All members of the myosin superfamily of actin-based molecular motors are thought to use the energy from ATP hydrolysis to generate force and motion. Some myosins are capable of forming parallel coiled-coil dimers by virtue of a coiled-coil domain that is C-terminal to the motor catalytic region (head) (1). This form of dimerization is thought to be essential for the hand-over-hand processive walking mechanism of myosins that function as intracellular transporters (2). In this walking mechanism the mechanochemical properties of the motors are such that the likelihood of at least one head of the dimer being attached to the actin filament is high, owing to a step size that closely matches the helical pitch of the actin filament and a strain-gating mechanism in which the enzymatic cycles of the two heads are coordinated (2). Processive walking allows for multiple successive steps and long-distance transport along a single actin filament. In a study presented in PNAS, Lu et al. (3) demonstrate an antiparallel dimerization mechanism in myosin X (MYO10) and suggest that this structural arrangement allows more efficient processive walking in the parallel actin bundles of filopodia. In addition, Lu et al. (3) demonstrate that formation of the antiparallel coiled coil enhances the ability of MYO10 to induce filopodia formation.
A highly debated topic has been how different classes of myosin function within the diverse actin-based structures (e.g., parallel bundles, antiparallel bundles, and branched/cross-linked networks) that exist in cells and whether there are specific domains of myosin required for efficient movement and/or force generation in these actin-based structures. Recently, work by Brawley and Rock (4) demonstrated that when single molecules of MYO5, MYO6, or MYO10 were exposed to Triton-extracted cytoskeletons of a number of cell types, the patterns of “traffic” for each myosin were different, suggesting motor specificity for different actin network architectures. Lu et al. propose that the antiparallel dimerization may be important for MYO10 to walk in a “straddle-like” fashion, with each head associated with an adjacent actin filament within the actin bundles, in addition to walking in a manner more similar to the single-filament mechanism described for class V myosins. Previous single-molecule studies by the Goldman laboratory support a straddle-like walking model: this work demonstrated that MYO10 takes ∼34-nm steps on single actin filaments and both 34-nm and 18-nm steps on fascin-bundled actin (5). The steps size in myosins is thought to be determined by the length of the lever arm domain (6), a region that binds calmodulin or calmodulin-like light chains. Interestingly, MYO10 only has three calmodulin-binding IQ motifs and produces steps of 34 nm, which is nearly the same as the 36-nm steps produced by MYO5 that contains six IQ motifs. Studies have demonstrated that MYO10 contains a single α-helical domain (SAH) that can extend the lever arm (7), although it is possible that the antiparallel coiled coil also plays a role in extending the lever arm. Work from the Rock group (8) also demonstrated that single molecules of purified MYO10 took steps of ∼20 nm and were capable of moving laterally across the actin bundle. Lateral steps have been hypothesized to allow for obstacle avoidance. Lu et al. emphasize that the previous single-molecule work with MYO10 clearly demonstrates that this motor can walk along a single or bundled actin structure. Therefore, they suggest that the antiparallel geometry of the MYO10 structure may allow it to walk more efficiently among the different actin structures it encounters in the cell. Many of the in vitro studies performed with MYO10 have used constructs containing a forced-dimer, GCN4, or MYO5 parallel coiled-coil domain at the C terminus (5, 8, 9). Therefore, single-molecule studies that can examine MYO10 stepping with and without disruption of the antiparallel dimerization region characterized by Lu et al. may further clarify the MYO10 walking mechanism and its importance for movement in filopodia.
