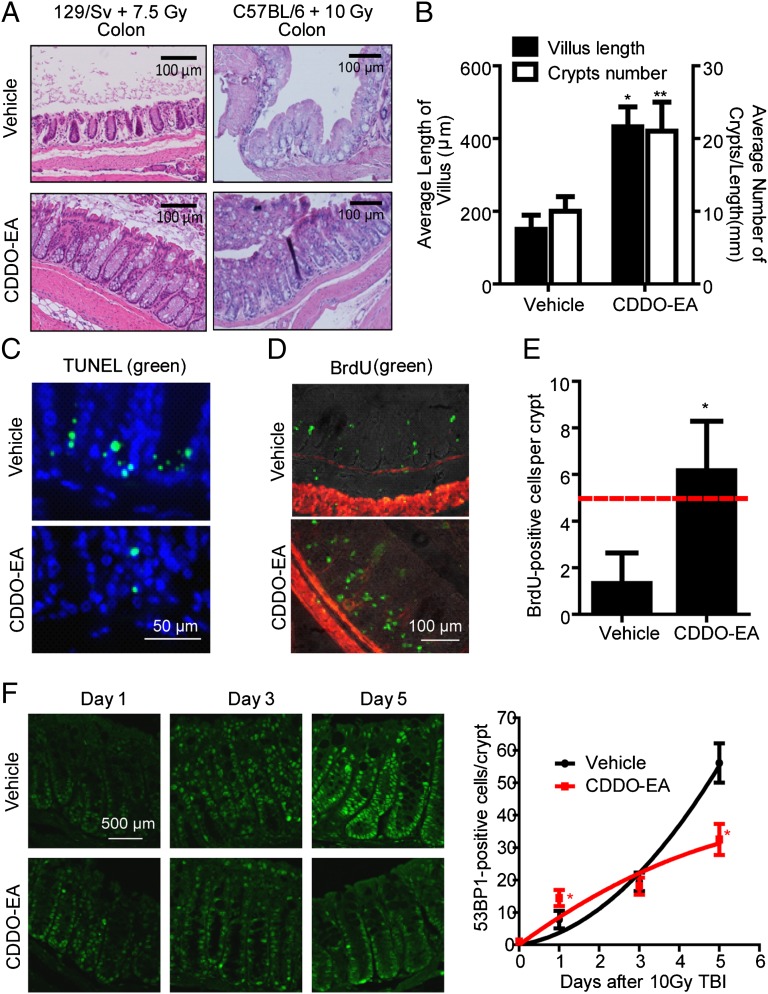
To investigate whether these synthetic triterpenoids have radioprotective activity in vivo, we used an ethyl amide derivative of a synthetic triterpenoid, CDDO-EA, which has been reported to have enhanced pharmacodynamic activity in mouse assays compared with BARD (14). We fed female WT 129/Sv mice (7–28 wk of age) a control diet (Lab Diet 5002; Purina Mills) or a diet containing CDDO-EA (400-mg/kg diet) (26). CDDO-EA treatment resulted in stabilization and activation of Nrf2 and increased phosphorylation of Nrf2 in unirradiated colonic tissue (Fig. 5A and Fig. S6). Treatment with CDDO-EA for 3 d before 7.5-Gy total body irradiation (TBI) significantly (95% confidence level) improved the median survival of mice from 13 to 21.5 d (Fig. 5B). To evaluate the radioprotective effect of CDDO-EA on the gastrointestinal (GI) tract further, colon tissues were examined 5 d after TBI from two different strains of mice (Fig. 6A). We found that mice fed with CDDO-EA were greatly protected from TBI-induced reduction in crypt size, number, cell density, and villus length in both the colon (Fig. 6A) and small intestine (Fig. 6B and Fig. S7). The protection of the GI tract was also quantitated by immunohistochemistry. Pretreatment of WT C57BL/6 mice with CDDO-EA for 3 d before 10-Gy TBI dramatically reduced the number of apoptotic cells (P = 0.0003 compared with vehicle control in the unpaired Student t test, n = 3) in colonic crypts (Fig. 6C). BrdU was injected 2 h before mice were killed to label proliferating crypt cells in 3-d postirradiated C57BL/6 mice (Fig. 6D). Control diet-fed mice showed a greater loss of proliferating cells compared with CDDO-EA diet-fed mice, which retained normal levels of proliferative crypt cells 3 d after 10-Gy TBI (Fig. 6E).
