Abstract
The mammalian intestine is home to a dense community of bacteria and its associated bacteriophage (phage). Virtually nothing is known about how phages impact the establishment and maintenance of resident bacterial communities in the intestine. Here, we examine the phages harbored by Enterococcus faecalis, a commensal of the human intestine. We show that E. faecalis strain V583 produces a composite phage (ϕV1/7) derived from two distinct chromosomally encoded prophage elements. One prophage, prophage 1 (ϕV1), encodes the structural genes necessary for phage particle production. Another prophage, prophage 7 (ϕV7), is required for phage infection of susceptible host bacteria. Production of ϕV1/7 is controlled, in part, by nutrient availability, because ϕV1/7 particle numbers are elevated by free amino acids in culture and during growth in the mouse intestine. ϕV1/7 confers an advantage to E. faecalis V583 during competition with other E. faecalis strains in vitro and in vivo. Thus, we propose that E. faecalis V583 uses phage particles to establish and maintain dominance of its intestinal niche in the presence of closely related competing strains. Our findings indicate that bacteriophages can impact the dynamics of bacterial colonization in the mammalian intestinal ecosystem.
Keywords: commensal bacteria, microbiota, enterococci, phage predation
The human gastrointestinal tract is colonized with a highly diverse population of bacteria (1). Many of these bacteria produce bacteriophage (phage), further increasing the complexity of this ecosystem. Recent studies of human intestinal viromes have shown that these viromes are dominated by lysogenic prophages that are integrated into the chromosomes of their bacterial hosts (2). In numerous other ecological systems, phages profoundly influence ecological networks by serving as reservoirs of genetic diversity (3, 4) and acting as predators of susceptible bacterial strains (5). However, little is known about how resident phages may influence the assembly and maintenance of bacterial communities in the mammalian intestine.
Enterococcus faecalis is an abundant member of the human intestinal microflora, and by adulthood, it can constitute as much as 0.5–0.9% of the total bacterial content of the intestinal tract (Table S1) (6). A Gram-positive facultative anaerobe, E. faecalis is a leading cause of antibiotic-resistant nosocomial bacteremia and endocarditis (7). Genomic sequencing has revealed a high degree of variation among E. faecalis genomes (8). Some of this variation can be attributed to an array of integrated prophage elements that encode components required for the production of phage particles.
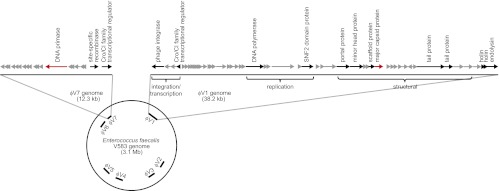
The first E. faecalis genome to be sequenced was strain V583, a clinical blood isolate that is vancomycin-resistant (9). The V583 chromosome harbors seven putative prophages designated prophages 1–7 (Fig. 1). At least two of these prophage elements seem to encode cryptic or satellite phage genomes (10) that, by themselves, do not produce functional phage particles but may encode accessory components that aid in the lytic cycle of other integrated prophages found in the V583 chromosome. Several homologs of V583 prophage sequences have been identified in the genomes of other E. faecalis strains, and the total number of integrated prophages varies among strains (8, 11). Although prophages are common in E. faecalis, their biological roles are poorly understood.
Fig. 1.
Schematic of the ϕV1 and ϕV7 prophage of E. faecalis V583. The locations of the seven putative prophages of E. faecalis V583 are indicated on the circular chromosome of E. faecalis V583. The ϕV1 and ϕV7 elements are magnified to show their gene organization. Arrows indicate predicted ORFs and are drawn to scale. The arrows highlighted in red show the genes EF2948 (ϕV7 DNA primase) and EF0339 (ϕV1 major capsid) that were mutagenized for this study.
Here, we show that E. faecalis V583 produces a composite phage, ϕV1/7, consisting of prophage 1 (ϕV1) and the satellite-like prophage 7 (ϕV7) DNA. We show that ϕV1 encodes structural components required for the assembly of ϕV1/7 particles, whereas ϕV7 encodes a DNA primase required for phage DNA replication and possibly also contains other factors important for host cell infection or lysis. We also show that the production of the composite phage ϕV1/7 is influenced by nutrient availability and provides an advantage to the host bacterium during competition with other E. faecalis strains in culture and the mammalian intestine. Our findings suggest that temperate prophages associated with resident intestinal bacteria influence the assembly of bacterial communities in the mammalian intestine.
