Abstract
Agaricus bisporus is the model fungus for the adaptation, persistence, and growth in the humic-rich leaf-litter environment. Aside from its ecological role, A. bisporus has been an important component of the human diet for over 200 y and worldwide cultivation of the “button mushroom” forms a multibillion dollar industry. We present two A. bisporus genomes, their gene repertoires and transcript profiles on compost and during mushroom formation. The genomes encode a full repertoire of polysaccharide-degrading enzymes similar to that of wood-decayers. Comparative transcriptomics of mycelium grown on defined medium, casing-soil, and compost revealed genes encoding enzymes involved in xylan, cellulose, pectin, and protein degradation are more highly expressed in compost. The striking expansion of heme-thiolate peroxidases and β-etherases is distinctive from Agaricomycotina wood-decayers and suggests a broad attack on decaying lignin and related metabolites found in humic acid-rich environment. Similarly, up-regulation of these genes together with a lignolytic manganese peroxidase, multiple copper radical oxidases, and cytochrome P450s is consistent with challenges posed by complex humic-rich substrates. The gene repertoire and expression of hydrolytic enzymes in A. bisporus is substantially different from the taxonomically related ectomycorrhizal symbiont Laccaria bicolor. A common promoter motif was also identified in genes very highly expressed in humic-rich substrates. These observations reveal genetic and enzymatic mechanisms governing adaptation to the humic-rich ecological niche formed during plant degradation, further defining the critical role such fungi contribute to soil structure and carbon sequestration in terrestrial ecosystems. Genome sequence will expedite mushroom breeding for improved agronomic characteristics.
Keywords: carbohydrate-active enzymes, humic substances, litter decay, wood decay fungi
Lignocellulose is the most abundant organic compound in the terrestrial environment, consisting of three main components: cellulose, hemicellulose, and lignin (1). These polymers are decayed primarily by wood and litter decomposers from the Agaricomycotina (mushroom-forming fungi). Comparative analyses of the “white-rot” fungi Phanerochaete chrysosporium and Schizophyllum commune (2, 3), the “brown-rots” Postia placenta and Serpula lacrymans (4, 5), and the coprophilous Coprinopsis cinerea (6) has provided considerable insight into the evolution of the wood-decomposition machinery in fungi. Much less is known about fungal decomposition of partially degraded plant material, particularly leaf litter, and about adaptation to humic-rich environments. Improving knowledge of processes governing this adaptation is critical to improving carbon management and predictive modeling of terrestrial carbon cycling.
The biomass of nonwoody litter in temperate woodlands can be four- to five times greater than that of woody litter (7). Concentrations of lignin and (hemi)cellulose in leaf litters of hardwood species range from 33–43% and 23–29%, respectively (8). After a succession of microbial colonisations, the litter is substantially modified by the removal of readily available carbon, nitrogen, and minerals, and the formation of humic substances. Humic substances originate from the decay of modified lignin and other recalcitrant aromatic compounds and microbial activity. These substances are chemically heterogeneous, complex, and difficult to define, consisting of relatively small molecules forming supramolecular associations by hydrophobic and hydrogen bonds, and sequester proteins (9, 10). Humic substances comprise up to 70–80% of organic compounds in mineral soils and their properties strongly influence the physical properties and structure of soil (9).
The basidiomycete Agaricus bisporus (Lange) Imbach is the favored model for adaptation, persistence, and growth in this humic-rich environment, where nutrition is not readily available to primary degrading fungi (11, 12). The ability to use humic proteins gives the fungus an advantage over other saprobes in this complex substrate. The machinery used by A. bisporus to exploit the diverse mixture of nutrient resources is, however, poorly understood.
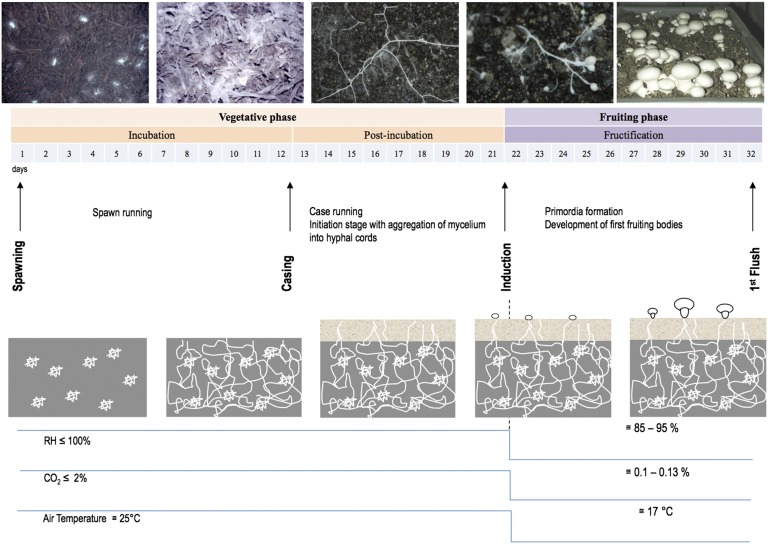
Aside from its ecological roles, A. bisporus is widely cultivated to produce mushrooms (Fig. 1) and is the basis of a multibillion dollar industry. This cultivation involves the large-scale (megaton) biotechnological conversion of agricultural lignocellulosic wastes to high-value food with obvious extrapolations for bioenergy and biorefining. In commercial production for mushroom cultivation, the humic-rich substrate is typically derived from composted wheat straw supplemented with gypsum and nitrogen-rich materials, such as chicken and horse manures.
Fig. 1.
