Abstract
The angiosperm order Malpighiales includes ∼16,000 species and constitutes up to 40% of the understory tree diversity in tropical rain forests. Despite remarkable progress in angiosperm systematics during the last 20 y, relationships within Malpighiales remain poorly resolved, possibly owing to its rapid rise during the mid-Cretaceous. Using phylogenomic approaches, including analyses of 82 plastid genes from 58 species, we identified 12 additional clades in Malpighiales and substantially increased resolution along the backbone. This greatly improved phylogeny revealed a dynamic history of shifts in net diversification rates across Malpighiales, with bursts of diversification noted in the Barbados cherries (Malpighiaceae), cocas (Erythroxylaceae), and passion flowers (Passifloraceae). We found that commonly used a priori approaches for partitioning concatenated data in maximum likelihood analyses, by gene or by codon position, performed poorly relative to the use of partitions identified a posteriori using a Bayesian mixture model. We also found better branch support in trees inferred from a taxon-rich, data-sparse matrix, which deeply sampled only the phylogenetically critical placeholders, than in trees inferred from a taxon-sparse matrix with little missing data. Although this matrix has more missing data, our a posteriori partitioning strategy reduced the possibility of producing multiple distinct but equally optimal topologies and increased phylogenetic decisiveness, compared with the strategy of partitioning by gene. These approaches are likely to help improve phylogenetic resolution in other poorly resolved major clades of angiosperms and to be more broadly useful in studies across the Tree of Life.
Malpighiales are one of the most surprising clades discovered in broad molecular phylogenetic studies of the flowering plants (1–3). The order contains ∼16,000 species and 42 families (2, 3) that exhibit remarkable morphological and ecological diversity. A few examples include cactus-like succulents (Euphorbiaceae), epiphytes (Clusiaceae), holoparasites (Rafflesiaceae), submerged aquatics (Podostemaceae), and wind-pollinated trees (temperate Salicaceae). The order is ecologically important: species in Malpighiales constitute up to 40% of the understory tree diversity in tropical rain forests worldwide (4). They also include many economically important species, such as Barbados nut (Jatropha curcas L., Euphorbiaceae), cassava (Manihot esculenta Crantz, Euphorbiaceae), castor bean (Ricinus communis L., Euphorbiaceae), coca (Erythroxylum coca Lam., Erythroxylaceae), flax (Linum usitatissimum L., Linaceae), the poplars (Populus spp., Salicaceae), and the rubber tree (Hevea brasiliensis Müll. Arg., Euphorbiaceae). Partially for this reason, genomic resources for Malpighiales are growing at a rapid pace and include whole-genome sequencing projects completed or near completion for Barbados nut (5), cassava, castor bean (6), flax, and poplar (7). Thus, a resolved phylogeny of Malpighiales is critical not only for evolutionary, ecological, developmental, and genomic investigations of flowering plants, but also for crop improvement.
Despite substantial progress in resolving the angiosperm Tree of Life during the last 20 y (1, 8–12), phylogenetic relationships within Malpighiales remain poorly resolved. Molecular studies (1, 4) using multiple gene regions from the plastid, mitochondrial, and nuclear genomes have confirmed the monophyly of Malpighiales and its component families with a high degree of confidence but have identified only a handful of well-supported multifamily clades. The most recent analysis by Wurdack and Davis (3) included 13 genes, totaling 15,604 characters, sampled across all three genomes from 144 Malpighiales. Their results indicated that all families are monophyletic, but interrelationships among the 16 major subclades remained unresolved. The difficulty in determining these deep relationships may result from the rapid rise of the order during the mid-Cretaceous (4).
We used phylogenomic approaches to resolve relationships within Malpighiales to provide a framework for studying their tempo and mode of diversification. Our core data set included 82 genes sampled from the plastomes of 58 species, 48 of which were newly sequenced for this study. We combined this core data set with the previously described taxon-rich data set of Wurdack and Davis (3). Our results greatly improve phylogenetic resolution within Malpighiales, highlight the value of a unique partitioning strategy for phylogenomic analyses, and reveal a dynamic history of shifts in net diversification rates across the order.
Results and Discussion
Taxon and Gene Sampling.
Our core data set, the 82-gene matrix, included 58 taxa (48 are newly sequenced; SI Appendix, Table S2) and 82 plastid genes common to most angiosperms (72,828 characters; 17% of the cells in the matrix were gaps or missing data; each taxon was represented by an average of 86% of the 82 genes; SI Appendix, Tables S2 and S3). The taxa were carefully selected to capture the basal nodes within deeply diverged families, such as Centroplacaceae and Euphorbiaceae (4); they represent 39 of the 42 families of Malpighiales (excluding Lophopyxidaceae, Malesherbiaceae, and Rafflesiaceae; SI Appendix, SI Materials and Methods) and relevant outgroups. To obtain the most comprehensive phylogenetic tree for the order, we used the 82-gene matrix as a scaffold to which we added the existing taxon-rich but character-sparse 13-gene matrix (186 taxa; 15,574 characters; 15% missing data) (3) to create our combined-incomplete matrix (Table 1). This matrix included 191 taxa and 91 genes (82 plastid genes, six mitochondrial genes, and three nuclear genes; 81,259 characters; 64% missing data). We also created a combined-complete matrix by reducing the taxon sampling in the combined-incomplete matrix to match the taxon sampling of the 82-gene matrix. This greatly reduced the percentage of missing data cells in our alignment from 64% to 12%. The combined-complete matrix included 58 taxa and 91 genes (81,117 characters). Finally, we reanalyzed the 13-gene matrix alone to determine the phylogenetic impact of adding the 82-gene matrix. Each of the four matrices was analyzed using four different data partitioning strategies that are described below.