Results
E. faecalis V583 Produces Composite Bacteriophage Particles.
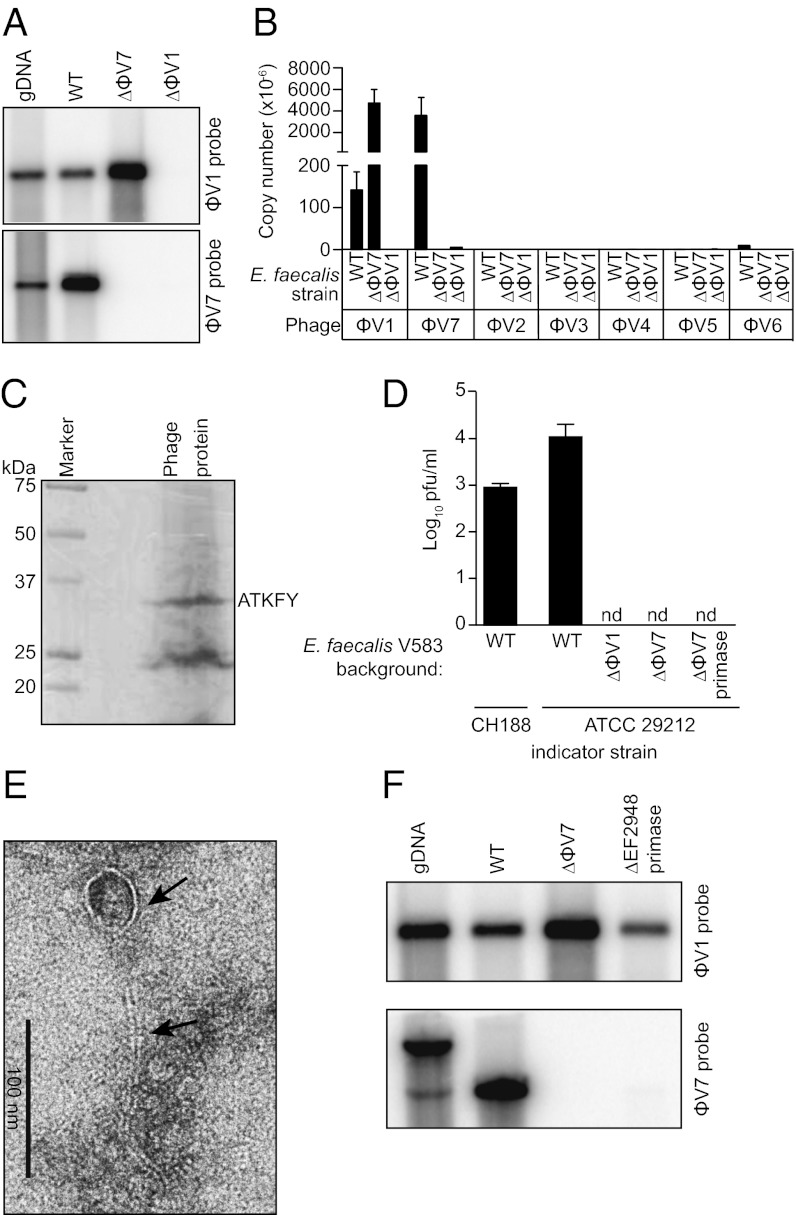
To determine whether E. faecalis V583 produces phage during growth in culture, we isolated phage particles from logarithmic phase E. faecalis culture supernatant by precipitation and purified the DNA. Southern blot analysis and quantitative PCR (qPCR) revealed that the majority of this DNA was derived from the ϕV1 and ϕV7 prophages (Fig. 2 A and B and Fig. S1A). A small amount of DNA from the ϕV5 and ϕV6 prophages and no DNA from prophages ϕV2–ϕV4 were detected (Fig. 2B and Fig. S1A). Total protein from isolated phage particles included a 32 kDa protein with an N-terminal amino acid sequence identical to residues 2–6 of the major capsid protein (EF0339) belonging to ϕV1 (Fig. 2C). Thus, E. faecalis phages ϕV1 and ϕV7 are produced during normal growth in culture.
Fig. 2.
E. faecalis V583 produces a ϕV1/7 composite bacteriophage. (A) Southern blot of phage DNA isolated from the culture supernatant of WT E. faecalis V583 (WT) or the isogenic mutant strains ΔϕV7 and ΔEF0339 (ΔϕV1 capsid protein). DNA was digested with NdeI (ϕV1 probe) or NsiI (ϕV7 probe). WT V583 genomic DNA (gDNA) is used as a control. (B) Quantification of phage DNA isolated from WT or ϕV1/7-deficient E. faecalis culture supernatant. The absolute copy number of each prophage was measured using qPCR. (C) Imido black staining of ϕV1/7 phage proteins separated by SDS/PAGE. N-terminal sequencing identified a ∼32 kDa protein as the EF0339 ϕV1 capsid protein. (D) Quantitative plaque assay measuring ϕV1/7 particles from E. faecalis V583 or the ΔϕV1, ΔϕV7, and ΔϕV7 primase mutants using E. faecalis CH188 or ATCC 29212 as indicator strains. nd, not detected. (E) Transmission electron micrograph of a ϕV1/7 particle. Arrows indicate the phage capsid and tail. (F) Southern blot of phage DNA isolated from the culture supernatant of the ϕV7 ΔEF2948 primase mutant E. faecalis strain. WT V583 genomic DNA and phage DNA from WT V583 and ΔϕV7 mutant cultures were probed for comparison. The genomic DNA band detected by the ϕV7 probe has a higher molecular weight because of a chromosomal NsiI site that is outside of the ϕV7 genome.