Fig. 5.
CDDO-EA increases mouse survival after an acute lethal dose of TBI. (A) CDDO-EA stabilizes and activates Nrf2 in mouse colon tissues. A CDDO-EA diet was provided to unirradiated WT mice for 1, 2, or 3 d, and the colon tissues were then lysed. Total Nrf2 and phospho-Nrf2 (p-Nrf2) were detected by Western blot analysis (Fig. S6; n = 3). (B) Groups of 129/Sv female mice were fed the CDDO-EA diet or control chow 3 d before 7.5-Gy TBI. Pooled results from two independent experiments are shown. Note a significant (within 95% confidence interval) increase in median survival in CDDO-EA–treated mice (21.5 d) compared with vehicle control (13 d).
Fig. 6.
CDDO-EA protects the GI tract from acute TBI. (A) Representative images of H&E staining of the colon tissue in different strains at 5 d after TBI with or without prior feeding of CDDO-EA chow. (B) Length of villi (left y axis) were measured in at least 40 complete, well-oriented villi cross-sections in the small intestine of 129/Sv mice 5 d after a 7.5-Gy dose of TBI. Crypt number per 1-mm length of the small intestine was counted (right y axis) (n = 3). (C–F) WT C57BL/6 mice were exposed to 10-Gy TBI 3 d after feeding of control or CDDO-EA chow. Representative TUNEL staining (green fluorescence) in the colon (C; 1 d postirradiation) and immunohistochemical detection of in vivo BrdU incorporation (green fluorescence) in the colon crypts (D; 3 d postirradiation) are shown. DAPI (blue) or smooth muscle actin (red fluorescence) was used for counterstaining. (E) BrdU-positive cells were counted in 15 complete, well-oriented crypt cross-sections (n = 3). The dashed line indicates the number of BrdU-positive cells considered critical for crypt survival (47). (F) Colon tissues were immunostained using anti-53BP1 antibodies 1, 3, or 5 d after a 10-Gy dose of TBI. (Left) Representative images show 53BP1-positive cells in colon tissues. DAPI was used for counterstaining (Fig. S7). (Right) Number of 53BP1-positive cells was counted in at least 60 complete, well-oriented crypt cross-sections in the colon (n = 3). *P < 0.0001 and **P = 0.0016 (compared with vehicle control) in the unpaired Student t test.
To explore DNA damage repair activity of CDDO-EA in vivo, we examined the appearance of 53BP1-positive cells in colonic crypts after TBI. Colonic tissues were fixed 1, 3, or 5 d after 10-Gy TBI. Paraffin sections were stained using a 53BP1-specific antibody, and 53BP1-positive cells were counted in each crypt (Fig. 6F). DAPI was used for counterstaining (Fig. S8). Consistent with the cell experiments, we observed that CDDO-EA treatment results in the rapid appearance of 53BP1-positive cells (day 1) and a shorter delay in DDRs (day 5). These results indicate that CDDO-EA also enhances the efficiency of HR repair of DSBs in vivo.
Discussion
This study has important implications for understanding new mechanistic insights of radioprotection activity through enhancing DNA damage sensing and subsequent repair. Here, we show that BARD pretreatment reduces IR-induced chromosome aberrations and cell killing by enhancing DNA damage sensing and repair in HCECs through Nrf2 activation. In addition, pretreatment with CDDO-EA increases mouse survival, which correlates with GI tract protection and DNA damage signaling promotion after exposure to a lethal dose of IR. BARD treatment before IR provides radioprotection, as determined by both in vivo and in vitro assays. Based on the established link between clonogenic survival and residual chromosome damage, we found that cells treated with BARD before irradiation have reduced IR-induced residual chromosome damage analyzed at metaphase. Because IR-induced NHEJ repair is predominant in G1-phase cells, we observed that BARD treatment has a significant effect on the G1-phase DNA DSB repair. In addition, the residual damage after BARD treatment was reduced in S- and G2-phase cells, arguing that BARD also activates the HR pathway to repair DNA DSB. This is based on demonstrating that cells treated with BARD had reduced amounts of tri- and quadriradials, in addition to the other chromatid types of aberrations. These observations suggest that BARD treatment before IR exposure may modify some aspect of cellular metabolism conducive to DNA DSB repair by both the NHEJ and HR pathways.