Table 1.
Characteristics of the four matrices and statistics of the best-scoring ML trees inferred from each of the four partitioning strategies
| Matrix | Taxa/characters/missing data % | Partitioning strategy | No. of partitions | Log-likelihood | AICc | ΔAICc | Coverage density | Fraction of triples | D | d | Terrace size |
| 82-gene | 58/72,828/17% | OnePart | 1 | −689042 | 1,378,328 | 166,322 | 1.00 | 1.00 | 1.00 | 1.00 | 1 |
| GenePart | 82 | −680357 | 1,362,435 | 150,429 | 0.88 | 1.00 | 1.00 | 1.00 | 1 | ||
| CodonPart | 4 | −680281 | 1,360,860 | 148,854 | 1.00 | 1.00 | 1.00 | 1.00 | 1 | ||
| MixtPart | 13 | −605772 | 1,212,006 | 0 | 1.00 | 1.00 | 1.00 | 1.00 | 1 | ||
| Combined-complete | 58/81,117/12% | OnePart | 1 | −739270 | 1,478,784 | 193,023 | 1.00 | 1.00 | 1.00 | 1.00 | 1 |
| GenePart | 91 | −728235 | 1,458,355 | 172,594 | 0.88 | 1.00 | 1.00 | 1.00 | 1 | ||
| CodonPart | 4 | −730551 | 1,461,401 | 175,640 | 1.00 | 1.00 | 1.00 | 1.00 | 1 | ||
| MixtPart | 15 | −642632 | 1,285,761 | 0 | 1.00 | 1.00 | 1.00 | 1.00 | 1 | ||
| Combined-incomplete | 191/81,259/64% | OnePart | 1 | −892791 | 1,786,362 | 234,881 | 1.00 | 1.00 | 1.00 | 1.00 | 1 |
| GenePart | 91 | −879681 | 1,761,794 | 210,313 | 0.36 | 0.93 | 0.00 | 0.97 | 14,025 | ||
| CodonPart | 4 | −883407 | 1,767,647 | 216,166 | 1.00 | 1.00 | 1.00 | 1.00 | 1 | ||
| MixtPart | 20 | −775178 | 1,551,481 | 0 | 1.00 | 1.00 | 1.00 | 1.00 | 1 | ||
| 13-gene | 186/15,574/15% | OnePart | 1 | −292212 | 585,198 | 47,256 | 1.00 | 1.00 | 1.00 | 1.00 | 1 |
| GenePart | 13 | −288145 | 577,294 | 39,352 | 0.93 | 1.00 | 1.00 | 1.00 | 1 | ||
| CodonPart | 4 | −289988 | 580,807 | 42,865 | 1.00 | 1.00 | 1.00 | 1.00 | 1 | ||
| MixtPart | 14 | −268460 | 537,942 | 0 | 1.00 | 1.00 | 1.00 | 1.00 | 1 |
The MixtPart partitioning strategy is highlighted in bold to indicate that it produced the best log-likelihood and AICc values in each case. The fraction of triples is the fraction of all possible triples of taxa for which sequence data are present for at least some partitions of the matrix; D is the probability that a pattern of taxon coverage is decisive for a random binary tree; d is the average number of edges distinguishable on a random binary tree (further details in SI Appendix, Materials and Methods).
Relationships in Malpighiales.
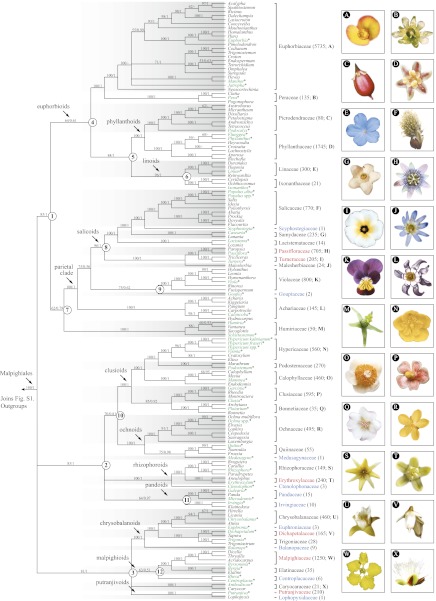
Our analyses produced a well-resolved phylogeny of Malpighiales (Fig. 1; relationships of outgroups provided in SI Appendix, Fig. S1). The maximum likelihood (ML) and Bayesian trees inferred from the combined-incomplete matrix are congruent with trees from the remaining three matrices (i.e., 82-gene, combined-complete, and 13-gene; SI Appendix, Figs. S1–S17 and S26–S28), using 75 ML bootstrap percentage (BP; as calculated using the standard bootstrap option in RAxML) and 0.95 Bayesian posterior probability (PP) thresholds. The 16 subclades of Malpighiales whose interrelationships were previously unresolved with respect to one another are resolved into three well-supported (>80 BP, 1.0 PP) clades. Moreover, we find comparable or greatly improved support for previously identified clades (3) and moderate to strong support for the 12 additional clades we identified (Fig. 1). Six of these clades were supported with >80 BP, 1.0 PP; two with ≥70 BP, >0.60 PP; and one with >60 BP, >0.95 PP. Importantly, each of the 12 clades is also united by morphological features (summarized in Table 2).