We next assessed whether the phage particles were infectious. We identified two E. faecalis strains, CH188 and ATCC 29212, which could be infected and lysed by the phage preparations (Fig. 2D and Fig. S1 B and C). Phage particles visualized by transmission EM after infection of E. faecalis ATCC 29212 resembled Siphoviridae phages with flexible, long, noncontractile tails and circular capsids (12) (Fig. 2E). Unlike ϕV1, the ϕV7 DNA sequence is devoid of any phage structural genes and therefore, resembles a cryptic or satellite phage genome that cannot produce phage particles. Because both ϕV1 and ϕV7 DNAs were detected in phage particles isolated from E. faecalis V583 culture supernatants, we hypothesized that the particles packaged both ϕV1 and ϕV7 DNA. To test this possibility, we generated two E. faecalis V583 deletion strains: one strain lacking the majority of the ϕV7 element (ΔϕV7) and another strain deficient in the ϕV1 capsid gene EF0339 (ΔϕV1) (Fig. 1). In the absence of either the ϕV1 capsid gene or ϕV7, no infectious phage particles were produced as indicated by plaque assay (Fig. 2D). However, unlike the ϕV7 mutant strain, which yielded packaged ϕV1 DNA, neither ϕV1 nor ϕV7 DNA was detected in the culture supernatant of the ϕV1 capsid deletion strain (Fig. 2A). This finding shows that ϕV7 requires the ϕV1 structural genes for its propagation.
The ϕV7 mutant strain produced phage particles that package ϕV1 DNA (Fig. 2A) but are noninfectious (Fig. 2D). This finding suggested that ϕV7 encodes a factor that promotes phage binding to or replication in target bacterial cells. The annotation of the ϕV7 genome revealed an ORF (EF2948) that is homologous to known phage DNA primases (Fig. 1). Such primases synthesize RNA primers used for nucleotide extension during lytic DNA replication (13). We generated an unmarked deletion of the EF2948 DNA primase gene in prophage ϕV7 on the E. faecalis chromosome. Deletion of the primase abolished production of infectious ϕV1/7 phage particles (Fig. 2D), suggesting an essential role in lytic replication. Similar to the whole ϕV7 deletion strain, culture supernatant from the ϕV7 DNA primase mutant strain contained packaged ϕV1 DNA but no packaged ϕV7 DNA (Fig. 2F). This finding shows that ϕV1 and ϕV7 DNAs are both required to generate an infectious lytic phage particle (Fig. S2).
We also noted that the smaller ϕV7 genome was present at a higher copy number than the larger ϕV1 genome in packaged phage particles (Fig. 2B). If we assume that the efficiency of ϕV1/7 DNA packaging is equivalent, then the number of genomes packaged into an individual capsid should be biased to the smaller ϕV7 genomes. This result would be consistent with a headful DNA packaging mechanism used by many tailed double-stranded DNA phages (14). However, it is also possible that the higher ϕV7 DNA copy number arises because the ϕV7 DNA packaging sites are bound with greater affinity by the phage DNA packaging machinery.
Amino Acid Availability Regulates ϕV1/7 Production.