Developmental stages of A. bisporus during the successive steps of its cultivation: spawning, casing, induction, and first fruiting-body flush. The vegetative (incubation, postincubation) and fructification phases are shown. Inoculation of compost is done with wheat kernels overgrown with mycelium (spawning). In 2 wk, mycelium has grown throughout the compost and induction of fruiting bodies is taking place 22 d after a change in aeration and addition of the casing layer. The first flush of fruiting bodies is observed at 32 d, with a switch from mycelium extension to the production of primordia (pinning). Key physicochemical factors (relative humidity, RH%, CO2 concentration and air temperature, T °C) are given at the bottom of the figure.
Our hypothesis has been that metabolic strategies and niche adaptations that might not be seen in the white-rot and brown-rot wood-decomposing fungi, nor in coprophilous fungi, such as C. cinerea, may have evolved in humicolous species, such as A. bisporus. Among the “detritophiles,” they may have a distinctly different deployment of substrate conversion enzymes or regulatory regimens in adaptation to their ecological niche, the partially degraded and humified plant litter.
Here, we report a draft 30-Mb genome and transcriptome sequence of A. bisporus H97, a European isolate obtained from a historically cultivated stock of var. bisporus, considered to represent a member of the “adapted” European population of the bisporic var. bisporus now associated with agricultural environments. We also sequenced the genome of the strain JB137-S8, belonging to the tetrasporic var. burnettii Kerrigan & Callac that is known only from the Sonoran desert of California, where it is associated with leaf litter in native stands of woody species. To identify A. bisporus-specific traits, we compared the H97 and JB137-S8 genomes with those of diverse fungi, including 12 newly sequenced species of white- and brown-rot Agaricomycotina (13). We focused annotation on gene families likely to be involved in litter decomposition and mushroom formation, and transcript profiling to reveal adaptation processes for growth on humic-rich substrates.
Results and Discussion
Genome Assembly and Gene Content.
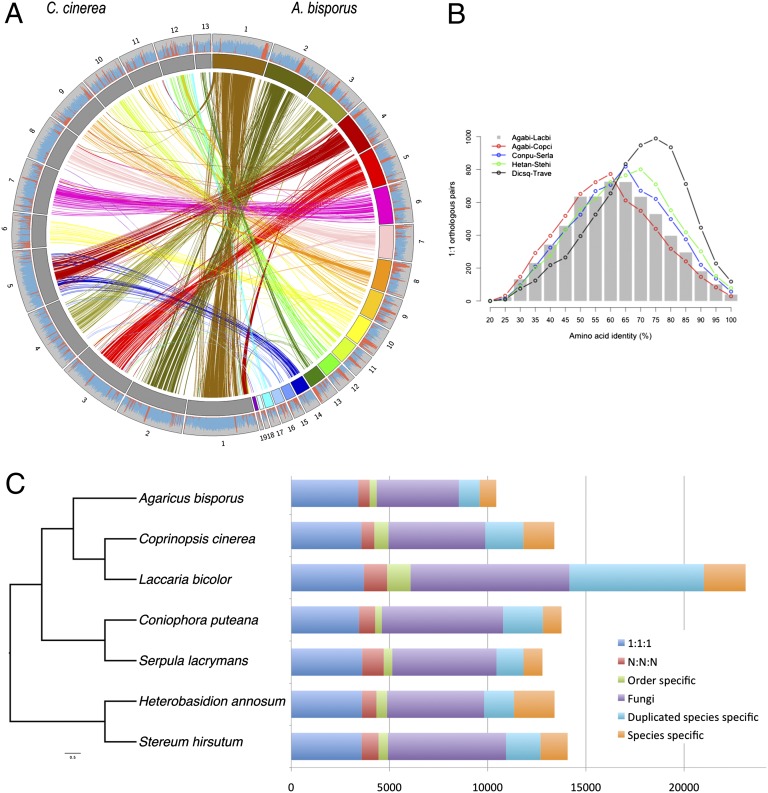
We sequenced and compared the genomes of the homokaryotic (haploid) strains H97 and JB137-S8. Sanger sequencing of the genomic DNA of strain H97 with 8.29× coverage resulted in a 30,387,844 base-pair genome assembly (SI Appendix, Table S1). Ninety-two genetic marker sequences selected along the 13 linkage groups (chromosomes) of A. bisporus (14, 15) were mapped on the H97 genome assembly to validate order and orientation of the 19 largest scaffolds (> 50 kbp) (SI Appendix, Fig. S1). The ratio of physical lengths to genetic distances averaged 33 kbp/cM. Syntenic regions between A. bisporus scaffolds and chromosomes of the taxonomically related agaric C. cinerea were apparent after aligning A. bisporus scaffolds with the 36-Mb chromosome assembly from C. cinerea (6) (Fig. 2A). The 19 longest scaffolds covered all 13 C. cinerea chromosomes, with the largest A. bisporus scaffolds (1 to 6) aligning with the entire length of C. cinerea chromosomes. The largest syntenic blocks occur in regions with low meiotic recombination rates, no transposable elements (TEs), and tight gene spacing, where orthologous single-copy genes are overrepresented.
Fig. 2.