Fig. 1.
ML 50% majority-rule bootstrap consensus tree of Malpighiales inferred from the combined-incomplete matrix using the MixtPart partitioning strategy. ML BPs/Bayesian PPs are indicated above each branch; a hyphen indicates that the node is not present in the Bayesian 50% majority-rule consensus tree. The 12 additional clades we identified are designated using the numbers corresponding to those in Table 2. Taxa included in the 82-gene matrix are highlighted in green and marked with asterisks; spp. = composite terminals compiled from multiple closely related species; the approximate number of accepted species for each family is given in parentheses to the right; lettered photographs depicting representative species from all included families are shown to the right. Clades exhibiting a shift in net species diversification are highlighted in red (acceleration) and blue (deceleration). More detailed results of diversification analyses are provided in the text and SI Appendix. Outgroup relationships are shown in SI Appendix, Fig. S1.
Table 2.
Morphological features for the 12 additional clades we identified in Malpighiales
| Clade | Morphological features |
| 1 | Androgynophore; ovules mostly crassinucellar |
| 2 | Tendency to incompletely tenuinucellar ovules |
| 3 | Tendency to (oblique) floral monosymmetry in chrysobalanoids and Malpighiaceae; tendency to bulging of ovaries; placentation mostly axile; inner integument thicker than outer |
| 4 | Tendency to unisexual, trimerous flowers with reduced petals (not in Ixonanthaceae and Linaceae); petals, if present, often contort; placentation mostly axile; ovules 2 (more rarely 1) per carpel; antitropous, pendant, with obturator; inner integument thicker than outer |
| 5 | Tendency to false septa in carpels; placentation axile; ovules 2 per carpel, antitropous, pendant, with obturator; inner integument thicker than outer |
| 6 | Flowers bisexual; mostly diplostemonous; carpels with false septa |
| 7 | Flowers mostly haplostemonous (not in Humiriaceae); carpels often 3 (5 in Humiria); placentation parietal (not in Humiriaceae); ovules often more than 2 per carpel, crassinucellar, without endothelium; seeds often with aril (not in Humiriaceae) |
| 8 | Corona present in some families; placentation parietal; ovules mostly more than 2 per carpel, crassinucellar; seeds often with aril |
| 9 | Flowers haplostemonous; anthers with conspicuous appendages; nectary, if present, at outer base of stamens; ovules more than 2 per carpel |
| 10 | Petals often contort; mostly polystemonous; placentation mostly axile; ovules often incompletely tenuinucellar, with endothelium |
| 11 | Placentation axile; ovule 1 per carpel, antitropous |
| 12 | Placentation axile; ovules crassinucellar, without endothelium; sepals persistent in fruit |
Data compiled from SI Appendix, SI refs. 24 and 28–34. Clades are labeled in Fig. 1 accordingly.
Clade 1 (85 BP, 1.0 PP) includes two major subclades: the euphorbioids (clade 4) and Humiriaceae + the parietal clade sensu Wurdack and Davis (3) (clade 7). Surprisingly, the euphorbioid clade (64 BP, 0.61 PP) reunites most of the former Euphorbiaceae (including Euphorbiaceae, Peraceae, Phyllanthaceae, and Picrodendraceae but excluding Putranjivaceae) (13, 14) with the well-supported (96 BP, 1.0 PP) linoid clade (clade 6; Ixonanthaceae + Linaceae) we identified. Within the euphorbioids, the linoids are well-supported (clade 5; 84 BP, 1.0 PP) as sister to the phyllanthoids (Phyllanthaceae + Picrodendraceae; 100 BP, 1.0 PP). The second major subclade identified here, clade 7 (62 BP, 0.79 PP), includes Humiriaceae and the parietal clade (100 BP, 1.0 PP). Within the parietal clade, Goupiaceae is sister to Violaceae (clade 9; 75 BP, 0.62 PP). Also within the parietal clade, (Malesherbiaceae (Passifloraceae + Turneraceae)) is sister to the salicoids [clade 8; (Lacistemataceae (Samydaceae (Salicaceae + Scyphostegiaceae))); 96 BP, 1.0 PP].