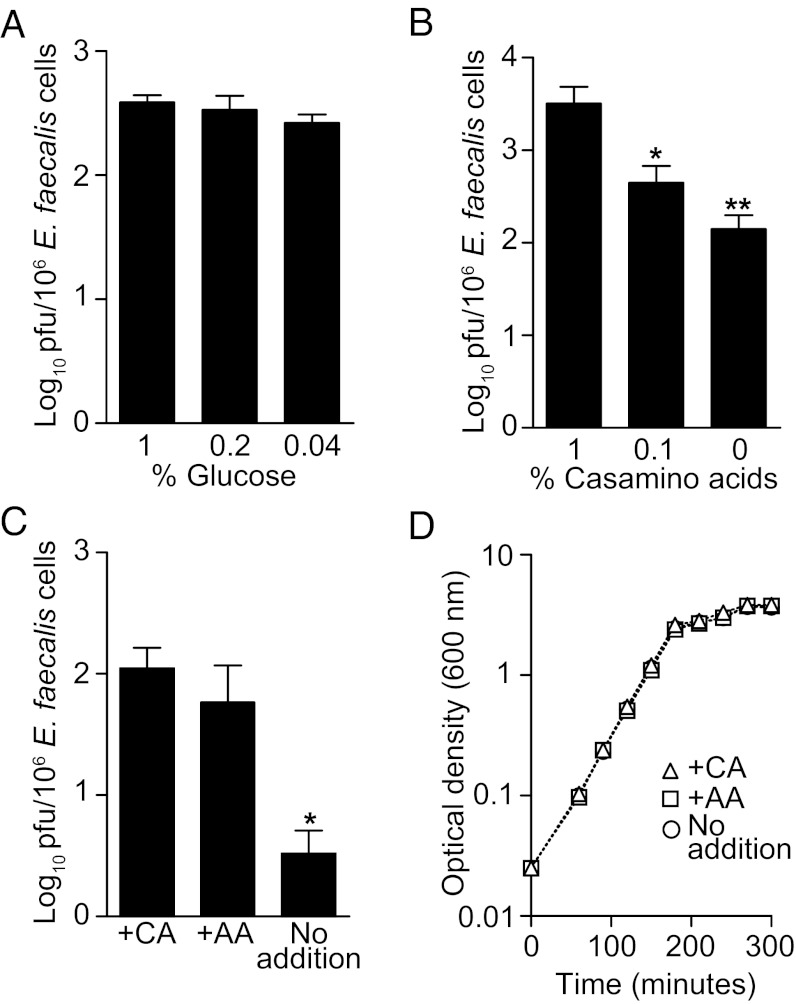
Environmental cues frequently govern the induction of prophage from bacterial chromosomes. These cues are diverse and include antibiotics, reactive oxygen species, and nutrient availability (15–17). A natural habitat of E. faecalis is the intestinal tract, where it likely encounters dynamic environmental signals, including carbohydrate and amino acid gradients. When grown in the complex medium Brain Heart Infusion (BHI), ϕV1/7 particle production was maximal during logarithmic growth, indicating that phage production is highest when the bacteria are actively dividing (Fig. S3A). To gain insight into whether growth substrate availability modulates induction of ϕV1/7, we first tested whether glucose concentrations influenced ϕV1/7 production. Increasing glucose concentrations had no significant effect on ϕV1/7 levels (Fig. 3A). However, when E. faecalis was grown in BHI or defined medium containing increasing concentrations of casamino acids, there was a dose-dependent increase in the number of ϕV1/7 particles produced (Fig. 3B and Fig. S3B). Furthermore, when BHI broth was supplemented with a mixture of 8 aa for which E. faecalis is auxotrophic, ϕV1/7 production was also increased (Fig. 3C). This increase was independent of differences in bacterial densities (Fig. 3D and Fig. S3C). These data suggest that enhancement of ϕV1/7 production is selectively dependent on amino acid availability.
Fig. 3.
Amino acids enhance the production of ϕV1/7. (A) ϕV1/7 production in defined medium containing varying concentrations of d-glucose as the sole carbon source. (B) ϕV1/7 levels determined by plaque assay in the presence of increasing concentrations of casamino acids in BHI. (C) Quantification of ϕV1/7 particles after the addition of 1% casamino acids (+CA), a defined mixture of purified amino acids (+AA), or no addition. (D) Cell density growth curve of E. faecalis grown in BHI or BHI supplemented with CA or AA. ϕV1/7 numbers are represented as pfus per 106 E. faecalis cells. Statistical analysis was performed using a two-tailed Student t test with a Mann–Whitney correction. Error bars ± SEM. *P < 0.05, **P < 0.005. n = 3–5 experiments performed in duplicate.
ϕV1/7 Is Produced in the Mouse Gastrointestinal Tract.
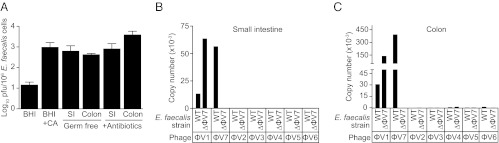
We next tested whether ϕV1/7 was produced by E. faecalis growing in the intestinal tract of mice. We used germ-free or antibiotic-treated mice for these studies, because the intestinal tracts of mice harboring a conventional microbiota often resist colonization by singly introduced bacterial species (18). We orally inoculated germ-free mice with E. faecalis V583 and enumerated E. faecalis cells and ϕV1/7 particles in the terminal small intestine (ileum) and colon 48 h later. Compared with growth in BHI medium, ϕV1/7 production was ∼200-fold higher in both the small intestine and colon (Fig. 4A). Similar results were obtained by inoculating E. faecalis into conventional mice that had been treated with broad-spectrum antibiotics to deplete the microbiota (Fig. 4A). We used qPCR to confirm that the isolated phage particles from these tissues were ϕV1/7 particles (Fig. 4 B and C). Interestingly, the number of ϕV1/7 particles produced per E. faecalis cell during growth in the intestine was similar to the number obtained for E. faecalis cells growing in BHI medium supplemented with 1% casamino acids (Fig. 4A). These data are consistent with the idea that ϕV1/7 production by E. faecalis is sensitive to nutrient availability.