Agaricomycete gene orthology and evolution. (A) Macrosynteny between A. bisporus var bisporus H97 scaffolds and C. cinerea Okayama7 chromosomes. A. bisporus scaffolds are depicted by the colored blocks and C. cinerea chromosomes are represented by gray blocks. Only regions larger than 5,000 bp are connected with links of colors matching those used for coloring A. bisporus scaffolds. The distributions of protein-coding regions and repetitive elements are shown in the outer circle, with protein-coding gene density in blue and repetitive sequences in red, with a window size of 0.1 Mb. Comparison between the two genomes sequences was performed with VISTA (http://genome.lbl.gov/vista). (B) The distribution of pair-wise amino acid identity. Histogram shows the distribution of sequence identity of 1:1 orthologs between A. bisporus and L. bicolor (diverged ∼85 million y ago) (12). To highlight the similar level of molecular divergence, 1:1 orthologs between A. bisporus and C. cinerea, 1:1 orthologs of two Boletales (S. lacrymans/Coniophora puteana), two Russulales (Heterobasidion annosum/Stereum hirsutum), and two Polyporales (Trametes versicolor/Dichomitus squalens) were plotted in red, blue, green, and black, respectively. (C) Orthology assignment of seven Agaricomycete genomes. Bars are subdivided to represent different types of orthology relationships. 1:1:1 indicates universal single-copy genes; N:N:N indicates other universal genes, but absence in a single genome within the different orders is tolerated. “Order specific” indicates Polyporales-, Russulales-, Boletales-, or Agaricales-specific genes; “Fungi” indicates Fungi-specific orthologs; “Duplicated species-specific” indicates species-specific duplicated genes; “Species specific” indicates species-specific genes (orphans).
The second genome, of A. bisporus var. burnettii strain JB137-S8, was sequenced using 454 pyrosequencing and Illumina HiSeq. The final assembly contained 52 scaffolds > 50 kbp (SI Appendix, Fig. S2 and Table S1).
Thousands of individual repeated TEs (class I and II transposons) belonging to 216 diverse families cover 11.2% of the genome (Fig. 2A and SI Appendix, Fig. S3). Estimated insertion times suggest a series of retrotransposition bursts of Copia-like and Gypsy-like long-terminal repeats at <1 million years ago (SI Appendix, Fig. S3). TEs are not uniformly spread across the genome, but are mainly clustered in telomeric and centromeric regions (Fig. 2A). Several TEs have been observed to be actively moving from site to site within the genome (16), and this provides one possible explanation for the development of anomalies, such as sectors or stroma from within otherwise healthy, stable cultures (17).
We estimated 10,438 and 11,289 protein-coding genes in the H97 and JB137-S8 genomes, respectively, by combining both homology-based and ab initio methods (SI Appendix, Tables S2 and S3), along with 1,140,000 expressed sequence tags (SI Appendix).
Agaricales Orthology and Evolution.
Within the Agaricomycetes, the protein-coding genes of Agaricales have similar levels of divergence as in the Boletales, Russulales, and Polyporales (Fig. 2B). Based on the sequence divergence between conserved protein sequences from A. bisporus and other sequenced Agaricales (e.g., L. bicolor, S. commune, C. cinerea), the split between the ancestors of these species has been estimated to have occurred at least 100 million y ago (SI Appendix, Fig. S4) (13). Nearly 83% of the predicted A. bisporus genes had homology with those in the public and JGI Mycocosm (18) databases, mostly to Agaricomycetes (Fig. 2C and SI Appendix, Table S4). Clustering of A. bisporus H97 proteins with those of other sequenced basidiomycete fungi revealed 5,058 clusters containing at least 10 protein members and two fungal taxa (SI Appendix, Fig. S5). The number of gene families exhibiting expansion was similar to those of related Agaricomycetes, such as C. cinerea, Pleurotus ostreatus, or S. commune, but was lower than of L. bicolor (SI Appendix, Fig. S5).
The A. bisporus proteome specializations are illustrated by the over- and underrepresentation of protein family (PFAM) domains compared with other fungi (SI Appendix, Tables S5–S7). Several families of well-known detoxification enzymes were found among the protein families in expansion [e.g., heme-thiolate peroxidase (HTP), methylmalonate semialdehyde dehydrogenase, β-etherase (glutathione-S-transferases cleaving β-aryl ether linkages), and pyranose dehydrogenase], suggesting a higher ability to metabolize derivatives of lignin and other polymers abundant in humicolous habitats.
Genes Involved in Lignocellulose Decomposition.
A. bisporus is a very poor competitor on fresh nondegraded plant wastes but competes well on partially decomposed plant litter on forest floors and grassland soils rich in humic substrates. A. bisporus is adapted to growing in this ecological niche, where it and other species of Agaricaceae can occur abundantly and even predominate based upon observed fructifications. To identify the genomic traits enabling A. bisporus to adapt to its biotope and to efficiently complete its life cycle, we have identified the repertoire and expression of genes known to be involved in organic matter degradation [carbohydrate-acting enzymes (CAZymes), lignin-related oxidoreductases, secreted proteases] and compared this arsenal to that of the white and brown wood-rotters, the coprophilous C. cinerea and the mycorrhizal symbiont L. bicolor (2–6, 13, 19). We also used custom microarrays to compare gene expression at four developmental stages, defined by mycelial cultures on agar-medium, mycelium colonizing the casing-soil layer, or the compost (a proxy for humic-rich composted material) and mature fruiting bodies (Fig. 1). Among the most highly up-regulated transcripts found in mycelium grown on compost were CAZymes, cutinases, oxidoreductases, and secreted proteases (SI Appendix, Fig. S6 and Table S8).
Carbohydrate-acting enzymes.
