Abstract
The endosomal sorting complexes required for transport (ESCRT) proteins have a critical function in abscission, the final separation of the daughter cells during cytokinesis. Here, we describe the structure and function of a previously uncharacterized ESCRT-III interacting protein, MIT-domain containing protein 1 (MITD1). Crystal structures of MITD1 reveal a dimer, with a microtubule-interacting and trafficking (MIT) domain at the N terminus and a unique, unanticipated phospholipase D-like (PLD) domain at the C terminus that binds membranes. We show that the MIT domain binds to a subset of ESCRT-III subunits and that this interaction mediates MITD1 recruitment to the midbody during cytokinesis. Depletion of MITD1 causes a distinct cytokinetic phenotype consistent with destabilization of the midbody and abscission failure. These results suggest a model whereby MITD1 coordinates the activity of ESCRT-III during abscission with earlier events in the final stages of cell division.
Keywords: HIV, multivesicular body, X-ray crystallography
The endosomal sorting complexes required for transport (ESCRTs) are present in all eukaryotes and they facilitate sorting of ubiquitinated membrane proteins to lysosomes via multivesicular endosomes (MVBs) (1–3). However, the most ancient ESCRT function is a role in cell division and cytokinesis because this function predates the divergence of eukaryotes from archaea (4–7). Four ESCRTs have been characterized: ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III, and all of these complexes have roles in endosomal sorting. In contrast, only ESCRT-I and ESCRT-III have demonstrated roles in eukaryotic cytokinesis (8).
Mammalian cytokinesis is a highly regulated process, requiring the concerted translocation of many functional components to the midbody, including the phosphoprotein CEP55, which subsequently recruits ESCRT-I and the ESCRT-associated protein ALIX. ALIX can then recruit ESCRT-III components to the midbody (4, 5, 9, 10). Adjacent to the midbody, cortical constriction zones are formed in which the diameter of the intercellular bridge is strongly reduced and where abscission ultimately occurs. This cortical constriction is thought to depend on the formation of 17-nm helical ESCRT-III filaments (11, 12), a model consistent with the ability of ESCRT-III to carry out membrane scission in vitro (13, 14). Besides abscission, ESCRT-III subunits are required for other cytokinetic events, such as proper function of centrosomes and spindle maintenance (15). Furthermore, ESCRT-III coordinates midbody resolution and the abscission checkpoint to prevent chromosomal damage (16).
The formation of ESCRT-III–dependent filaments in the constriction zone adjacent to the midbody is spatially and temporally coordinated with recruitment of other factors essential for cytokinesis. CHMP1-3 recruit the AAA-ATPase VPS4, which disassembles ESCRT-III filaments deposited on membranes, an activity that is required for all ESCRT functions including abscission (17–19). hIST1 and CHMP1B recruit the AAA-ATPase Spastin, which severs the microtubule bundle spanning the length of the intercellular bridge, an activity essential for abscission (20, 21). hIST1 further recruits Spartin, which also is involved in cytokinesis (22, 23). VPS4, Spastin, and Spartin all have in common a microtubule-interacting and trafficking (MIT) domain that mediates recruitment to the midbody by recognizing defined sequence motifs referred to as MIT interacting motifs (MIMs), present in the C-terminal region of certain ESCRT-III components (17, 18).
Here, we functionally and structurally characterize MIT-domain containing protein 1 (MITD1), a human gene of unknown function. The crystal structures of MITD1 revealed a dimer that, in addition to the predicted MIT domain at the N terminus, has a unique domain at the C terminus with an unanticipated phospholipase d fold (PLD fold) that binds avidly to phosphoinositide-containing membranes. We demonstrate that MITD1 recognizes a subset of ESCRT-III subunits through a typical MIT-MIM1 readout and that these interactions mediate recruitment of MITD1 to the midbody during cell division. Accordingly, we show that depletion of MITD1 has a distinct effect on cytokinesis that is consistent with both midbody instability and abscission defects. Taken together, these data suggest that MITD1 coordinates abscission with earlier stages of cytokinesis.
Results
MITD1 MIT Domain Interacts with a Subset of ESCRT-III MIM.