Fig. 4.
ϕV1/7 is produced during E. faecalis V583 growth in the mouse intestinal tract. (A) The abundance of ϕV1/7 production was measured from E. faecalis cells grown in broth culture or recovered from the small intestines and colons of germ-free and antibiotic-treated C57BL/6 mice. ϕV1/7 levels were normalized to the total bacterial load from each environment. qPCR was used to measure the absolute abundance of ϕV1/7 DNA and any other E. faecalis V583 prophage DNA from the small intestines (B) or colons (C) of germ-free mice. n = 6–8 for A and n = 2 groups of five pooled mouse intestinal samples for B and C.
ϕV1/7 Enhances E. faecalis V583 Colonization in a Competitive Ecosystem.
One potential benefit to phage production by a bacterial cell would be to limit colonization of an environmental niche by invading competitors. This situation could arise when related bacterial strains in a mixed microbial population compete for limited nutrients. To test whether ϕV1/7 confers a competitive advantage to its parental strain, we performed coculture experiments. The cocultures included the ϕV1/7-sensitive E. faecalis strain CH188 and either the E. faecalis V583 WT strain (ϕV1/7-producing) or the ϕV7 mutant strain that does not produce infectious phage particles. When WT V583 was mixed in equal numbers with CH188 and cultured, the ratio of WT V583 cells to CH188 cells was greater than when the ϕV7 deletion strain was mixed with CH188 (Fig. 5A). Similar results were obtained when using a second ϕV1/7-sensitive E. faecalis strain, ATCC 29212 (Fig. S4A), or BHI supplemented with 1% casamino acids (Fig. S4B).
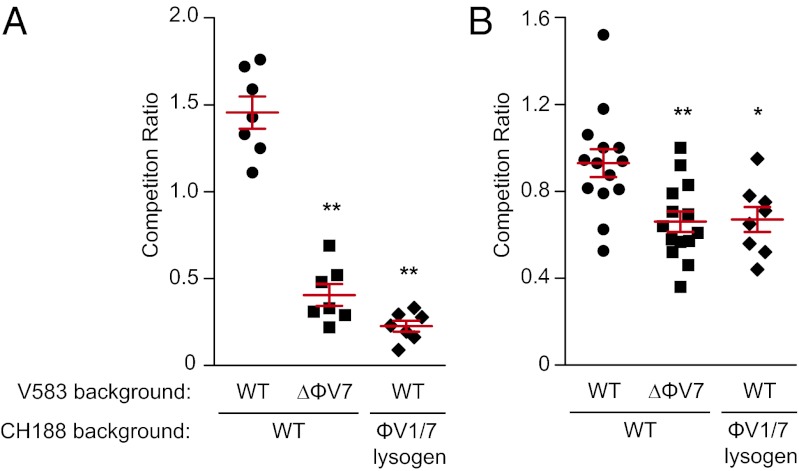
Fig. 5.
ϕV1/7 enhances E. faecalis V583 colonization in a competitive ecosystem. (A) Competition between WT E. faecalis V583 or the ΔϕV7 strain in mixed coculture with E. faecalis CH188 in BHI broth. V583 was also competed with ϕV1/7-lysogenized CH188, which is resistant to ϕV1/7-mediated lysis (Fig. S6C). (B) Competition between WT E. faecalis V583 or ΔϕV7 and the susceptible strain CH188 from the feces of germ-free mice 24 h after cocolonization. Competition ratio is the ratio of the parental strain (V583 background) to the susceptible or lysogenized strain. Each data point represents one independent culture or individual mouse. Statistical analysis was done by a two-tailed Student t test. Error bars ± SEM *P < 0.05, **P < 0.005.
We next determined whether ϕV1/7 also enhances V583 colonization in vivo. Germ-free C57BL/6 mice were orally inoculated with a mixture of CH188 and either WT V583 or the ϕV7 mutant. Feces were collected after 24 h of colonization, and each bacterial strain was enumerated by selective plating. The ΔϕV7 mutant showed reduced relative abundance compared with WT V583, suggesting that ϕV1/7 enhanced the ability of V583 to colonize in a mixed strain microbial population (Fig. 5B). The ϕV1 capsid deletion strain produced a similar phenotype in the intestinal cocolonization model (Fig. S5). These findings indicate that phage production allows E. faecalis V583 to compete better with related enterococcal strains and suggest that phage predation enhances the success of V583 during competitive growth.
To further test the role of phage predation during in vitro and in vivo competition, we generated strains that are resistant to ϕV1/7 infection because of lysogeny (Figs. S1 B and C and S6 A and B). Similar to V583, which is a ϕV1/7 double lysogen, CH188 and ATCC 29212 strains lysogenized for both ϕV1 and ϕV7 resisted superinfection by ϕV1/7 (Fig. S6C) and generated phage particles that produced plaques on the parental host strain (Fig. S6D). When ϕV1/7-lysogenized CH188 was used in competition experiments, WT V583 did not show an increased abundance, similar to the ΔϕV7 mutant in vitro and in vivo (Fig. 5). This result supports the idea that phage predation enhances the ability of V583 to compete with related enterococcal strains.