A. bisporus is a generalist with respect to polysaccharide degradation when grown in laboratory culture on minimal medium and specific carbon sources (SI Appendix, Fig. S7). A. bisporus grows better (relative to glucose) on xylan than do other basidiomycetes, such as S. commune and L. bicolor. Xylan is the second most abundant polysaccharide in plant cell walls (PCW), comprising 7–12% of plant dry mass (20), and in wheat straw (21, 22). Although A. bisporus grows well on cellulose, it is less efficient than S. commune and C. cinerea. This catabolic ability concurs with the presence of a large set of genes encoding CAZymes (23) acting on plant, fungal, and bacterial cell wall polysaccharides [including 188 glycoside hydrolases (GH), 59 polysaccharide lyases (PL), and 10 carbohydrate esterases (CE)]. Clustering of CAZyme profiles in a large set of sequenced fungi, including white- and brown-rots, plant and animal parasites, and an ectomycorrhizal symbiont showed that the CAZyme profiles deviate from species phylogeny. The total CAZyme repertoire for A. bisporus is similar to that of white- and brown-rot basidiomycetes (Fig. S8) rather than the more closely taxonomically related C. cinerea and L. bicolor. This pattern likely reflects the adaptation of A. bisporus to PCW polysaccharide-rich leaf litters (8). The up-regulation of transcripts with high similarity to PCW-degrading GHs (e.g., GH5, GH6, GH7, GH12, GH61, GH105) from basidiomycetes implicated in wood decay confirms that A. bisporus has the generic potential to break down PCW polymers by deploying a complete suite of enzymes degrading crystalline cellulose and xylans (SI Appendix, Fig. S8, S9, and Table S10). Notably, families GH6 and GH7 (SI Appendix, Fig. S9A), which include cellobiohydrolases that are involved in the attack of crystalline cellulose (24, 25), are present in all white-rot lineages and A. bisporus, but they are absent in brown-rot lineages (except Boletales) and L. bicolor (Fig. S8).
Profiling of CAZyme transcripts from mycelium growing in compost demonstrated that 115 (51%) of GHs, PLs, and CEs present in A. bisporus were up-regulated (from 10- to 1,450-fold) in compost, contrasting sharply with only 17% up-regulated transcripts in both the differentiating casing-soil mycelium and the fruiting body (SI Appendix, Fig. S6A and Table S9). Four genes (two CE5 acetyl xylan esterases and two GH12 cellulases) showed the highest up-regulation compared with agar-grown mycelium (SI Appendix, Table S9). A rhamnogalacturonyl hydrolase (GH105) was induced 836-fold. Notably, growth on compost was accompanied by the up-regulation of all 16 genes encoding cellulose-binding motif 1-containing proteins. Although significantly up-regulated, the pectin digestion machinery was comparatively less prominent than that for cellulose and xylan (SI Appendix, Fig. S6A and Table S9). The transcript profile of the mycelium growing on compost is therefore compatible with a prevailing substrate preference for xylan and cellulose, a medium activity on pectin and a slight activity on mannan. This digestion pattern is well-matched to the known composition of grasses and straw (20, 21) and sequential changes in carbohydrates during composting and mushroom growth (22).
Lignin-converting oxidoreductase genes.
To gain access to cellulose, wood-decaying white-rots use fungal class II lignolytic peroxidases (PODs) to degrade lignin (13). We searched sequenced Agaricomycotina genomes for 27 gene families encoding oxidoreductases and CAZymes that have been implicated in wood decay (SI Appendix, Fig. S10). A. bisporus has a distinctive pattern that is not seen in white- and brown-rotters, C. cinerea and L. bicolor. Comparative analysis of the distribution of genes encoding lignolytic PODs [lignin peroxidase, manganese peroxidase (MnP), and versatile peroxidase] revealed that A. bisporus retains a limited POD machinery to degrade lignin (SI Appendix, Table S11). Of the genes encoding lignolytic PODs, which have been shown to be important for lignin degradation in wood decayers (13, 26, 27), only two MnP genes were found in the A. bisporus genome. In compost, transcript levels of one of these MnPs (MNP1, JGI ID#221245) were significantly up-regulated relative to agar medium (SI Appendix, Fig. S6B). Thus, the repertoire of A. bisporus lignolytic PODs differs from P. chrysosporium and other sequenced white-rot fungi, which feature 6–26 POD genes (SI Appendix, Fig. S11) (13). The number and types of lignolytic PODs is similar to brown-rot lineages and C. cinerea having a single POD (13) (SI Appendix, Table S11). In contrast, the A. bisporus genome contains the largest set (24 members) of HTP genes, including aromatic peroxygenases (APOs) and classic chloroperoxidases (CPOs), a significant expansion relative to wood-decay fungi (Fig. 3 and SI Appendix, Table S11) (13). Typical APOs and CPOs are secreted, versatile enzymes with multiple catalytic activities with organic hydrocarbons and lignin-like aromatic compounds, including peroxidative oxidation, epoxidation, hydroxylation, and oxygen transfer reactions (27). Moreover, 16 of these HTP genes were significantly up-regulated in compost relative to defined medium, including an APO-induced 1,492-fold (SI Appendix, Fig. S6B and Table S12). Several transcripts coding for β-etherases, one of the A. bisporus expanding family (SI Appendix, Table S7), were also significantly up-regulated in compost relative to agar medium (SI Appendix, Fig. S6B). Possibly, the number and expression patterns of A. bisporus HTPs and β-etherases are related to the heterogeneous nature of humic-rich lignocellulosic materials, such as compost (22, 25). To estimate patterns of duplication and loss of genes encoding HTPs in the organismal phylogeny, we performed gene-tree/species-tree reconciliation analyses using Notung (28). This analysis suggests that the ancestor of the Agaricomycotina possessed six HTP gene copies, and that the number of HTP paralogs has been more or less steady throughout the evolution of most Agaricomycetes (Fig. 3 and SI Appendix, Fig. S14). However, an abrupt expansion of HTPs is reconstructed in the lineage leading from the common ancestor of Agaricus, Coprinopsis, and Laccaria, which is reconstructed as having seven HTP gene copies, and Agaricus, which has 24 HTP gene copies (Fig. 3 and SI Appendix, Fig. S14). Expansion of HTPs could have been an adaption for decomposition of nonwoody plant matter and humic substances in soil, which is a common substrate for species of Agaricaceae.