The observations that cells treated with BARD have a faster disappearance of CtIP or 53BP1 foci associated with repairosomes suggest that BARD treatment may modulate chromatin structure conducive for DNA DSB repair by HR. A key feature of HR is DNA strand invasion, which is catalyzed by the Rad51 protein coating single-stranded DNA and the formation of distinct nuclear foci in response to IR (27). The kinetics of signaling (53BP1) and resection (CtIP) have been well-established in the repair of DNA damage. The frequency of cells with such foci was found to be higher in BARD-treated cells or CDDO-EA–dieted mouse colon tissue, supporting the role of BARD in facilitating DNA DSB repair. Although repairosomes appear on recognition of DNA damage, treatment with BARD resulted in the rapid disappearance of repairosomes, providing further support for BARD-induced acceleration of DNA damage repair. These results are also consistent with the analysis of single DNA fibers, where cells treated with BARD have a higher frequency of new replication origins after HU-induced DNA damage, which is the result of enhanced damage repair. HU does result in DNA DSB poststalled replication forks, which is close to the DSBs induced by other agents. However, HU-induced DSBs almost exclusively arise in S-phase cells, whereas IR-induced DSBs arise in all cell cycle phases.
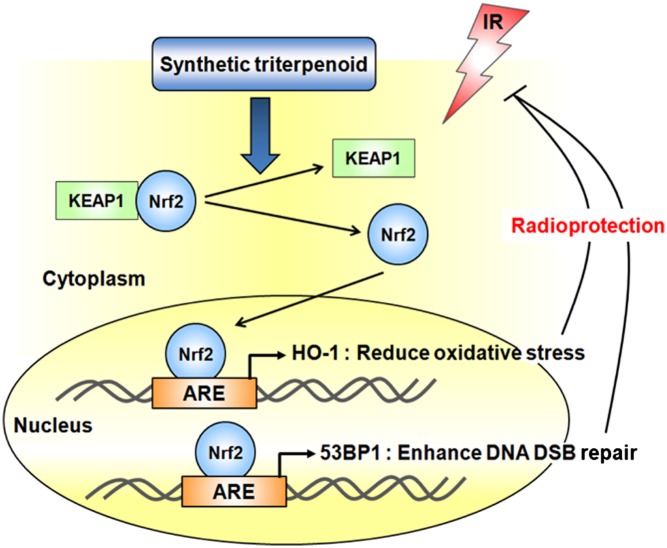
We have demonstrated that Nrf2 is required for the radioprotective activity of BARD. Nrf2 knockdown failed to increase clonogenic cell survival or to enhance the kinetics of repairosome formation with BARD treatment. Nrf2 is a known target for synthetic triterpenoids, which binds to AREs and induces antioxidative enzyme expression (13). Increased Nrf2/ARE signaling by pretreatment with synthetic triterpenoids before irradiation is sufficient for reduction of oxidation-mediated DNA damage and radioprotection. However, the mechanism of Nrf2 functions in repairosome formation and if synthetic triterpenoids enhance the frequency of DNA DSB repair were previously unknown. Here, we provide evidence that Nrf2 activation by BARD directly regulates 53BP1 transcription. We found that the 53BP1 promoter region has three ARE core sequences and that Nrf2 binds to all these AREs, resulting in an increase in 53BP1 expression. Nrf2 knockdown cells may have a slightly higher background of oxidative stress due to lack of Nrf2, and, as previously reported, oxidative stress also increases 53BP1 levels (28). However, we emphasize that BARD treatment increases 53BP1 protein levels only in nonknockdown (control) cells and not in Nrf2 knockdown cells (Fig. 4D).