Clade 2 (83 BP, 1.0 PP) includes three subclades in a trichotomy. Its first major subclade, clade 10 (70 BP, 0.81 PP), includes the previously identified (6, 15) clusioid clade [((Bonnetiaceae + Clusiaceae) (Calophyllaceae (Hypericaceae + Podostemaceae))); 100 BP, 1.0 PP] plus their sister group the ochnoids [(Ochnaceae (Medusagynaceae + Quiinaceae)); 100 BP, 1.0 PP]. The second subclade in clade 2 is the recently identified (3) rhizophoroids [(Ctenolophonaceae (Erythroxylaceae + Rhizophoraceae)); 100 BP, 1.0 PP]. The third subclade is the pandoids (clade 11; Irvingiaceae + Pandaceae; 64 BP, 0.97 PP).
Clade 3 (81 BP, 1.0 PP) consists of four subclades, three of which have been previously identified (3), in a polytomy. These four subclades are the chrysobalanoids [(Balanopaceae ((Chrysobalanaceae + Euphroniaceae) (Dichapetalaceae + Trigoniaceae))); 100 BP, 1.0 PP], the malpighioids [clade 12; (Centroplacaceae (Elatinaceae + Malpighiaceae)); 63 BP, 0.51 PP], the putranjivoids (Lophopyxidaceae + Putranjivaceae; 100 BP, 1.0 PP), and Caryocaraceae.
Improved Phylogenetic Resolution Results from a Posteriori Data Partitioning and Better Taxon Sampling.
Several previous phylogenomic studies of angiosperms have applied a single substitution matrix in ML analyses to multiple-gene concatenated data sets (OnePart; e.g., refs. 8, 16, and 17). More recently, to better accommodate evolutionary rate heterogeneity across different characters, alignments have been partitioned a priori by gene (GenePart; e.g., refs. 11 and 12) or by codon position (CodonPart; e.g., refs. 11 and 18). The GenePart approach creates a partition for each gene and estimates the substitution rate matrix parameters separately for each partition, resulting in up to 83 partitions for many plastid data sets. The CodonPart approach partitions characters according to codon position, with a fourth partition added for noncoding regions (if present). These partitioning strategies are somewhat arbitrary, assuming for example that all third codon positions evolve rapidly or that gene boundaries define a class of sites that are expected to share a similar model of molecular evolution. As an alternative, we explored the use of an a posteriori partitioning strategy for ML analyses based on the partitions inferred from Bayesian searches of the matrix using a mixture model approach (19). Using a reversible-jump implementation, the Bayesian mixture model estimates the number of substitution rate matrices that best fit an alignment by allowing the fitting of multiple rate matrices to each character separately (20). We used this approach to find the optimal number of partitions for each matrix and then defined the characters in each class as a partition for subsequent ML analyses (MixtPart).
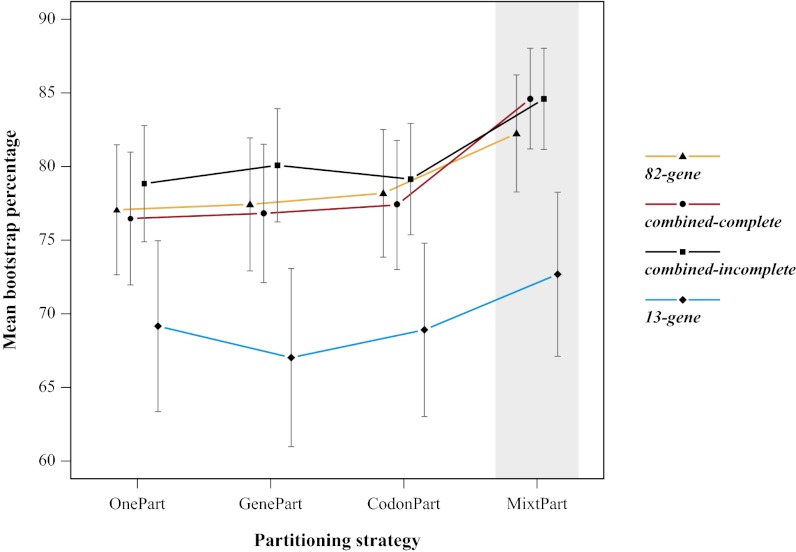
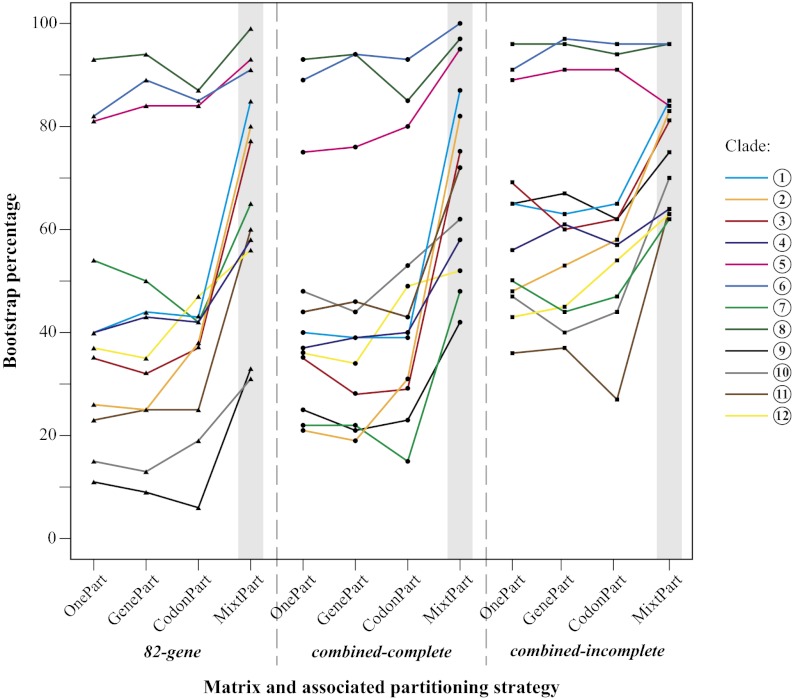
Using MixtPart, we found that the optimal number of partitions was 13 for the 82-gene matrix, 15 for the combined-complete matrix, and 20 for the combined-incomplete matrix. Thus, in all cases, defining the partitions on the basis of the mixture model search reduced the number of partitions substantially (from 82 for the 82-gene and from 91 for the two combined matrices using GenePart). Notably, our results show that using MixtPart substantially improved the likelihood of the best-scoring ML tree as measured by the corrected Akaike information criterion (AICc) (21) for all four matrices (Table 