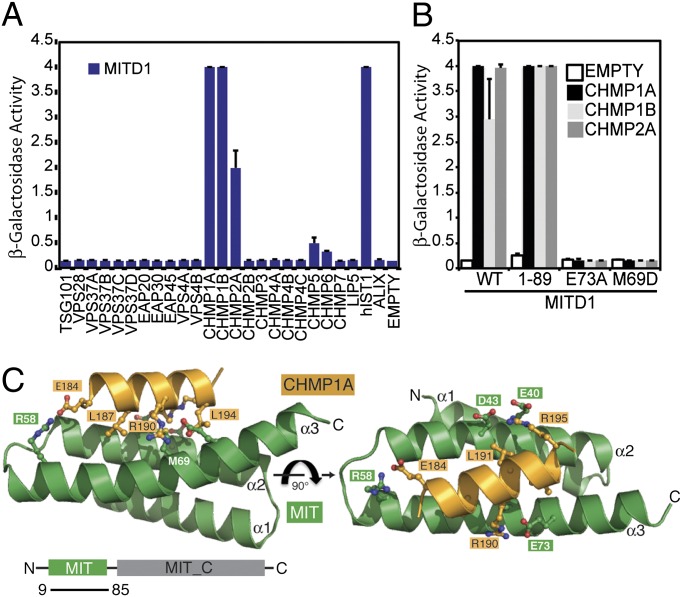
MITD1 has recently been identified as an ESCRT-III–binding protein (24, 25). To further investigate whether MITD1 binds other components of the ESCRT machinery, the human subunits of ESCRT-I, ESCRT-II, ESCRT-III, and ESCRT-III–associated proteins were tested in a directed yeast two-hybrid screen. In addition to the published interactions with CHMP2A, CHMP1B and hIST1 an interaction with CHMP1A was observed (Fig. 1A), whereas no binding to ESCRT-I and ESCRT-II could be detected, thus indicating a specific interaction of MITD1 with ESCRT-III subunits. These results were confirmed by using a GST coprecipitation procedure in which 293T cells were transfected with a GST-MITD1 fusion plasmid together with plasmids encoding CHMP1A, CHMP1B, CHMP2A, or hIST1 fused to YFP (Fig. S1A). As negative controls, binding of the ESCRT-III proteins to GST alone was undetectable and GST-MITD1 did not interact with YFP-CHMP2B.
Fig. 1.
MITD1 interacts with a subset of ESCRT-III proteins. (A) Human MITD1 was tested for interactions with human ESCRT-I, ESCRT-II, and ESCRT-III subunits by yeast two-hybrid assays. MITD1 binds selectively CHMP1A, CHMP1B, CHMP2A, and hIST1. Error bars indicate the SD from the mean of triplicate measurements. (B) Mutations of either M69 or E73 in MITD1 abolish interactions with ESCRT-III subunits in yeast two-hybrid assays. Error bars indicate the SD from the mean of triplicate measurements. (C) Structure of the MITD1 MIT domain in complex with the C-terminal tail of CHMP1A (184–196; orange). Critical interacting residues in MIT domain (M69 and E73) and key MIM1 residues in CHMP1A are shown as ball-and-stick.
Further analysis by yeast two-hybrid showed that MITD1 binds to ESCRT-III through its N-terminal MIT domain (MITD1 1-89; Fig. 1B). Because all four ESCRT-III interacting partners of MITD1 have a MIM1 motif in their C-termini, we undertook a structural approach to gain insight into the MITD1–MIM1 interaction and determined the crystal structure of a complex formed between MITD19–85 and CHMP1A184–196 (Fig. 1C and Table S1). The crystals contain two MIT/CHMP1A complexes in the asymmetric unit, with essentially identical conformations. The MITD1 MIT domain has a fold consisting of three helices arranged as a right-handed solenoid (Fig. 1C) and is most similar to the MIT domains from orthologs of the AAA-ATPase VPS4, with which it shares 26% sequence identity (Fig. S1). Accordingly, an internal salt bridge between MITD1 D26 and R71 at one end of the wide groove separating helices α1 and α3 is conserved in even distant orthologs of VPS4, such as the archaeal VPS4 (7, 17).