Taken together, our results suggest that E. faecalis V583 can use the ϕV1/7 composite phage as a weapon, infecting and lysing related strains of E. faecalis during colonization of the intestinal tract. We propose that this result confers an advantage to the phage-producing strain by reducing competition for nutrients from related bacterial strains that have similar metabolic requirements.
Discussion
The bacteria that colonize the human intestine harbor a large number of prophage, but the biological significance is unclear (2). E. faecalis genomes, in particular, have retained a number of integrated prophages, although the E. faecalis genome is rapidly evolving (8, 9). With many new E. faecalis phages being identified and the fact that many of the sequenced E. faecalis isolates contain one or more prophages (8, 9, 19), it is likely that these elements are integrally important to E. faecalis physiology.
Our studies have provided initial insight into the biological role of E. faecalis prophages. Here, we have shown that E. faecalis V583 produces an unusual composite lytic phage derived from two prophages, ϕV1 and ϕV7 (Fig. S2). Each of these prophages contributes elements that are critical for the production of phage particles. Our findings indicate that ϕV1 provides the structural components for phage particle biosynthesis, whereas the ϕV7 prophage encodes accessory genes used during lytic replication in an infected host cell. Previous studies have uncovered examples of cooperation between two or more distinct phage elements. For example, Staphylococcus aureus uses a temperate helper phage to transduce pathogenicity island DNA to target bacteria (20). In another example, the DNA element toxin-linked cryptic uses the morphogenesis genes of the filamentous phage fs2ϕ to form transducing phage particles. When this phage is integrated into the Vibrio cholerae chromosome by lysogeny, it restores the attachment site for integration of the cholera toxin-producing phage CTXϕ (21, 22). Similarly, the ϕV7 prophage uses the structural genes of ϕV1 for its transmission in concert with transmission of ϕV1 DNA. In turn, ϕV7 supplies a protein that is essential for phage DNA replication within a host cell.
Nutrient availability is known to influence prophage induction and lytic replication (23–25). E. faecalis V583 responds to enhanced amino acid availability in culture by inducing ϕV1/7, suggesting that amino acid availability acts as an important environmental cue governing E. faecalis prophage induction. This finding is consistent with the fact that E. faecalis strains are amino acid auxotrophs that cannot endogenously synthesize 9 of 20 essential amino acids (9). Furthermore, on introduction into the mouse intestine, E. faecalis produces ϕV1/7 to a level similar to the level observed during in vitro growth in the presence of elevated amino acid concentrations. Although we cannot rule out that prophage production in the intestine occurs in response to other environmental cues, the presence of millimolar concentrations of free amino acids in the intestine (26) suggests that amino acids may constitute one environmental factor that determines production of ϕV1/7 in vivo. Sensing amino acid concentration gradients within the intestinal tract and inducing lytic ϕV1/7 production may allow E. faecalis V583 to compete with related enterococci that are also amino acid auxotrophs. This idea is supported by our finding that E. faecalis cells that produce ϕV1/7 are more competitive in a mixed enterococcal coculture than their isogenic counterparts that lack the ability to produce infectious ϕV1/7 particles.
We have identified homologs of ϕV1/7 genes in two independent intestinal metagenomic studies, suggesting that similar prophages are present in the human microbiome (Fig. S7). Homologs of ORFs ϕV1 EF0348 (phage tail protein) and ϕV7 EF2948 (DNA primase) were identified in two independent public intestinal metagenome databases (SI Materials and Methods) (27, 28). Although the sequences were of low abundance, it is clear that homologs of these phage genes are present in the intestinal microbiome. Owing to the low-coverage sequencing of human intestinal microbiomes, the presence of homologs of these phage genes indicates that similar temperate prophage may influence bacterial colonization dynamics in the human intestine.
In summary, we have shown that two E. faecalis V583 prophage elements converge to produce the composite phage ϕV1/7. ϕV1/7 is beneficial to E. faecalis cells during competition in mixed-strain microbial populations. Phage production may be one way in which E. faecalis strains maintain dominance of their habitat in the presence of metabolically similar strains. These findings contribute initial insight into the role of temperate phages in the gastrointestinal ecosystem and provide a framework for future studies on how phages impact microbiota ecology and host biology.
Materials and Methods
Bacterial Growth Conditions.