1). For example, compared with the GenePart approach, the MixtPart approach increased the AICc values by 7–12%. MixtPart also outperformed the OnePart, GenePart, and CodonPart approaches with respect to improving the branch support as measured by BP values. To compare these BP values among trees with different taxon sets, the bipartition trees inferred from the combined-incomplete and 13-gene matrices (SI Appendix, Figs. S10–S17) were pruned to match the taxon sampling of the 82-gene and combined-complete matrices (SI Appendix, Figs. S18–S25). This revealed that the use of MixtPart resulted in an increase in mean BP values by 5–11% (Fig. 2 and SI Appendix, Table S4) and most strikingly a mean increase in BP values by 20–49% for the 12 clades we identified (Fig. 3). It should be noted that the addition of our 82-gene matrix alone was insufficient to resolve the deeper nodes of Malpighiales. Although it was helpful [e.g., mean BP values increased by 13% when comparing between the 82- vs. 13-gene MixtPart analyses (Fig. 2)], only 4 of these 12 clades were supported with >50 BP using OnePart, GenePart, and CodonPart, vs. 10 clades that were resolved with >50 BP using MixtPart (Fig. 3). This indicates that the use of MixtPart results in substantial improvement.
Fig. 2.
Mean ML BPs of the bipartition trees inferred using ML for each of the four matrices and four partitioning strategies. SEs around the means are indicated, and the MixtPart partitioning strategy is highlighted in gray. The bipartition trees inferred from the combined-incomplete and 13-gene matrices were pruned to match the taxon sampling of the 82-gene and combined-complete matrices.
Fig. 3.
ML BPs of the 12 additional clades we identified in Malpighiales (Fig. 1) inferred from three matrices and four partitioning strategies. The MixtPart partitioning strategy is highlighted in gray.
Our results also suggest that for the commonly used partitioning strategies, particularly for OnePart and GenePart, increased taxon sampling improves branch support, regardless of the increase in missing data. For example, despite its much higher percentage of missing data (64% vs. 12%), analyses of the 191-taxon combined-incomplete matrix yielded a better-supported phylogeny than the 58-taxon combined-complete matrix: the mean BP values increased by 3% and 4% for OnePart and GenePart, respectively (Fig. 2). Although this improvement might seem relatively small when comparing mean BP values, it is much more impressive for the 12 clades we identified, which showed an average increase of BP values by 34% and 36% for the OnePart and GenePart analyses, respectively (Fig. 3). These results provide empirical support for conclusions that increased taxon sampling improves phylogenetic accuracy (22–24), even when the amount of missing data increases (25–27). Theoretical studies (e.g., refs. 28 and 29) have shown that it is the number of complete characters rather than the number of empty cells that determines the impact of missing data on phylogenetic accuracy. The improved branch support we observed in trees from the combined-incomplete matrix likely results from our strategic scaffold approach, in which we ensured that critical nodes were deeply sampled for most characters. A similar scaffold approach was advocated by Wiens et al. (30) and more recently applied using large amounts of genomic data to successfully resolve relationships of butterflies and moths (31).