Fig. 3.
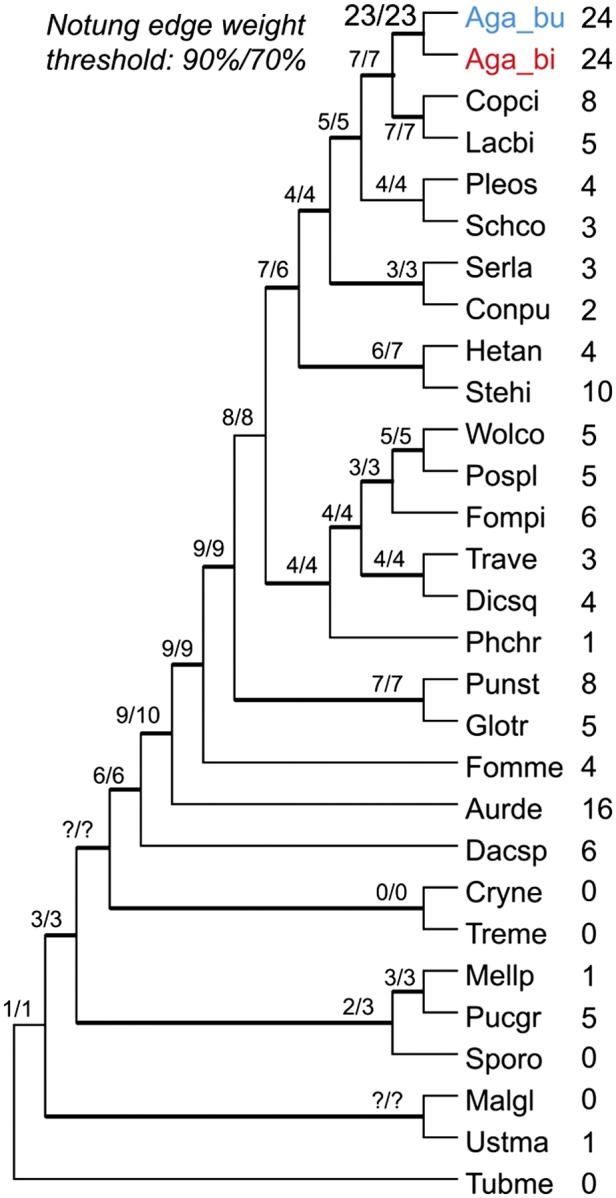
The expansion of HTPs in A. bisporus. HTP copy numbers at internal nodes in the Agaricomycetes as reconstructed by gene-tree species-tree reconciliations in Notung (28) under two edge-weight threshold values. Numbers after species names denote the extant copy numbers of HTPs found in the genomes of these species.
Substrate complexity may also explain the presence of three compost-induced glyoxal oxidase-encoding genes. These copper radical oxidases are thought to play a role in production of extracellular H2O2 from simple aldehydes, such as glyoxal and methylglyoxal, and they are typically associated with class II PODs (SI Appendix, Tables S11 and S12). Catalytically distinct from the copper radical oxidases, at least 12 laccases sensu stricto were also confidently predicted (SI Appendix, Table S11). Three A. bisporus laccase-encoding genes were significantly up-regulated (>10-fold, P < 0.05) in compost relative to agar medium (SI Appendix, Fig. S6B and Table S12). Other potential H2O2-generating extracellular enzymes include various glucose-methanol-choline oxidoreductases that include a likely aryl alcohol oxidase (JGI ID#185801) and a methanol oxidase (JGI ID#195553). The gene coding for the latter enzyme is highly up-regulated (SI Appendix, Fig. S6B and Table S12).
Intracellular metabolism of lignin metabolites and related compounds is poorly understood but cytochrome P450s (CYPs) are generally thought to play an important role. The A. bisporus genome contains a relatively low number of CYPs (109 genes) (SI Appendix, Table S11), but many were highly expressed and regulated. Specifically, six CYP64-encoding genes were up-regulated >10-fold in compost relative to defined medium (SI Appendix, Table S12).
Induction of protease genes on compost.
Litter decomposers share the ability to use proteins as a sole source of carbon and nitrogen (26). The “casing soil” layer used in cultivation is extremely nutrient-poor, and induces A. bisporus hyphae to aggregate and form a corded morphology where nutrient transport and proteinase activity is important. In line with this view, the two sequenced A. bisporus genomes reflect a remarkable metabolic capability for protein degradation and remobilization through amino acid and peptide transporters. Using the MEROPS protease nomenclature, which is based on intrinsic evolutionary and structural relationships (29), we identified 111 genes coding for proteases that possessed a secretion signal (SI Appendix, Table S13). We did not detect in A. bisporus any protease unique to this species. However, several transcripts coding for serine proteases (S8, S9, S10, and S12 families) were among the most highly up-regulated transcripts (>50-fold in comparison with agar-grown mycelium) in compost-grown mycelium (SI Appendix, Fig. S6C and Table S14). From the microarray data, a gene cluster coding for three serine proteases (located on chromosome IX) showed high expression on compost compared with other conditions. The most abundantly expressed of these was SPR1 (JGI ID# 194648), showing a 100-fold induction in mycelium colonizing compost, which is consistent with a major role in nutrient acquisition (11). A. bisporus serine proteinase has been shown to be synthesized specifically in response to humic-protein complex; transgenic analysis of the SPR1 promoter elements confirmed that the promoter is able to regulate mycelial serine proteinase production in response to specific nitrogen sources (11, 30). The abundance and substrate-induction of active proteinases may enable niche specialization in A. bisporus.