There is an immediate need to develop easily administrable radioprotectants or mitigators with fewer side effects for first responders at nuclear accidents (29). It is also urgent to provide a safe radioprotector for astronauts for future long-term space missions (30, 31). In the past decade, a number of compounds have been extensively investigated for their cytoprotective capabilities against radiation. Among them, amifostine is the only drug registered by the US Food and Drug Administration as a cytoprotective agent (32). The DMF of amifostine was calculated to be 1.59 when injected into C57BL/6J mice (33). Because amifostine is associated with multiple side effects, including nausea, vomiting, sneezing, sleepiness, and hypotension (32, 34), there is a need to identify safer compounds that are capable of protecting cells from the adverse effects of radiation with fewer side effects. The synthetic triterpenoids, BARD and CDDO-EA, belong to a novel class of noncytotoxic, multifunctional, and orally available drugs that have been studied as anti-inflammatory modulators and as anticancer agents in vitro and in vivo (10–12). BARD is currently in a phase 3 clinical trial known as BEACON (bardoxolone methyl evaluation in patients with chronic kidney disease and type 2 diabetes: the occurrence of renal events) that involves 1,600 patients in over 300 sites worldwide (http://clinicaltrials.gov/ct2/show/NCT01351675?term=CDDO&rank=7). There are already robust patient data that BARD taken long term is safe, and therefore may have utility in other diseases associated with oxidative damage or inflammation. It also has been reported that treatment with BARD before exposure to heavy ions reduced oxidative stress and transformation in HCECs (15). In this study, we demonstrate that cells treated with BARD show an increased appearance of the number of foci, suggesting that the DDR is efficient in such cells. Furthermore, such cells show fast disappearance of foci, supporting the argument that repair is efficient. For CtIP and 53BP1, the residual foci are reduced in BARD-treated cells, consistent with our chromosomal data. Thus, the enhanced survival in CDDO-EA–treated mice and improved histology of the GI tract are likely due to an enhanced repair mechanism. Collectively, these studies imply that BARD and CDDO-EA are potent radioprotectors. The ease of administration (orally available) and relative lack of side effects support their further evaluation for clinical applications.
Materials and Methods
Cell Culture, Drug Treatments, and Irradiation.
Immortalized HCEC CT7s (15) were maintained as described previously (35). To establish an Nrf2 knockdown cell line, we infected HCEC CT7s with a lentiviral vector expressing a shRNA against Nrf2 (Open Biosystems) in the presence of 2 μg/mL Polybrene (Sigma). BARD (Reata Pharmaceuticals, Inc.) was dissolved in DMSO and treated at a concentration of 50 nM 18 h before irradiation. In some experiments, almost confluent cultures were exposed to γ-irradiation using a 137Cs source at a dose rate of 243.08 cGy/min at the University of Texas Southwestern Medical Center. In other experiments, trypsinized cells were seeded for colony formation assays in 100-mm dishes and irradiated for 48 h after plating. Fourteen days later, colonies were stained with a mixture of 6.0% (vol/vol) glutaraldehyde and 0.5% crystal violet and counted. Colonies were defined as clusters of >50 cells. Cell survival measurements were fitted by a linear quadratic equation [SF = exp (−αD − βD2)] (SF: surviving fraction; D: radiation dose in Gy) using GraphPad Prism.
Animals and Irradiation.
All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center. WT 129/Sv or C57BL/6 female mice, which were bred and housed in our facilities, were irradiated with IR using an X-RAD 320 irradiator (Precision X-ray, Inc.). Mice that were not anesthetized were held in ventilated 50-mL falcon tubes and placed 50 cm from the radiation source [source–skin distance (SSD)]. A 5-cm diameter brass collimator was placed in the collimator holder. Mice were irradiated at a dose rate of 111 cGy/min with 250 kV(peak), using 6 mA. Because C57BL/6 mice are more radioresistant than 129/Sv mice, we exposed 129/Sv mice to 7.5-Gy doses of radiation and C57BL/5 mice to 10-Gy doses of radiation. In addition, based on several published references, 7.5-Gy doses of radiation were used to determine survival after radiation exposure and 10-Gy doses of radiation were used to see acute intestinal toxicity and development of radiation-induced GI syndrome. Control (Lab Diet 5002) and CDDO-EA (400 mg/kg food) chow (provided by Reata Pharmaceuticals and Michael Sporn, Dartmouth Medical School, Hanover, NH) was prepared into chow pellets by Purina Mills (36).
Immunohistochemistry.