Despite the apparent successes of the scaffold approach, recent studies (32, 33) have shown that partial taxon coverage (whereby sequences from some partitions are missing for some taxa) can result in a vast terrace of phylogenetic trees that have different topologies but the same optimality score. In cases where complete taxon coverage for a partition is achieved the data set is expected to be decisive for all trees (32), and under these circumstances the problem of terraces does not arise (33). This is likely to be rare for large phylogenomic data sets, however, which sacrifice completeness for the additional taxa and characters. This problem was most clearly illustrated in the recent analysis of a 298-taxon grass data set with 34% missing data that produced a terrace including 61 million optimal trees (33). We found that different partitioning strategies induced different patterns of taxon coverage. Notably, the use of GenePart reduced taxon coverage density in all cases, and in the case of the combined-incomplete matrix it resulted in a pattern of taxon coverage that was indecisive and the best-scoring ML tree was on a terrace of 14,025 trees, whereas the use of MixtPart was decisive for all trees (Table 1). Despite this lack of decisiveness, the BUILD tree (i.e., the Adams consensus of the 14,025 trees) includes only four polytomies, all of which are restricted to subfamily relationships (SI Appendix, Fig. S29). Thus, our scaffold approach yielded a matrix that is resilient to reduced coverage density. Together our results suggest that there may be cases, depending on the patterns of taxon coverage, in which GenePart would be a poor choice for partitioning concatenated matrices.
Patterns of Species Diversification in Malpighiales.
Studies of diversification patterns across angiosperms have not previously detected shifts in net species diversification rates (speciation minus extinction) in Malpighiales (34, 35), possibly because a well-resolved, taxon-dense phylogeny was not available for the order. We used our 191-taxon, combined-incomplete matrix to test the hypothesis that net diversification rates have been constant throughout the history of Malpighiales. This matrix was originally constructed to include the deepest phylogenetic splits within each family (3, 4) and is an excellent foundation for exploring the tempo of evolution in the order. We first used the approach implemented in MEDUSA (36) to detect shifts of diversification rate using a time-calibrated Malpighiales phylogeny (SI Appendix, Fig. S30) that accounts for unsampled taxonomic diversity (SI Appendix, Fig. S31). This method sequentially adds break points to a multirate birth-and-death model fitting the given branch lengths and terminal diversities until subsequent break points do not improve the AICc values. Using MEDUSA we found significant decelerations in five clades (Goupiaceae, Lophopyxidaceae, Medusagynaceae, Scyphostegiaceae, and Irvingiaceae + Pandaceae) and acceleration in one clade (Passifloraceae + Turneraceae) (Fig. 1 and SI Appendix, Fig. S31).
Additionally, we used a method that models diversification as a stochastic, time-homogeneous birth-and-death process (34). This method does not use the phylogeny directly but considers stem or crown group ages within clades of interest and the survival of each lineage to the present. The results were similar to those from the phylogeny-based MEDUSA analysis, with the main difference being the detection of an additional four rate decelerations and four accelerations. Assuming a constant birth-and-death model, eight clades (Balanopaceae, Centroplacaceae, Ctenolophonaceae, Euphroniaceae, Goupiaceae, Lophopyxidaceae, Medusagynaceae, and Scyphostegiaceae) experienced decelerations, and five clades (Dichapetalaceae, Erythroxylaceae, Malpighiaceae, Passifloraceae, and Putranjivaceae) experienced accelerations (Fig. 1 and SI Appendix, Fig. S32).
These overlapping results, together with a well-resolved phylogeny, provide an improved foundation for exploring the mechanisms that have led to such substantial diversity within Malpighiales. In some cases (e.g., Malpighiaceae and Passifloraceae), specialized plant–pollinator mutualisms (37–39) may account for all or part of their exceptional diversification rates. These and other hypotheses can now be tested in more detailed studies of phylogeny, morphology, ecology, and biogeography.
Conclusions
Our phylogeny of Malpighiales provides a critical context for future comparative studies of plant species that are economically and ecologically important. Although the increasing ease of genome-scale sampling may render moot the long-standing argument about whether it is better to add taxa or characters (40), the question remains important. Given the amount of biodiversity remaining to be discovered, described, and classified, the goal should be to maximize taxonomic sampling for phylogenetic study, but to do so in the most effective way possible. Our analyses confirm that one efficient and economical way to resolve difficult clades is to construct a scaffold using phylogenetically critical placeholders sampled for many characters augmented by many more taxa sampled for a modest number of characters. Most importantly, our analyses indicate that searching with a Bayesian mixture models leads to an optimal, a posteriori data partitioning strategy, which not only improves the branch support of phylogenetic trees but also minimizes the impact of missing data on phylogenetic decisiveness. Its use is likely to help resolve several remaining poorly resolved, major clades of angiosperms (e.g., Euasterids I and II and Ericales) (12) and to be more broadly useful in studies across the Tree of Life.
Materials and Methods
See SI Appendix, SI Materials and Methods for details on plastome sequencing, sequence alignment, and analyses of phylogenetic decisiveness, divergence time, and species diversification.
Phylogenetic Analyses.