One of the cellular roles of MYO10 that has been well characterized is its ability to stimulate the formation of filopodia (10). These structures contain a core of fascin-bundled parallel actin and can protrude from the surface of the cell either a short distance or many microns, depending upon the cell type. The seminal work by Berg and Cheney (11) demonstrated that although full-length GFP-MYO10 and truncated forms of MYO10 containing the putative dimerization region could localize to existing filopodia, the full-length form of MYO10 was required for stimulation of filopodial formation in COS7 cells (cells that lacked high numbers of filopodia), and that these filopodia were also longer than those from control cells. Additionally, ectopic expression of a MYO10 construct consisting of just the dimerization region resulted in a decrease in the number of filopodia in HeLa cells, a cell line with a high density of filopodial structures (12). By engineering versions of MYO10 with a variety of dimerization mechanisms and others with alterations that would prevent antiparallel dimerization, Lu et al. demonstrate that GFP-tagged versions of MYO10 that contain the tail domain and endogenous ability to form antiparallel dimers show the strongest localization to filopodial tips in HeLa and COS7 cells. Additionally, it was only the wild-type GFP construct that was able to increase the number of filopodia per cell in both cell types, which is in agreement with the work from the Cheney laboratory (11, 12). The work by Tokuo et al. (13) demonstrated that MYO10 constructs forced to dimerize by fusion to FK506 binding protein (FKBP, which dimerizes in the presence of a specific small molecule) were also capable of enhancing the formation of actin-based protrusions in COS7 cells via a mechanism that was sensitive to lever-arm length. Although these data may seem contradictory, a possible explanation may be that the mechanisms of dimerization used by Tokuo created a dimer with a molecular geometry and spacing more similar to the native geometry formed by the antiparallel dimer than what was achieved with the GCN4 forced dimer. Additionally, the work by Tokuo et al. (13) reported an increase in the number of “microspikes,” or short filopodia-like structures. Constructs lacking the myosin tail homology 4 (MyTH4) domain have been shown to lack the ability to stimulate the formation of dorsal filopodia (12). Expression of FERM domain-deleted MYO10 still resulted in increased numbers of filopodia; however, those filopodia were shorter than filopodia from cells expressing full-length MYO10 (14). Similarly, the truncated, MYO10 forced-dimer constructs may be able to stimulate
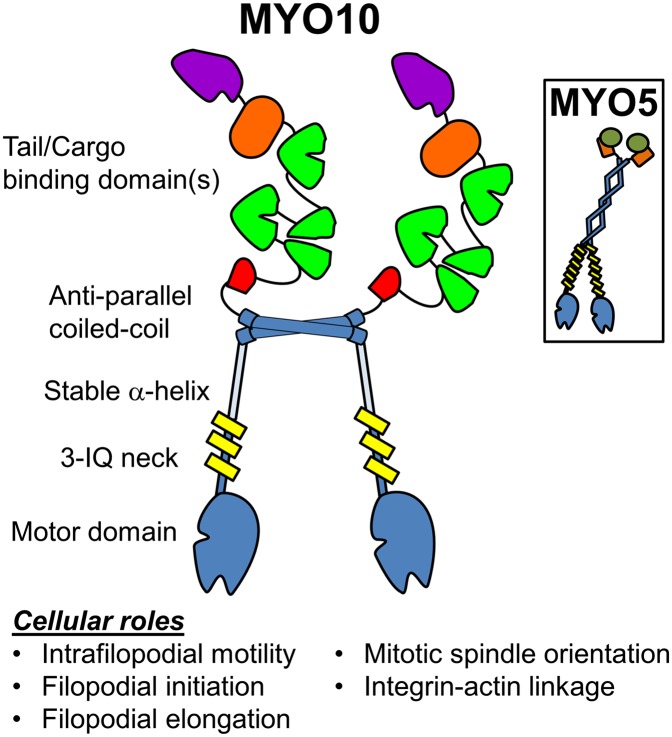
The geometry of MYO10 dimers may relate to its motility and function in parallel actin bundles and other actin-based structures.
the filopodial initiation event by stimulating actin filament convergence at sites of filopodial initiation, but because they lack the tail domain these constructs have an impaired ability to support elongation of nascent filopodia through interactions with either cell surface receptors that may stabilize the filopodial tip as a point of future elongation (15, 16), or with the actin regulatory machinery itself (17, 18).
The geometry of MYO10 and mechanism of proposed motility raise questions relating to how this structural information relates to the function of MYO10 (Fig. 1). (i) Can full length MYO10 form either antiparallel or parallel dimers, as was investigated in scallop myosin II (19)? (ii) How does the geometry of MYO10 relate to its roles in other cellular architecture, such as phagocytic cups (20), osteoclast podosomes (21), junctional assemblies in epithelial cells (22), and mitotic spindles (23, 24)? (iii) Do MYO10 dimers contain a strong bias toward which head dissociates from the track during stepping (as is the case with dimeric kinesins and MYO5), or is there a weaker strain gating as has been demonstrated with dynein (25)? (iv) Does the antiparallel dimerization play a role in cargo binding or phospholipid-dependent regulation (26)?
Fig. 1.
Cartoon representation of the proposed MYO10 dimer formed by the antiparallel coiled-coil domain, which differs from the parallel coiled coil of motors like MYO5 (Inset). The tail domain of MYO10 contains several subdomains, including a PEST region (red), three PH domains (green), a myosin tail homology 4 domain (orange), and a FERM domain (purple). The geometry of MYO10 dimers may relate to its motility and function in parallel actin bundles and other actin-based structures.
Studies into the structure of MYO10 have already broadened our understanding of the variety of functional domains that can be found in myosin molecules (27), and the work by Lu et al. continues to build on that heritage. As our understanding of the diversity of myosin molecules continues to grow (28), detailed study of the nanomechanics of these yet-to-be-characterized molecular motors are bound to yield other novel structure–function relationships, enhancing our understanding of the inner workings of motile mechanisms in cells.
Footnotes
The authors declare no conflict of interest.
See companion article on page 17388.