The CHMP1A MIM forms a single helix and binds parallel to helix α2 in the shallow groove between helices α2 and α3. The accessible surface area buried in the interface between the MITD1 MIT domain and the CHMP1A peptide is 1,323 Å2. The CHMP1A–MITD1 interaction is similar to that of the VPS4 MIT domain with CHMP1A (18) and yeast Vps4p MIT domain with Vps2, the yeast homolog of CHMP2 (17). This interaction involves several highly conserved residues in MITD1 and residues that comprise the MIM1 motif (D/E)xxLxxRLxxL(K/R) (Fig. S1) (17, 18). All three hydrophobic residues from this MIM1 sequence fit in the α2/α3 groove of MITD1, and all three charged residues form salt bridges with oppositely charged residues in MITD1 (CHMP1AR195:MITD1E40, CHMP1AR184:MITD1E58, and CHMP1AR190:MITD1E70) (Fig. 1C). Mutations of either a hydrophobic residue (MITD1 M69D) or a salt bridge at the MITD1/MIM1 interface (MITD1 E73A) abolish binding to MIM1-containing ESCRT-III subunits as determined by yeast two-hybrid (Fig. 1B), underscoring the importance of both types of interactions.
Interestingly, in the structure of the MIM1/MITD1 complex, the slot between MITD1 helices α1 and α3 has a bound monomethyl ether PEG 2000 in one of the two MIT domains in the asymmetric unit (Fig. S1). This site is analogous to the region where the VPS4A MIT domain binds the MIM2 peptide of CHMP6 (26). Although by yeast two-hybrid we did not observe binding between MITD1 and CHMP6, it is possible the interaction is very weak, similar to the VPS4B MIM2 binding site, which binds the CHMP6 MIM2 only very weakly (∼800 μM affinity) (26). Alternatively, this slot might bind a distinct MIM2-related sequence in some unknown partner.
MITD1 Functions in Cytokinesis.
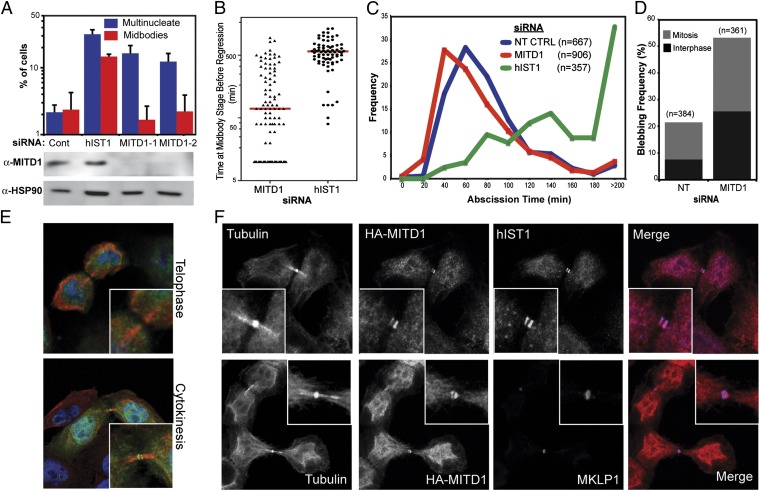
To examine whether MITD1 has a role in the canonical ESCRT-mediated functions we conducted gene-silencing experiments. Depletion of endogenous MITD1 with two independent small interfering RNAs (siRNAs) resulted in phenotypes consistent with cytokinesis failure, as indicated by a six- to eightfold increase of multinucleated cells compared with control siRNA-transfected cells (Fig. 2A). As a positive control for cytokinesis failure, depletion of hIST1 resulted in a 14-fold accumulation of multinucleated cells (24, 27). Intriguingly, depletion of MITD1 does not result in increased number of dividing cells connected with intercellular bridges, as opposed to hIST1-depleted cells (Fig. 2A).
Fig. 2.
MITD1 has a role in cytokinesis. (A) HeLa cells were transfected with two different siRNAs against MITD1 or a luciferase control and scored for multinucleation or visible midbodies. Depletion of hIST1 was used as a positive control. Depletion of MITD1 was verified by Western blot using anti-MITD1 antibody. Anti-HSP90 was used as a loading control. Error bars indicate the SD from the mean for three independent measurements. (B–D) Asynchronous cultures of HeLa cells stably expressing mCherry-Tubulin were transfected with the indicated siRNA and imaged live. (B) Quantification of the time before a dividing cell coalesces into a multinucleated cell. The red line represents the median time. (C) The median abscission time for cells that divided without becoming multinucleated was calculated across six independent experiments [Non Targeting Control (NT CTRL): 80 ± 44.4 min, n = 667; MITD1-1: 70 ± 56.9 min, n = 906; hIST1: 160 ± 111.2 min, n = 357]. The differences in abscission times are significant (P < 0.0001) as determined by a Mann–Whitney u test. (D) Quantification of the percentage of cells that displayed pronounced plasma membrane blebbing. (E and F) Confocal fluorescence images of HeLa cells stably expressing HA-MITD1 alone (E) or in combination with YFP-Tubulin (F). Cells were doubly stained with anti-HA and either anti-tubulin (E), anti-hIST1 (F, Upper) or anti-MKLP1 (F, Lower) antibodies. In the overlays, DNA is shown in blue, MITD1 in green, and tubulin in red (E) or MITD1 in red and hIST1/MKLP1 in blue (F). In all cases, the inset shows a magnification of the midbody.