Bayesian and ML analyses were performed on four matrices (Table 1) as described above. The Bayesian analyses were implemented with the parallel version of BayesPhylogenies v2.0 (19) using a reversible-jump implementation of the mixture model as described by Venditti et al. (20). This approach allows the fitting of multiple models of sequence evolution to each character in an alignment without a priori partitioning. Two independent Markov chain Monte Carlo (MCMC) analyses were performed, and the consistency of stationary-phase likelihood values and estimated parameter values was determined using Tracer v1.5. We ran each MCMC analysis for 10 million generations, sampling trees and parameters every 1,000 generations. Bayesian PPs were determined by building a 50% majority-rule consensus tree from two MCMC analyses after discarding the 20% burn-in generations (Fig. 1 and SI Appendix, Figs. S1 and S26–S28).
The ML analyses were conducted using RAxML v7.2.8 (41) with the GTR+Γ model. The best-scoring ML tree was obtained for each matrix using the rapid hill-climbing algorithm (41), and 1,000 bootstrap replicates were estimated using the standard bootstrap option. The BPs were summarized from all 1,000 bootstrap trees, and the bipartition tree was obtained by mapping these BPs to the best-scoring ML tree (SI Appendix, Figs. S2–S17) (42). We used four different partitioning strategies for our data analyses described above in Results and Discussion: OnePart (single data partition), GenePart (partitioned by gene), CodonPart (partitioned by codon), and MixtPart (described below). For the MixtPart approach, the data partitions identified in the Bayesian analyses were extracted from the output using a custom Perl script (SI Appendix, SI Script). This script selected the partition with the highest probability for each character. The matrices were then partitioned accordingly in RAxML.
Supplementary Material
Acknowledgments
We thank D. Barua, J. Beaulieu, M. Clements, R. Cronn, M. Ethier, D. Goldman, M. Guisinger-Bellian, R. Jansen, M. Kent, M. McMahon, A. Meade, M. Moore, M. Sanderson, A. Stamatakis, and members of the C.C.D. and S.M. laboratories for technical assistance. This work was supported by Brazil Conselho Nacional de Desenvolvimento Científico e Tecnológico Grant 563548/10-0 (to A.M.A.), Swiss National Science Foundation Grant 129804 (to P.K.E.), US National Science Foundation (NSF) Assembling the Tree of Life Grants DEB-0622764 and DEB-1120243 (to C.C.D.), and NSF Doctoral Dissertation Enhancement Project Grant OISE-0936076 (to C.C.D. and B.R.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.K.J. is a guest editor invited by the Editorial Board.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. JX661767–JX665032).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205818109/-/DCSupplemental.
References
- 1.Chase MW, et al. Phylogenetics of seed plants: An analysis of nucleotide sequences from the plastid gene rbcL. Ann Mo Bot Gard. 1993;80:528–580. [Google Scholar]
- 2.APG An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc. 2003;141:399–436. [Google Scholar]
- 3.Wurdack KJ, Davis CC. Malpighiales phylogenetics: Gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am J Bot. 2009;96(8):1551–1570. doi: 10.3732/ajb.0800207. [DOI] [PubMed] [Google Scholar]
- 4.Davis CC, Webb CO, Wurdack KJ, Jaramillo CA, Donoghue MJ. Explosive radiation of Malpighiales supports a mid-cretaceous origin of modern tropical rain forests. Am Nat. 2005;165(3):E36–E65. doi: 10.1086/428296. [DOI] [PubMed] [Google Scholar]
- 5.Sato S, et al. Sequence analysis of the genome of an oil-bearing tree, Jatropha curcas L. DNA Res. 2011;18(1):65–76. doi: 10.1093/dnares/dsq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan AP, et al. Draft genome sequence of the oilseed species Ricinus communis. Nat Biotechnol. 2010;28(9):951–956. doi: 10.1038/nbt.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuskan GA, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313(5793):1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 8.Jansen RK, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 2007;104(49):19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci USA. 2007;104(49):19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, et al. Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc Natl Acad Sci USA. 2009;106(10):3853–3858. doi: 10.1073/pnas.0813376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA. 2010;107(10):4623–4628. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soltis DE, et al. Angiosperm phylogeny: 17 genes, 640 taxa. Am J Bot. 2011;98(4):704–730. doi: 10.3732/ajb.1000404. [DOI] [PubMed] [Google Scholar]
- 13.Cronquist A. The Evolution and Classification of Flowering Plants. 2nd Ed. Bronx, NY: New York Botanical Garden; 1988. [Google Scholar]
- 14.Webster GL. Classification of the Euphorbiaceae. Ann Mo Bot Gard. 1994;81:3–32. [Google Scholar]
- 15.Ruhfel BR, et al. Phylogeny of the clusioid clade (Malpighiales): Evidence from the plastid and mitochondrial genomes. Am J Bot. 2011;98(2):306–325. doi: 10.3732/ajb.1000354. [DOI] [PubMed] [Google Scholar]