Colonic tissues were collected and fixed in 10% natural buffered formalin (NBF), embedded in paraffin, and sectioned. Apoptotic cells in the colonic crypts were detected 1 d after 10-Gy TBI in paraffin-embedded specimens. Apoptotic cells were stained by the TUNEL method using an ApopTag Fluorescein In Situ Apoptosis Detection Kit (green, catalog no. S7110; Millipore). Nuclei were visualized by DAPI staining (blue). To determine proliferating crypt cells 3 d after 10-Gy TBI, BrdU incorporation was performed. Proliferating cells were labeled by i.p. injection of BrdU (120 mg/kg) into mice 2 h before euthanasia. Colonic tissue was dissected, fixed in 10% (vol/vol) NBF, embedded in paraffin, and sectioned. BrdU-incorporating cells were visualized using anti-BrdU antibodies (catalog no. ab6326; Abcam) and secondary anti-rat antibodies labeled with FITC (Jackson Laboratories). Cy3-conjugated anti-smooth muscle actin antibodies (Sigma) were used for counterstaining.
Western Blot Analysis.
Colonic tissues were homogenized in lysis buffer [50 mM Tris⋅HCl (pH 7.5), 120 mM NaCl, 1% Triton X-100, and protease inhibitors] and clarified by centrifugation. Cells were lysed in Laemmli SDS reducing buffer [50 mM Tris⋅HCl (pH 6.8), 2% SDS, and 10% glycerol], boiled, and separated by SDS/PAGE. The following antibodies were used: anti-Nrf2 (Santa Cruz Biotechnology), anti–HO-1 (BD Transduction Laboratories), anti-phospho Nrf2 (S40; Abcam), anti-Rad51 (Biomeda), and anti–β-actin (Sigma) antibodies.
Subcellular Fractionation.
Cells were scraped off the dish and collected by centrifugation at 2,000 × g for 5 min. Cells were resuspended in 500 μL of cell lysis buffer [50 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 1 mM MgCl2, 2 mM EDTA, and protease inhibitors], allowed to swell on ice for 10 min, and passaged five times through a 27-gauge syringe. Nuclei were collected by centrifugation at 500 × g for 10 min, and the supernatant was saved for cytosolic extracts. The nuclei were resuspended in 50 μL of nuclear extraction buffer [20 mM Hepes (pH 7.9), 1.5 mM MgCl2, 25% (vol/vol) glycerol, 400 mM KCl, 0.5 mM DTT, and 0.2 mM PMSF], stirred on ice for 30 min, and then centrifuged at 20,000 × g for 5 min. The supernatant was collected for the nuclear extract. Protein concentration was determined using a Pierce BCA Protein Assay Kit with BSA as the standard.
Immunofluorescence.
Cells were cultured in chamber slides, fixed, and immunostained as previously described (19, 37, 38). Sections through nuclei were captured, and fluorescent images of foci were obtained by projection of the individual sections as recently described (39). The results shown are from three independent experiments.
Assay for Chromosomal Aberrations at Metaphase.
All three stage-specific chromosomal aberrations were analyzed at metaphase after exposure to IR. G1-type chromosomal aberrations were assessed in cells exposed to 5 Gy of IR and incubated for 14 h. Cells were then subcultured, and metaphases were collected (40, 41). S-phase–specific chromosome aberrations were assessed after exponentially growing cells (pulse-labeled with BrdU), which were irradiated with 4 Gy of IR. Metaphases were harvested following 4 h of irradiation, and S-phase types of chromosomal aberrations were scored. For G2-specific aberrations, cells were irradiated with 1 Gy and metaphases were collected minutes posttreatment (42). Chromosome spreads were prepared after hypotonic treatment of cells, fixed in acetic acid⋅methanol, and stained with Giemsa (43). The categories of G1-type asymmetrical chromosome aberrations that were scored include dicentrics, centric rings, interstitial deletions/acentric rings, and terminal deletions. S-phase chromosome aberrations were assessed by counting both the chromosome and chromatid aberrations, including triradial and quadriradial exchanges per metaphase as previously described (40, 41). G2-phase chromosomal aberrations were assessed by counting chromatid breaks and gaps per metaphase as previously described (40, 41). Fifty metaphases were scored for each postirradiation time point.
DNA Fiber Assay.