Bacterial strains are listed in Table S2. Information on the construction of bacterial strains can be found in SI Materials and Methods. E. faecalis was grown at 37 °C in BHI broth (Becton Dickinson), BHI supplemented with casamino acids and 50 µg/mL l-tryptophan, or complex-defined medium containing glucose (29). In some experiments, a mixture of purified amino acids (200 µg/mL each) of the l-forms of lysine, tryptophan, histidine, arginine, isoleucine, leucine, serine, and valine was added to BHI. Escherichia coli was grown in LB broth (Sigma) at 37 °C. Antibiotics were used at the following concentrations (per 1 mL): 10 µg vancomycin, 15 µg chloramphenicol, 15 µg tetracycline, and 100 µg gentamicin for E. faecalis and 8 µg chloramphenicol and 100 µg ampicillin for E. coli.
Mice.
Conventional and germ-free C57BL/6 mice were bred at the University of Texas Southwestern Medical Center. Germ-free C57BL/6 mice were reared in sterile isolators as previously described (30). In some experiments, mice were administered an antibiotic mixture (per 1 mL) of 1 mg streptomycin, 1 mg metronidazole, 1 mg neomycin, 1 mg ampicillin, and 0.5 mg vancomycin daily for 6 d through oral gavage and ad libitum in their drinking water before colonization with E. faecalis. Animal protocols were approved by the Institutional Animal Care and Use Committees of the University of Texas Southwestern Medical Center.
Bacteriophage Isolation.
E. faecalis V583 was subcultured and grown to an OD600 of 0.5. The cells were subcultured a second time into 1 L BHI and grown to an OD600 of 1. The bacterial cells were pelleted by centrifugation, and the supernatant was filtered (0.45 µm). For intestinal contents, a 9 cm length of distal small intestine and the entire colon from five mice were flushed with 5 mL SM buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris⋅HCl, pH 7.5), and the samples were pooled. Supernatant or intestinal flushes were treated with 5 µg/mL both DNase and RNase for 1 h at room temperature. Phages were precipitated with 1 M NaCl and 10% wt/vol PEG-8000 on ice overnight, pelleted at 7,025 × g for 20 min, and resuspended in 1 mL SM buffer. Phage DNA was extracted according to a published protocol, and phages were enumerated by agar overlay (31). SI Materials and Methods has a detailed description of phage protein sequencing and phage particle visualization by transmission EM.
qPCR.
Total phage DNA was isolated as described above; >200-bp regions of each of the seven prophages were amplified by PCR and topoisomerase (TOPO) cloned into pCR2.1 (Invitrogen). Absolute phage DNA copy number was calculated using these vectors to generate standard curves of known DNA concentrations using SYBR green dye (Invitrogen).
Southern Blot.
For Southern blot analysis, ϕV1- and ϕV7-specific DNA probes were generated by PCR with incorporation of [α-32P]-dATP (Perkin-Elmer) using the RadPrime DNA Labeling Kit (Invitrogen); 2 µg genomic DNA or 1 µg total phage DNA were digested to completion with NdeI (ϕV1 probe) or NsiI (ϕV7 probe) restriction enzymes. The DNA fragments were separated on a 0.7% agarose gel, and Southern blotting was performed using standard methods (31).
Competition Assays.
For in vitro competition assays, E. faecalis strains were grown in BHI broth to an OD600 of 0.5, and ∼1 × 106 cells of each strain were subcultured into a BHI culture with or without casamino acids. The cocultures were grown for 3 h at 37 °C with shaking. The cultures were measured for the total abundance of each strain by dilution plating on BHI agar containing vancomycin or tetracycline for V583, CH188, and CHLys3.2 and gentamicin or chloramphenicol for V583 and ATCC 29212. The competition ratio was calculated as the final cfu ratio (parental/susceptible or lysogenized) after normalization to the initial starting inoculum for each strain.
In vivo competition assays were done using germ-free C57BL/6 mice colonized with ∼5 × 107 cfu both E. faecalis V583 and CH188, the E. faecalis ϕV7 mutant strain and CH188, or V583 and CHLys3.2. After 24 h, feces were collected from individual mice, and the total number of each bacterial strain was determined by dilution plating on Enterococcosel agar (Becton Dickinson) containing either vancomycin or tetracycline. The competition ratio was calculated as the final cfu ratio (parental/susceptible or lysogenized).
Supplementary Material
Acknowledgments
We thank S. Kuss, K. Palmer, X. Yu, and C. Behrendt for their assistance with this work. We thank S. Mukherjee and H. Zheng for assistance with EM and L. Hancock and M. Gilmore for providing bacterial strains and reagents. This work was supported by Ruth L. Kirschstein National Research Service Award F32 DK089718 (to B.A.D.), The Howard Hughes Medical Institute (L.V.H.), National Institutes of Health Grant R01 DK070855 (to L.V.H.), and a Burroughs Wellcome Foundation Investigators in the Pathogenesis of Infectious Diseases award (to L.V.H.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206136109/-/DCSupplemental.