- 16.Cai ZQ, et al. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: Implications for the phylogenetic relationships of magnoliids. BMC Evol Biol. 2006;6:77. doi: 10.1186/1471-2148-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen DR, et al. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae) Mol Phylogenet Evol. 2007;45(2):547–563. doi: 10.1016/j.ympev.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Moore MJ, et al. Phylogenetic analysis of the plastid inverted repeat for 244 species: Insights into deeper-level angiosperm relationships from a long, slowly evolving sequence region. Int J Plant Sci. 2011;172:541–558. [Google Scholar]
- 19.Pagel M, Meade A. A phylogenetic mixture model for detecting pattern-heterogeneity in gene sequence or character-state data. Syst Biol. 2004;53(4):571–581. doi: 10.1080/10635150490468675. [DOI] [PubMed] [Google Scholar]
- 20.Venditti C, Meade A, Pagel M. Phylogenetic mixture models can reduce node-density artifacts. Syst Biol. 2008;57(2):286–293. doi: 10.1080/10635150802044045. [DOI] [PubMed] [Google Scholar]
- 21.Hurvich CM, Tsai CL. Regression and time series model selection in small samples. Biometrika. 1989;76:297–307. [Google Scholar]
- 22.Pollock DD, Zwickl DJ, McGuire JA, Hillis DM. Increased taxon sampling is advantageous for phylogenetic inference. Syst Biol. 2002;51(4):664–671. doi: 10.1080/10635150290102357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zwickl DJ, Hillis DM. Increased taxon sampling greatly reduces phylogenetic error. Syst Biol. 2002;51(4):588–598. doi: 10.1080/10635150290102339. [DOI] [PubMed] [Google Scholar]
- 24.Hedtke SM, Townsend TM, Hillis DM. Resolution of phylogenetic conflict in large data sets by increased taxon sampling. Syst Biol. 2006;55(3):522–529. doi: 10.1080/10635150600697358. [DOI] [PubMed] [Google Scholar]
- 25.McMahon MM, Sanderson MJ. Phylogenetic supermatrix analysis of GenBank sequences from 2228 papilionoid legumes. Syst Biol. 2006;55(5):818–836. doi: 10.1080/10635150600999150. [DOI] [PubMed] [Google Scholar]
- 26.Heath TA, Hedtke SM, Hillis DM. Taxon sampling and the accuracy of phylogenetic analyses. J Syst Evol. 2008;46:239–257. [Google Scholar]
- 27.Burleigh JG, Hilu KW, Soltis DE. Inferring phylogenies with incomplete data sets: a 5-gene, 567-taxon analysis of angiosperms. BMC Evol Biol. 2009;9:61. doi: 10.1186/1471-2148-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiens JJ. Missing data, incomplete taxa, and phylogenetic accuracy. Syst Biol. 2003;52(4):528–538. doi: 10.1080/10635150390218330. [DOI] [PubMed] [Google Scholar]
- 29.Wiens JJ. Can incomplete taxa rescue phylogenetic analyses from long-branch attraction? Syst Biol. 2005;54(5):731–742. doi: 10.1080/10635150500234583. [DOI] [PubMed] [Google Scholar]
- 30.Wiens JJ, Fetzner JW, Parkinson CL, Reeder TW. Hylid frog phylogeny and sampling strategies for speciose clades. Syst Biol. 2005;54(5):778–807. doi: 10.1080/10635150500234625. [DOI] [PubMed] [Google Scholar]
- 31.Cho S, et al. Can deliberately incomplete gene sample augmentation improve a phylogeny estimate for the advanced moths and butterflies (Hexapoda: Lepidoptera)? Syst Biol. 2011;60(6):782–796. doi: 10.1093/sysbio/syr079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanderson MJ, McMahon MM, Steel M. Phylogenomics with incomplete taxon coverage: The limits to inference. BMC Evol Biol. 2010;10:155. doi: 10.1186/1471-2148-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanderson MJ, McMahon MM, Steel M. Terraces in phylogenetic tree space. Science. 2011;333(6041):448–450. doi: 10.1126/science.1206357. [DOI] [PubMed] [Google Scholar]
- 34.Magallón S, Sanderson MJ. Absolute diversification rates in angiosperm clades. Evolution. 2001;55(9):1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- 35.Smith SA, Beaulieu JM, Stamatakis A, Donoghue MJ. Understanding angiosperm diversification using small and large phylogenetic trees. Am J Bot. 2011;98(3):404–414. doi: 10.3732/ajb.1000481. [DOI] [PubMed] [Google Scholar]
- 36.Alfaro ME, et al. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc Natl Acad Sci USA. 2009;106(32):13410–13414. doi: 10.1073/pnas.0811087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson WR. Floral conservatism in neotropical Malpighiaceae. Biotropica. 1979;11:219–223. [Google Scholar]
- 38.Neff JL. The passionflower bee: Anthemurgus passiflorae. J Newsl Passiflora Soc Int. 2003;13:7–9. [Google Scholar]
- 39.Zhang W, Kramer EM, Davis CC. Floral symmetry genes and the origin and maintenance of zygomorphy in a plant-pollinator mutualism. Proc Natl Acad Sci USA. 2010;107(14):6388–6393. doi: 10.1073/pnas.0910155107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graybeal A. Is it better to add taxa or characters to a difficult phylogenetic problem? Syst Biol. 1998;47(1):9–17. doi: 10.1080/106351598260996. [DOI] [PubMed] [Google Scholar]
- 41.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 42.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.