We performed DNA fiber spreads using previously described procedures (44) with the following minor modifications. In brief, exponentially growing cells were labeled for sites of ongoing replication with IdU (50 μM) for 20 min, followed by exposure to HU (4 mM). HU was removed by washing cells four times with PBS before labeling with media containing CIdU (50 μM) to mark the sites of replication recovery. After trypsinization, cells were washed with cold PBS, resuspended in lysis buffer (0.5% SDS, 50 mM EDTA, and 200 mM Tris⋅HCl), and spread on tilted glass slides, facilitating the spread of genomic DNA into single-molecule DNA fibers by gravity. Next, the slides were fixed in acetic acid and methanol (1:3 ratio). DNA was then denatured by treating fibers with 2.5 M HCl, neutralized, washed first with 1× PBS (pH 8.0), and subsequently washed three times with 1× PBS (pH 7.4). This was followed by blocking with 10% goat serum and 0.1% Triton-X in PBS (60 min) and incubation with primary antibodies against IdU (BD Biosciences) and CIdU (Novus Biologicals), followed by incubation with secondary antibodies for 1 h each. Fibers were analyzed using ImageJ software (National Institutes of Health).
ChIP-qPCR.
ChIP was performed as previously described (45, 46). Briefly, cells were fixed with formaldehyde 18 h after vehicle or BARD treatment. Chromatin was sheared and immunoprecipitated with anti-Nrf2 antibody (Santa Cruz Biotechnology). ChIP-qPCR primers for the AREs of the HO-1 or 53BP1 promoter region are described in Table S2. qPCR was performed using SsoFast EvaGreen Supermix (Bio-Rad) with the following cycling parameters: 95 °C for 5 min and 45 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. Samples were run in triplicate, and data were normalized to vehicle-treated control after subtraction of signals obtained from antibody isotype control and input control. ChIP-qPCR was repeated twice to confirm the reproducibility of results.
Statistical Analysis.
Results are described as mean ± SEM. Comparisons of different groups for statistical significance were analyzed using a two-tailed, unpaired Student t test. A P value of 0.05 or less was considered significant.
Supplementary Material
Acknowledgments
We thank Reata Pharmaceuticals, Inc., and Michael Sporn for providing reagents and advice for these studies and Richard Baer (Health Science Division, Columbia University, New York) for providing the CtIP antibody. We also thank Gail Fasciani, Richard MacDonnell, and Summer Barron for technical help. This work was supported by National Aeronautics and Space Administration Grants NNX11AC15G, NNJ05HD36G, and NNX09AU95G (to J.W.S.) and National Institutes of Health National Cancer Institute Grants R01CA123232, R01CA129537, R01CA154320, and U19A1091175 (to T.K.P.).
Footnotes
Conflict of interest statement: J.W.S. is on the Science Advisory Board (SAB) of Reata Pharmaceuticals.
This article is a PNAS Direct Submission.
See Author Summary on page 17327 (volume 109, number 43).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207718109/-/DCSupplemental.
References
- 1.Szumiel I. Epidermal growth factor receptor and DNA double strand break repair: The cell’s self-defence. Cell Signal. 2006;18(10):1537–1548. doi: 10.1016/j.cellsig.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 3.Pitcher RS, Wilson TE, Doherty AJ. New insights into NHEJ repair processes in prokaryotes. Cell Cycle. 2005;4(5):675–678. doi: 10.4161/cc.4.5.1676. [DOI] [PubMed] [Google Scholar]