Metabolites released during litter (and compost) decay can efficiently be taken up by A. bisporus mycelium colonizing this substrate, as several genes coding for monosaccharide, cellodextrins, nicotinic acid, and amino acid transporters also were strikingly up-regulated on compost (cluster IV in SI Appendix, Fig. S12).
To identify the regulatory regions involved in adaptation to humic rich environments, transcriptomic data were compared with promoter sequences. Expression ratios were calculated and ranked from the microarray data comparing A. bisporus growing in humic (compost) vs. nonhumic (agar-medium) environments. A conserved sequence motif (TC[CA][TG]G[AT][GTA]A[AC]AATCTC) in the promoters of 23 of the top 33 compost-induced genes occurs at a much higher rate than in orthologs of C. cinerea and L. bicolor (SI Appendix, Fig. S12 and Table S15A), and exists in compost-induced genes of both A. bisporus genomes at a frequency significantly greater than random chance (SI Appendix, Table S15B). It is interesting to speculate (and to ultimately determine) whether this motif, which occurs more frequently in promoters of genes very highly expressed in humic-rich substrates, forms a common regulatory mechanism governing adaptation to the ecological niche.
Mushroom Development.
The fruiting bodies of A. bisporus are the most commonly sold mushrooms in Europe and North America. However, some aspects of the sexual life cycle are poorly understood, although they are critical factors for breeding and other industrial applications. Fruiting body formation is a highly complex developmental process that has been studied in the model basidiomycetes S. commune and C. cinerea (31–33). After the substrate has been colonized and the application of specific environmental signals (reduced temperature and levels of CO2 and 8-carbon volatiles), hyphae differentiate into the fruiting body where meiosis occurs (Fig. 1). The mating-type loci are the master regulators of fruiting-body development in fungi (31, 33). The sexually unifactorial A. bisporus contains only a single mating-type locus (34). The locus coding for homeodomain proteins typical for the A mating type in bifactorial species, such as C. cinerea and S. commune, and for the single mating-type locus in other unifactorial Agaricomycetes (35) was found on chromosome I (scaffold 1), supporting earlier studies (36). On the other hand, the genes encoding conserved pheromone and pheromone receptor genes are dispersed across the genome and the unifactorial A. bisporus has dispensed with the B mating-type specificity deployed by bifactorial species.
To identify genes associated with fruiting-body formation, we compared transcript profiles of undifferentiated mycelium grown on agar medium, compost, or casing-soil, and of fruiting bodies (SI Appendix, Fig. S6, and Tables S8, S9, S12, S14, and S16). Of the 7,538 transcripts detected in fruiting bodies, 613 (8%) were significantly up-regulated (P < 0.05; false-discovery rate < 0.05) in fruiting bodies in comparison with undifferentiated mycelium grown on agar medium or compost. Among the 50 most highly induced genes in fruiting bodies, 40% are coding for orphan, lineage-specific genes. Genes coding for hydrophobins, lectins, tyrosinases, and transcriptional factors were among the most highly induced genes. Hydrophobins and tyrosinases are known to accumulate in mushroom caps during A. bisporus fruit-body development (37, 38). We compared genes induced during the fruiting-body formation in A. bisporus, L. bicolor, and S. commune to identify common developmental gene networks (SI Appendix, Table S16A). Only 35 and 22 homologous genes were significantly up-regulated (P < 0.05) in both A. bisporus/L. bicolor fruiting bodies or A. bisporus/S. commune fruiting bodies, respectively. Only 13 genes were significantly up-regulated (P < 0.05) in the fruiting bodies of these three species (e.g., aromatic-ring hydroxylase, GH16, FAD-linked oxidase, fatty acid desaturase), suggesting that genes induced during mushroom development are mainly clade-specific.
Recently, a set of transcription factors has been identified in S. commune that act downstream of the mating-type loci (3, 39). These transcription factor genes have orthologs in A. bisporus (SI Appendix, Table S16B). Based on transcript profiling, several of these transcription-factor genes may be involved in the regulation of mushroom formation in A. bisporus. The DNA binding protein pcc1 gene, which leads to A-regulated sexual morphogenesis in C. cinerea (40), was highly expressed in A. bisporus mycelium grown on agar medium, compost, or casing, and in fruiting bodies. The expression profiles of the orthologs of c2h2, fst3, fst4, and hom1 were similar in S. commune and A. bisporus (i.e., up-regulation of gene expression in mushrooms compared with mycelium), suggesting that these agarics share similar master developmental switches for fruiting-body formation, making these genes primary targets for a functional analysis. Studying the regulatory mechanisms underlying fructification in A. bisporus would allow the control of mushroom-pin formation, considered by mushroom growers as the most important step in managing the mushroom crop.