References
- 1.Duerkop BA, Vaishnava S, Hooper LV. Immune responses to the microbiota at the intestinal mucosal surface. Immunity. 2009;31:368–376. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischetti VA. In vivo acquisition of prophage in Streptococcus pyogenes. Trends Microbiol. 2007;15:297–300. doi: 10.1016/j.tim.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Hambly E, Suttle CA. The viriosphere, diversity, and genetic exchange within phage communities. Curr Opin Microbiol. 2005;8:444–450. doi: 10.1016/j.mib.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Valera F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 6.Tannock GW, Cook G. Enterococci as members of the intestinal microflora of humans. In: Gilmore MS, editor. Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance. Washington, DC: ASM Press; 2002. pp. 101–132. [Google Scholar]
- 7.Klare I, Werner G, Witte W. Enterococci. Habitats, infections, virulence factors, resistances to antibiotics, transfer of resistance determinants. Contrib Microbiol. 2001;8:108–122. doi: 10.1159/000060406. [DOI] [PubMed] [Google Scholar]
- 8.Palmer KL, et al. Comparative genomics of enterococci: Variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. MBio. 2012;3:e00318-11. doi: 10.1128/mBio.00318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulsen IT, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 10.Casjens S. Prophages and bacterial genomics: What have we learned so far? Mol Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 11.Yasmin A, et al. Comparative genomics and transduction potential of Enterococcus faecalis temperate bacteriophages. J Bacteriol. 2010;192:1122–1130. doi: 10.1128/JB.01293-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ackermann HW. 5500 Phages examined in the electron microscope. Arch Virol. 2007;152:227–243. doi: 10.1007/s00705-006-0849-1. [DOI] [PubMed] [Google Scholar]
- 13.Hamdan SM, van Oijen AM. Timing, coordination, and rhythm: Acrobatics at the DNA replication fork. J Biol Chem. 2010;285:18979–18983. doi: 10.1074/jbc.R109.022939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 15.Allen HK, et al. Antibiotics in feed induce prophages in swine fecal microbiomes. MBio. 2011;2:e00260-11. doi: 10.1128/mBio.00260-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDaniel L, Paul JH. Effect of nutrient addition and environmental factors on prophage induction in natural populations of marine synechococcus species. Appl Environ Microbiol. 2005;71:842–850. doi: 10.1128/AEM.71.2.842-850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeMarini DM, Lawrence BK. Prophage induction by DNA topoisomerase II poisons and reactive-oxygen species: Role of DNA breaks. Mutat Res. 1992;267:1–17. doi: 10.1016/0027-5107(92)90106-c. [DOI] [PubMed] [Google Scholar]
- 18.Barthel M, et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazaheri Nezhad Fard R, Barton MD, Heuzenroeder MW. Novel Bacteriophages in Enterococcus spp. Curr Microbiol. 2010;60:400–406. doi: 10.1007/s00284-009-9555-z. [DOI] [PubMed] [Google Scholar]
- 20.Novick RP, Christie GE, Penadés JR. The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol. 2010;8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faruque SM, et al. RS1 element of Vibrio cholerae can propagate horizontally as a filamentous phage exploiting the morphogenesis genes of CTXphi. Infect Immun. 2002;70:163–170. doi: 10.1128/IAI.70.1.163-170.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassan F, Kamruzzaman M, Mekalanos JJ, Faruque SM. Satellite phage TLCφ enables toxigenic conversion by CTX phage through dif site alteration. Nature. 2010;467:982–985. doi: 10.1038/nature09469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen LW. Delayed lysis with salmonella bacteriophage p22: Induction of lysis by addition of cysteine or histidine to the growth medium. J Virol. 1969;4:214–218. doi: 10.1128/jvi.4.3.214-218.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lunde M, Aastveit AH, Blatny JM, Nes IF. Effects of diverse environmental conditions on ϕLC3 prophage stability in Lactococcus lactis. Appl Environ Microbiol. 2005;71:721–727. doi: 10.1128/AEM.71.2.721-727.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smit E, Wolters AC, Lee H, Trevors JT, vanElsas JD. Interactions between a genetically marked Pseudomonas fluorescens strain and bacteriophage ϕR2f in soil: Effects of nutrients, alginate encapsulation, and the wheat rhizosphere. Microb Ecol. 1996;31:125–140. doi: 10.1007/BF00167859. [DOI] [PubMed] [Google Scholar]
- 26.Schott K, Huether G, Neuhoff V. Free amino acid concentrations in the gut lumen of developing rats. Biochem Med. 1983;29:285–292. doi: 10.1016/0006-2944(83)90064-9. [DOI] [PubMed] [Google Scholar]
- 27.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang JC, LaSarre B, Jimenez JC, Aggarwal C, Federle MJ. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog. 2011;7:e1002190. doi: 10.1371/journal.ppat.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch EF, Maniatis T, editors. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.