References
- 1.Peckham M. Coiled coils and SAH domains in cytoskeletal molecular motors. Biochem Soc Trans. 2011;39(5):1142–1148. doi: 10.1042/BST0391142. [DOI] [PubMed] [Google Scholar]
- 2.Hammer JA, 3rd, Sellers JR. Walking to work: Roles for class V myosins as cargo transporters. Nat Rev Mol Cell Biol. 2012;13(1):13–26. doi: 10.1038/nrm3248. [DOI] [PubMed] [Google Scholar]
- 3.Lu Q, Ye F, Wei Z, Wen Z, Zhang M. Antiparallel coiled-coil–mediated dimerization of myosin X. Proc Natl Acad Sci USA. 2012;109:17388–17393. doi: 10.1073/pnas.1208642109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brawley CM, Rock RS. Unconventional myosin traffic in cells reveals a selective actin cytoskeleton. Proc Natl Acad Sci USA. 2009;106(24):9685–9690. doi: 10.1073/pnas.0810451106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y, et al. Single-molecule stepping and structural dynamics of myosin X. Nat Struct Mol Biol. 2010;17(4):485–491. doi: 10.1038/nsmb.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyska MJ, Mooseker MS. Myosin-V motility: These levers were made for walking. Trends Cell Biol. 2003;13(9):447–451. doi: 10.1016/s0962-8924(03)00172-7. [DOI] [PubMed] [Google Scholar]
- 7.Baboolal TG, et al. The SAH domain extends the functional length of the myosin lever. Proc Natl Acad Sci USA. 2009;106(52):22193–22198. doi: 10.1073/pnas.0909851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ricca BL, Rock RS. The stepping pattern of myosin X is adapted for processive motility on bundled actin. Biophys J. 2010;99(6):1818–1826. doi: 10.1016/j.bpj.2010.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy S, et al. A myosin motor that selects bundled actin for motility. Proc Natl Acad Sci USA. 2008;105(28):9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerber ML, Cheney RE. Myosin-X: A MyTH-FERM myosin at the tips of filopodia. J Cell Sci. 2011;124(Pt 22):3733–3741. doi: 10.1242/jcs.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berg JS, Cheney RE. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat Cell Biol. 2002;4(3):246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- 12.Bohil AB, Robertson BW, Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci USA. 2006;103(33):12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tokuo H, Mabuchi K, Ikebe M. The motor activity of myosin-X promotes actin fiber convergence at the cell periphery to initiate filopodia formation. J Cell Biol. 2007;179(2):229–238. doi: 10.1083/jcb.200703178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe TM, Tokuo H, Gonda K, Higuchi H, Ikebe M. Myosin-X induces filopodia by multiple elongation mechanism. J Biol Chem. 2010;285(25):19605–19614. doi: 10.1074/jbc.M109.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, et al. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat Cell Biol. 2004;6(6):523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- 16.Almagro S, et al. The motor protein myosin-X transports VE-cadherin along filopodia to allow the formation of early endothelial cell-cell contacts. Mol Cell Biol. 2010;30(7):1703–1717. doi: 10.1128/MCB.01226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quintero OA, et al. Dynamics of myosin-X (Myo10) and VASP at the filopodial tip. Mol Biol Cell. 2003;14s:1010. [Google Scholar]
- 18.Tokuo H, Ikebe M. Myosin X transports Mena/VASP to the tip of filopodia. Biochem Biophys Res Commun. 2004;319(1):214–220. doi: 10.1016/j.bbrc.2004.04.167. [DOI] [PubMed] [Google Scholar]
- 19.Málnási-Csizmadia A, et al. Dimerization of the head-rod junction of scallop myosin. Biochem Biophys Res Commun. 1998;252:595–601. doi: 10.1006/bbrc.1998.9603. [DOI] [PubMed] [Google Scholar]
- 20.Cox D, et al. Myosin X is a downstream effector of PI(3)K during phagocytosis. Nat Cell Biol. 2002;4(7):469–477. doi: 10.1038/ncb805. [DOI] [PubMed] [Google Scholar]
- 21.McMichael BK, Cheney RE, Lee BS. Myosin X regulates sealing zone patterning in osteoclasts through linkage of podosomes and microtubules. J Biol Chem. 2010;285(13):9506–9515. doi: 10.1074/jbc.M109.017269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu KC, Jacobs DT, Dunn BD, Fanning AS, Cheney RE. Myosin-X functions in polarized epithelial cells. Mol Biol Cell. 2012;23(9):1675–1687. doi: 10.1091/mbc.E11-04-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber KL, Sokac AM, Berg JS, Cheney RE, Bement WM. A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature. 2004;431(7006):325–329. doi: 10.1038/nature02834. [DOI] [PubMed] [Google Scholar]
- 24.Woolner S, O’Brien LL, Wiese C, Bement WM. Myosin-10 and actin filaments are essential for mitotic spindle function. J Cell Biol. 2008;182(1):77–88. doi: 10.1083/jcb.200804062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu W, et al. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat Struct Mol Biol. 2012;19(2):193–200. doi: 10.1038/nsmb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umeki N, et al. Phospholipid-dependent regulation of the motor activity of myosin X. Nat Struct Mol Biol. 2011;18(7):783–788. doi: 10.1038/nsmb.2065. [DOI] [PubMed] [Google Scholar]
- 27.Knight PJ, et al. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J Biol Chem. 2005;280(41):34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 28.Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8(9):R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]