Our genomic and transcriptomic data suggest that A. bisporus has the decay machinery to decompose lignocellulosic material; yet to our knowledge, it has not been shown to decompose wood in nature. Although there are similarities in genome composition, A. bisporus fits neither brown-rot nor white-rot classifications. We hypothesized that a humicolous fungus adapted to growth in a humic-rich environment is atypical of classic wood-degrading fungi, and transcriptome expression data support this view. The wide repertoire of HTP, β-etherases, multicopper oxidase, and CYP450 oxidoreductases and their striking up-regulation in mycelium-colonizing compost suggest a broad mode of attack on decaying lignin and related metabolites, and an adaptation to challenges posed by complex composts. The large gene repertoire of compost-induced CAZymes and oxidoreductases, together with high protein degradation and nitrogen-scavenging abilities, are key features of A. bisporus adaptation to humic-rich ecosystems. This study reveals genetic and enzymatic mechanisms governing adaptation of A. bisporus to a humic-rich ecological niche created by primary degradation of plant material, demonstrating the critical role such fungi contribute to soil structure and carbon sequestration.
Materials and Methods
Genome Sequencing, Assembly, and Annotation.
The homokaryotic A. bisporus var bisporus H97 and A. bisporus var. burnettii JB137-S8 strains were sequenced by whole-genome sequencing and were assembled into predicted 30.2-Mb and 32.6-Mb genomes, respectively (SI Appendix). The protein-coding genes were predicted with a combination of automated gene callers, ESTs produced from each A. bisporus strain, and filtering dubious genes with similarity to transposable elements (SI Appendix). In total, the gene sets included 10,438 and 11,289 predicted genes for H97 and JB137-S8, respectively; these were the basis for multigene family analyses. The A. bisporus H97 genome sequence can be accessed at http://jgi.doe.gov/Abisporus_var_bisporus and the A. bisporus var. burnettii JB137-S8 genome sequence can be accessed at http://genome.jgi-psf.org/Agabi_varbur_1.
Microarray Analysis of Gene Expression.
Gene expression was assessed in mycelium grown on defined medium, casing substrate and compost, and fruiting bodies, using specific custom 60-mer Agilent microarrays (as described in SI Appendix).
Supplementary Material
Acknowledgments
We thank W. W. Lilly, J. L. Lavin, J. J. Mes, J. A. Oguiza, V. Garcia-Tagua, S. Bastian, L. Elbourne, W. Gao, E. Battaglia, B. Gruben, M. Nadal, J. van den Brink, R. Finkers, and A. D. Wiebenga for the annotation of genomic features not presented in this article and their technical assistance. The work conducted by the US Department of Energy Joint Genome Institute is supported by the Office of Science of the US Department of Energy under Contract DE-AC02-05CH11231. This work was also supported by grants from the National Institute of Agricultural Research and the Région Lorraine Council (to F.M.); and Horticultural Development Company, United Kingdom grants for Agaricus bisporus custom array development and transcriptomics.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AEOK00000000 and AEOL00000000).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206847109/-/DCSupplemental.
References
- 1.Sánchez C. Lignocellulosic residues: Biodegradation and bioconversion by fungi. Biotechnol Adv. 2009;27(2):185–194. doi: 10.1016/j.biotechadv.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Martinez D, et al. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol. 2004;22(6):695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- 3.Ohm RA, et al. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol. 2010;28(9):957–963. doi: 10.1038/nbt.1643. [DOI] [PubMed] [Google Scholar]
- 4.Martinez D, et al. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci USA. 2009;106(6):1954–1959. doi: 10.1073/pnas.0809575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eastwood DC, et al. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science. 2011;333(6043):762–765. doi: 10.1126/science.1205411. [DOI] [PubMed] [Google Scholar]
- 6.Stajich JE, et al. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus) Proc Natl Acad Sci USA. 2010;107(26):11889–11894. doi: 10.1073/pnas.1003391107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boddy L, Watkinson SC. Wood decomposition, higher fungi, and their role in nutrient redistribution. Can J Bot. 1995;73:S1377–S1383. [Google Scholar]
- 8.Sariyildiz T, Anderson JM. Variation in the chemical composition of green leaves and leaf litters from three deciduous tree species growing on different soil types. For Ecol Manage. 2005;210:303–319. [Google Scholar]
- 9.Piccolo A. The supramolecular structure of humic substances. A novel understanding of humus chemistry and implications in soil science. Adv Agron. 2002;75:57–134. [Google Scholar]
- 10.Tomaszewski JE, Schwarzenbach RP, Sander M. Protein encapsulation by humic substances. Environ Sci Technol. 2011;45(14):6003–6010. doi: 10.1021/es200663h. [DOI] [PubMed] [Google Scholar]
- 11.Burton KS, Smith JF, Wood DA, Thurston CF. Extracellular proteinases from the mycelium of the cultivated mushroom (Agaricus bisporus) Mycol Res. 1997;101:1341–1347. [Google Scholar]
- 12.Fermor TR, Wood DA. Degradation of bacteria by Agaricus bisporus and other fungi. J Gen Microbiol. 1981;126:377–387. [Google Scholar]