- 4.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283(1):1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 5.Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132(2):387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonoda E, Takata M, Yamashita YM, Morrison C, Takeda S. Homologous DNA recombination in vertebrate cells. Proc Natl Acad Sci USA. 2001;98(15):8388–8394. doi: 10.1073/pnas.111006398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265(5176):1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 8.You Z, et al. CtIP links DNA double-strand break sensing to resection. Mol Cell. 2009;36(6):954–969. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kousholt AN, et al. CtIP-dependent DNA resection is required for DNA damage checkpoint maintenance but not initiation. J Cell Biol. 2012;197(7):869–876. doi: 10.1083/jcb.201111065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thimmulappa RK, et al. Preclinical evaluation of targeting the Nrf2 pathway by triterpenoids (CDDO-Im and CDDO-Me) for protection from LPS-induced inflammatory response and reactive oxygen species in human peripheral blood mononuclear cells and neutrophils. Antioxid Redox Signal. 2007;9(11):1963–1970. doi: 10.1089/ars.2007.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petronelli A, Pannitteri G, Testa U. Triterpenoids as new promising anticancer drugs. Anticancer Drugs. 2009;20(10):880–892. doi: 10.1097/CAD.0b013e328330fd90. [DOI] [PubMed] [Google Scholar]
- 12.Vannini N, et al. The synthetic oleanane triterpenoid, CDDO-methyl ester, is a potent antiangiogenic agent. Mol Cancer Ther. 2007;6(12 Pt 1):3139–3146. doi: 10.1158/1535-7163.MCT-07-0451. [DOI] [PubMed] [Google Scholar]
- 13.Liby K, et al. The synthetic triterpenoids, CDDO and CDDO-imidazolide, are potent inducers of heme oxygenase-1 and Nrf2/ARE signaling. Cancer Res. 2005;65(11):4789–4798. doi: 10.1158/0008-5472.CAN-04-4539. [DOI] [PubMed] [Google Scholar]
- 14.Liby K, et al. The synthetic triterpenoids CDDO-methyl ester and CDDO-ethyl amide prevent lung cancer induced by vinyl carbamate in A/J mice. Cancer Res. 2007;67(6):2414–2419. doi: 10.1158/0008-5472.CAN-06-4534. [DOI] [PubMed] [Google Scholar]
- 15.Eskiocak U, et al. CDDO-Me protects against space radiation-induced transformation of human colonic epithelial cells. Radiat Res. 2010;173:27–36. doi: 10.1667/RR2155.1. [DOI] [PubMed] [Google Scholar]
- 16.Pike MC, Alper T. A method for determining dose-modification factors. Br J Radiol. 1964;37:458–462. doi: 10.1259/0007-1285-37-438-458. [DOI] [PubMed] [Google Scholar]
- 17.Moore RC, Bender MA. Chromosomal aberration types in cells at the second division after irradiation in G1 or G2. Int J Radiat Biol. 1993;63(6):731–741. doi: 10.1080/09553009314552131. [DOI] [PubMed] [Google Scholar]
- 18.Gupta A, et al. Involvement of human MOF in ATM function. Mol Cell Biol. 2005;25(12):5292–5305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandita TK. Role of mammalian Rad9 in genomic stability and ionizing radiation response. Cell Cycle. 2006;5(12):1289–1291. doi: 10.4161/cc.5.12.2862. [DOI] [PubMed] [Google Scholar]
- 20.Henry-Mowatt J, et al. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol Cell. 2003;11(4):1109–1117. doi: 10.1016/s1097-2765(03)00132-1. [DOI] [PubMed] [Google Scholar]
- 21.Bianchi V, Pontis E, Reichard P. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J Biol Chem. 1986;261(34):16037–16042. [PubMed] [Google Scholar]
- 22.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37(4):492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlacher K, et al. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145(4):529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 1991;266(18):11632–11639. [PubMed] [Google Scholar]
- 25.Lee HH, et al. Piceatannol induces heme oxygenase-1 expression in human mammary epithelial cells through activation of ARE-driven Nrf2 signaling. Arch Biochem Biophys. 2010;501(1):142–150. doi: 10.1016/j.abb.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 26.Stack C, et al. Triterpenoids CDDO-ethyl amide and CDDO-trifluoroethyl amide improve the behavioral phenotype and brain pathology in a transgenic mouse model of Huntington’s disease. Free Radic Biol Med. 2010;49(2):147–158. doi: 10.1016/j.freeradbiomed.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarsounas M, Davies AA, West SC. RAD51 localization and activation following DNA damage. Philos Trans R Soc Lond B Biol Sci. 2004;359(1441):87–93. doi: 10.1098/rstb.2003.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, Yang J, Huang H. Oxidative stress induces H2AX phosphorylation in human spermatozoa. FEBS Lett. 2006;580(26):6161–6168. doi: 10.1016/j.febslet.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 29.Bhattacharjee Y. Devastation in Japan. Candidate radiation drugs inch forward. Science. 2011;331(6024):1505. doi: 10.1126/science.331.6024.1505. [DOI] [PubMed] [Google Scholar]