The distinct cytokinetic phenotype in MITD1-depleted cells suggests a role for MITD1 in early events of cytokinesis, perhaps during ingression of the cleavage furrow or in midbody formation/stability. To distinguish between these possibilities, we analyzed cell division by live imaging of asynchronous HeLa cells stably expressing mCherry-Tubulin. MITD1-depleted cells progressed normally to late stages of mitosis, and cleavage furrow formation and ingression was comparable to control cells (Movies S1 and S2). However, a fraction of the cells showed failed abscission that resulted in the formation of multinucleated cells in agreement with the results shown in Fig. 2A (10.3% of the MITD1-treated cells vs. 3.1% of the control or 20.7% of the hIST1-treated cells). The majority of these cells coalesced shortly after midbody formation, before the median abscission time observed in normally dividing cells (5.9% of the total cells vs. 1.3% in control or 1.4% in hIST1 treated cells) (Fig. 2B and Movies S3 and S4), suggesting a defect in midbody stability. In the remaining 4.4% of cells, the two daughter cells were connected by an intercellular bridge for a prolonged period before regressing and forming a multinucleated cell, resembling the phenotype in hIST1-depleted cells (19.3% in hIST1 treated cells vs. 1.8% in control), (Fig. 2B and Movies S5 and S6). Taken together, these data suggest that MITD1 has a dual role in cytokinesis: stabilizing the intercellular bridge and facilitating abscission in concert with components of the ESCRT pathway.
Interestingly, the MITD1-depleted cells that divided without becoming multinucleated, exhibited premature abscission and resolved their midbodies 10 min faster than control treated cells (Fig. 2C and Fig. S2A, median abscission time of 70 min versus 80 min for control treated cells; P < 0.0001, Mann–Whitney u test). This result is in contrast to the result observed in hIST1-depleted cells, that showed a pronounced delay in abscission times compared with control treated cells (median abscission time of 160 min; P < 0.0001, Mann–Whitney u test), once again showing that the defect caused in cell division is different between these two ESCRT-associated proteins. The faster abscission time observed in cells that lack MITD1 is not related to the recently reported role of ESCRT-III in the AuroraB-dependent abscission checkpoint (NoCut) (16), because the checkpoint activation was not prevented by codepletion of MITD1 with NUP153 (Fig. S2B).
Additionally, approximately 53% of the dividing MITD1-depleted cells exhibited excessive cortical blebbing (Fig. 2D and Movies S7 and S8). Although some blebbing occurred in control cells (21.6%), it was much less pronounced than the membrane aberrations observed upon MITD1 ablation and was predominantly observed in cells in mitosis. Intriguingly, the plasma membrane instabilities observed in the absence of MITD1 did not necessarily culminate in cytokinesis failure and occurred equally in cells in interphase or in mitosis.
Down-regulation of MHC Class I by Kaposi's sarcoma-associated herpesvirus (KSHV) K3 (28) or degradation of the antiviral protein tetherin by KSHV K5 (29) (Fig. S3 A and B) were not rescued by MITD1 depletion, demonstrating that MITD1 is not required for ESCRT-dependent sorting of endosomal cargo. A negative result was also observed for HIV-1 budding (Fig. S3C). Altogether these results show that MITD1 is specifically needed for cytokinesis.