- 13.Floudas D, et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336(6089):1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 14.Sonnenberg AS, et al. Isolation of expressed sequence tags of Agaricus bisporus and their assignment to chromosomes. Appl Environ Microbiol. 1996;62(12):4542–4547. doi: 10.1128/aem.62.12.4542-4547.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foulongne-Oriol M, et al. Comparative linkage mapping in the white button mushroom Agaricus bisporus provides foundation for breeding management. Curr Genet. 2011;57(1):39–50. doi: 10.1007/s00294-010-0325-z. [DOI] [PubMed] [Google Scholar]
- 16.Sonnenberg ASM, Baars JJP, Mikosch TSP, Schaap PJ, Van Griensven LJLD. Abr1, a transposon-like element in the genome of the cultivated mushroom Agaricus bisporus (Lange) Imbach. Appl Environ Microbiol. 1999;65(8):3347–3353. doi: 10.1128/aem.65.8.3347-3353.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horgen PA, Carvalho D, Sonnenberg A, Li A, Van Griensven LJLD. Chromosomal abnormalities associated with strain degeneration in the cultivated mushroom, Agaricus bisporus. Fungal Genet Biol. 1996;20:229–241. [Google Scholar]
- 18.Grigoriev IV, et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012;40(Database issue):D26–D32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin F, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452(7183):88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- 20.Fry SC. The Growing Plant Cell Wall: Chemical and Metabolic Analysis. Caldwell, NJ: Blackburn; 1988. p. 352. [Google Scholar]
- 21.Lawther JM, Sun R, Banks WB. Extraction, fractionation, and characterization of structural polysaccharides from wheat straw. J Agric Food Chem. 1995;43:667–675. [Google Scholar]
- 22.Iiyama K, Stone BA, Macauley BJ. Compositional changes in compost during composting and growth of Agaricus bisporus. Appl Environ Microbiol. 1994;60(5):1538–1546. doi: 10.1128/aem.60.5.1538-1546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coughlan MP, Hazlewood GP. β-1,4-D-xylan-degrading enzyme systems: Biochemistry, molecular biology and applications. Biotechnol Appl Biochem. 1993;17(Pt 3):259–289. [PubMed] [Google Scholar]
- 25.Baldrian P, Valásková V. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol Rev. 2008;32(3):501–521. doi: 10.1111/j.1574-6976.2008.00106.x. [DOI] [PubMed] [Google Scholar]
- 26.Baldrian P. Enzymes of saprotrophic Basidiomycetes. In: Boddy L, Frankland JC, van West P, editors. Ecology of Saprotrophic Basidiomycetes. London: Academic Press; 2008. pp. 19–41. [Google Scholar]
- 27.Hofrichter M, Ullrich R, Pecyna MJ, Liers C, Lundell T. New and classic families of secreted fungal heme peroxidases. Appl Microbiol Biotechnol. 2010;87(3):871–897. doi: 10.1007/s00253-010-2633-0. [DOI] [PubMed] [Google Scholar]
- 28.Vernot B, Stolzer M, Goldman A, Durand D. Reconciliation with non-binary species trees. J Comput Biol. 2008;15(8):981–1006. doi: 10.1089/cmb.2008.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rawlings ND, Barrett AJ, Bateman A. MEROPS: The peptidase database. Nucleic Acids Res. 2010;38(Database issue):D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heneghan MN, et al. Characterisation of serine proteinase expression in Agaricus bisporus and Coprinopsis cinerea using GFP and the A. bisporus SPR1 promoter. Appl Environ Microbiol. 2009;75:792–801. doi: 10.1128/AEM.01897-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kües U. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev. 2000;64(2):316–353. doi: 10.1128/mmbr.64.2.316-353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Umar MH, Van Griensven LJLD. Morphological studies on the life span, developmental stages, senescence and death of fruit bodies of Agaricus bisporus. Mycol Res. 1997;101:1409–1422. [Google Scholar]
- 33.Raudaskoski M, Kothe E. Basidiomycete mating type genes and pheromone signaling. Eukaryot Cell. 2010;9(6):847–859. doi: 10.1128/EC.00319-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J, Kerrigan RW, Horgen PA, Anderson JB. Localization of the mating type gene in Agaricus bisporus. Appl Environ Microbiol. 1993;59(9):3044–3049. doi: 10.1128/aem.59.9.3044-3049.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kües U, James TY, Heitman J. Mating type in Basidiomycetes: Unipolar, bipolar, and tetrapolar patterns of sexuality. In: Pöggeler S, Wöstemeyer J, editors. The Mycota XIV: Evolution of Fungi and Fungal-Like Organisms. Berlin: Springer; 2011. pp. 97–160. [Google Scholar]
- 36.Li Y, Challen M, Elliott T, Casselton L. Molecular analysis of breeding behaviour in Agaricus species. Mushr Sci. 2004;16:103–109. [Google Scholar]
- 37.De Groot PWJ, Schaap PJ, Sonnenberg AS, Visser J, Van Griensven LJLD. The Agaricus bisporus hypA gene encodes a hydrophobin and specifically accumulates in peel tissue of mushroom caps during fruit body development. J Mol Biol. 1996;257(5):1008–1018. doi: 10.1006/jmbi.1996.0219. [DOI] [PubMed] [Google Scholar]
- 38.Soler-Rivas C, Möller AC, Arpin N, Olivier JM, Wichers HJ. Induction of a tyrosinase mRNA in Agaricus bisporus upon treatment with a tolaasin preparation from Pseudomonas tolaasii. Physiol Mol Plant Pathol. 2001;58(2):95–99. [Google Scholar]
- 39.Ohm RA, de Jong JF, de Bekker C, Wösten HAB, Lugones LG. Transcription factor genes of Schizophyllum commune involved in regulation of mushroom formation. Mol Microbiol. 2011;81(6):1433–1445. doi: 10.1111/j.1365-2958.2011.07776.x. [DOI] [PubMed] [Google Scholar]
- 40.Murata Y, Fujii M, Zolan ME, Kamada T. Molecular analysis of pcc1, a gene that leads to A-regulated sexual morphogenesis in Coprinus cinereus. Genetics. 1998;149(4):1753–1761. doi: 10.1093/genetics/149.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.