- 30.Shay JW, Cucinotta FA, Sulzman FM, Coleman CN, Minna JD. From mice and men to earth and space: Joint NASA-NCI Workshop on lung cancer risk resulting from space and terrestrial radiation. Cancer Res. 2011;71(22):6926–6929. doi: 10.1158/0008-5472.CAN-11-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durante M, Cucinotta FA. Heavy ion carcinogenesis and human space exploration. Nat Rev Cancer. 2008;8(6):465–472. doi: 10.1038/nrc2391. [DOI] [PubMed] [Google Scholar]
- 32.Andreassen CN, Grau C, Lindegaard JC. Chemical radioprotection: A critical review of amifostine as a cytoprotector in radiotherapy. Semin Radiat Oncol. 2003;13(1):62–72. doi: 10.1053/srao.2003.50006. [DOI] [PubMed] [Google Scholar]
- 33.Yuhas JM. Biological factors affecting the radioprotective efficiency of S-2-[2-aminopropylamino] ethylphosphorothioic acid (WR-2721). LD50(3)) doses. Radiat Res. 1970;44(3):621–628. [PubMed] [Google Scholar]
- 34.Hensley ML, et al. American Society of Clinical Oncology clinical practice guidelines for the use of chemotherapy and radiotherapy protectants. J Clin Oncol. 1999;17(10):3333–3355. doi: 10.1200/JCO.1999.17.10.3333. [DOI] [PubMed] [Google Scholar]
- 35.Roig AI, et al. Immortalized epithelial cells derived from human colon biopsies express stem cell markers and differentiate in vitro. Gastroenterology. 2010;138(3):1012–1021.e1–e5. doi: 10.1053/j.gastro.2009.11.052. [DOI] [PubMed] [Google Scholar]
- 36.Neymotin A, et al. Neuroprotective effect of Nrf2/ARE activators, CDDO ethylamide and CDDO trifluoroethylamide, in a mouse model of amyotrophic lateral sclerosis. Free Radic Biol Med. 2011;51(1):88–96. doi: 10.1016/j.freeradbiomed.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agarwal M, et al. Inhibition of telomerase activity enhances hyperthermia-mediated radiosensitization. Cancer Res. 2008;68(9):3370–3378. doi: 10.1158/0008-5472.CAN-07-5831. [DOI] [PubMed] [Google Scholar]
- 38.Sharma GG, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;30(14):3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta A, et al. Cell cycle checkpoint defects contribute to genomic instability in PTEN deficient cells independent of DNA DSB repair. Cell Cycle. 2009;8(14):2198–2210. doi: 10.4161/cc.8.14.8947. [DOI] [PubMed] [Google Scholar]
- 40.Hunt CR, et al. Genomic instability and enhanced radiosensitivity in Hsp70.1- and Hsp70.3-deficient mice. Mol Cell Biol. 2004;24(2):899–911. doi: 10.1128/MCB.24.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma GG, et al. Human heterochromatin protein 1 isoforms HP1(Hsalpha) and HP1(Hsbeta) interfere with hTERT-telomere interactions and correlate with changes in cell growth and response to ionizing radiation. Mol Cell Biol. 2003;23(22):8363–8376. doi: 10.1128/MCB.23.22.8363-8376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhar S, Squire JA, Hande MP, Wellinger RJ, Pandita TK. Inactivation of 14-3-3sigma influences telomere behavior and ionizing radiation-induced chromosomal instability. Mol Cell Biol. 2000;20(20):7764–7772. doi: 10.1128/mcb.20.20.7764-7772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandita TK. Effect of temperature variation on sister chromatid exchange frequency in cultured human lymphocytes. Hum Genet. 1983;63(2):189–190. doi: 10.1007/BF00291543. [DOI] [PubMed] [Google Scholar]
- 44.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140(6):1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98(5):675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 46.Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7(4):592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 47.Cohn SM, Schloemann S, Tessner T, Seibert K, Stenson WF. Crypt stem cell survival in the mouse intestinal epithelium is regulated by prostaglandins synthesized through cyclooxygenase-1. J Clin Invest. 1997;99(6):1367–1379. doi: 10.1172/JCI119296. [DOI] [PMC free article] [PubMed] [Google Scholar]