Because of the role of MITD1 in cytokinesis, we examined by immunofluorescence the localization of a stably expressed HA-tagged MITD1 during different stages of the cell cycle in HeLa cells. No obvious accumulation of the protein was seen until cytokinesis, when HA-MITD1 localizes to the midbody (Fig. 2E), adjacent to the central Flemming body, as seen by MKLP1 staining (Fig. 2F, Lower). This distribution matches the patterns previously seen for several ESCRT-III proteins (5, 11, 12, 24, 27) and, in fact, HA-MITD1 colocalized with endogenous hIST1 at the midbody (Fig. 2F, Upper). Because MITD1 binds to hIST1, CHMP1A, CHMP1B, and CHMP2A, we asked whether these interactions mediate recruitment to the midbody. The individual ESCRT-III proteins were depleted by siRNA, HA-MITD1 was visualized by immunofluorescence, and cells clearly in cytokinesis were scored for the presence of MITD1. We found that only depletion of CHMP2A prevented MITD1’s localization at the midbody (Fig. S2C). ESCRT-III is recruited to the midbody by the centrosome protein 55 kDa (CEP55) (4, 5) so we examined the functional relationship between CEP55 and MITD1. Midbodies were scored as described above for the presence of HA-MITD1 or YFP-CEP55 after siRNA treatment of cells stably expressing these tagged proteins. CEP55 depletion prevented MITD1 recruitment to the midbody (Fig. S4A) whereas in cells lacking MITD1, CEP55 was always found at the midbody (Fig. S4B). Analysis of cell division by live imaging of cells lacking both MITD1 and CEP55 showed that abscission failed in a similar way to that observed for CEP55 depletion alone (Fig. S4C), suggesting that the rapid regression induced by MITD1 depletion requires either CEP55 or additional factors recruited by CEP55.
Structure of MITD1 C-Terminal Domain Reveals a PLD-Like Fold.
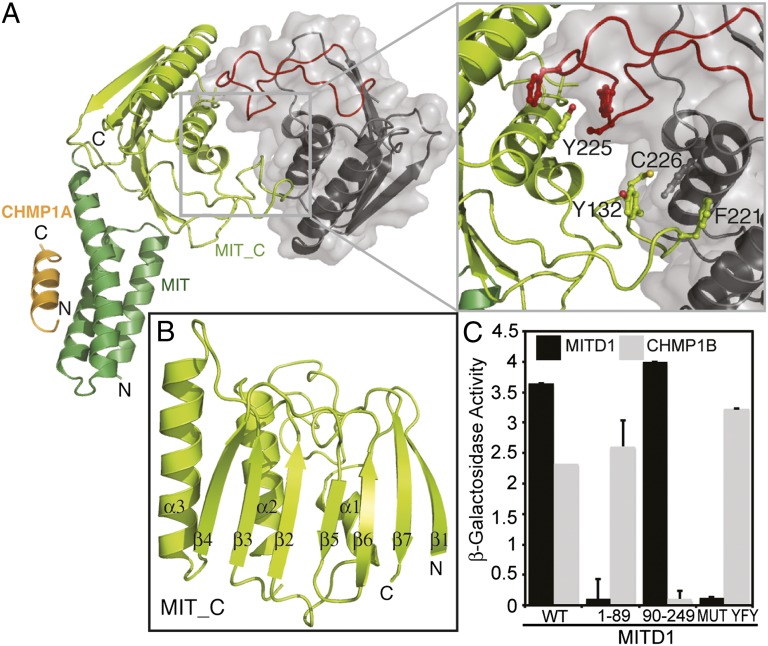
The C-terminal “effector” region of MITD1 is highly conserved in eukaryotes with the exception of plants and yeast. The domain has been annotated as a MIT_C domain in the Conserved Domain Database (CDD), but it has no known structure or function. In addition to MITD1, the MIT_C domain is also found in a class of Lon-type protease in bacteria. To get more insight into the function of this region, we determined the structures of both the full-length MITD1 in complex with a CHMP1A peptide and the C-terminal domain alone (MIT_C, residues 91–242) (Fig. 3 A and B and Table S1). The structure of the C-terminal domain was determined by using multiple wavelength anomalous dispersion methods with seleno-methionine–substituted protein, whereas full-length MITD1 was solved by molecular replacement by using the high-resolution structures of the N- and C-terminal domains.
Fig. 3.
Crystal structure of human MITD1. (A) A cartoon representation of the full-length MITD1 dimer in a complex with the CHMP1A MIM1-containing C-terminal tail. Only one MIT domain in the dimer is ordered. MITD1 MIT_C domain dimer has a twofold symmetry. One MIT_C protomer is colored lemon, whereas the other is shown in gray. The dimer is viewed from the top looking down the twofold symmetry axis. Inset shows residues at the interface that are critical for dimer formation (Y132, F221, Y225, and C226) depicted as ball and stick. The elaborate β6/β7 loop involved in dimerization is colored red. (B) A cartoon representation of the MITD1 MIT_C domain protomer (residues 91–242) with labeled secondary structure elements. (C) MITD1 dimer formation tested by yeast two-hybrid analysis. MITD1 dimerizes through its C-terminal domain. Mutation of three residues at the interface (Y132/F221/Y225) to alanine abolishes dimerization.
Crystals of both the full-length MITD1 and the C-terminal domain contain four molecules in the asymmetric unit that are arranged as two dimers, with 2,600 Å2 of accessible surface area buried in the dimer interface. Each monomer of MIT_C consists of a seven-stranded mixed β-sheet and three α-helices (Fig. 3B and Fig. S5). The helices are packed at the dimer interface where they interact with a long, elaborate loop (between strands β6/β7) of the opposing monomer. This loop is conserved in all MIT_C domains (Fig. S5). In the full-length MITD1 crystals, only one molecule per dimer has an ordered MIT domain (Fig. 3A). The orientation of the ordered MIT domain relative to MIT_C is essentially identical in both dimers in the asymmetric unit. The interface between MIT and MIT_C is small (860 Å2 of buried accessible surface area) but defined, involving both hydrophobic interactions and a salt bridge (Fig. S6A). Small angle X-ray scattering shows a good fit to a model with both MIT domains positioned as observed in the crystal structure (Fig. S6B). Taken together, our data suggest that the conformation observed in the crystal exists in solution, although some flexibility of the MIT domains likely does occur.
Consistent with the crystal structures, both the C-terminal domain and full-length MITD1 formed monodisperse dimers in solution as detected by multi-angle laser light scattering (MALLS) (Fig. S6C). In addition, mutation C226D at the observed dimer interface results in an altered mobility and molar mass (Fig. S6C). Similar results were obtained by using a yeast two-hybrid approach, which revealed that full-length MITD1 binds to itself or to MITD191–242 but not to the MIT domain (MITD11–89) (Fig. 3C). Furthermore, mutation of the interface residues Y132, F221, and Y225 to A (MUT YFY) abrogated dimerization without affecting ESCRT-III binding (Fig. 3 A and C).
Despite the MITD1 C-terminal region having no sequence similarity with domains of known function, the crystal structure shows a fold similar to the phospholipase D (PLD) superfamily. This family includes enzymes like PLD, polyphosphate kinase, and the nuclease Nuc, all of which share a common catalytic mechanism involving two copies of a signature motif HxK present either in a single polypeptide or at a homodimer interface (for example, in bacterial nuclease Nuc) (30, 31). The MITD1 C-terminal domain has no HxK motif, but instead it has a single, highly conserved HxR motif (residues 193–195) (Fig. S5) located in an analogous region of the PLD fold (Fig. S7 A and C), suggesting that MITD1 might have an enzymatic activity. However, we were unable to detect PLD activity toward phosphatidylcholine as a substrate (Fig. S7B).
MITD1 C-Terminal Domain Binds to Membranes.
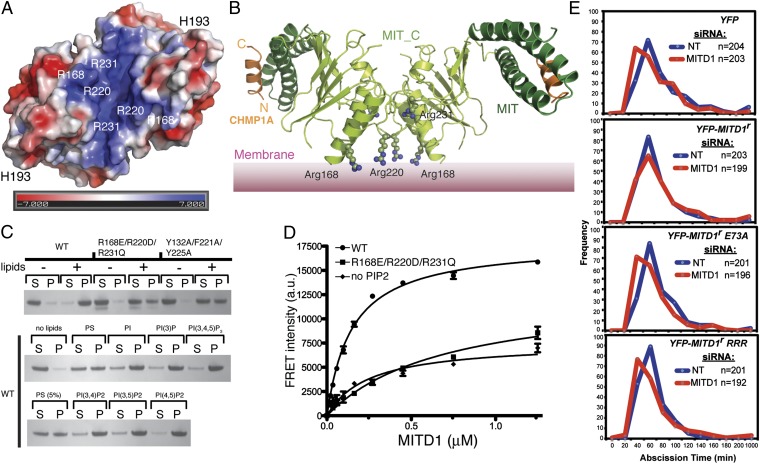
ESCRTs associate on membranes to mediate the scission of membrane bud necks. Accordingly, many components of the ESCRT machinery have an intrinsic affinity for membranes. The surface of MITD1 has a distinct basic patch close to the dimer interface that could mediate binding to acidic lipids (Fig. 4 A and B). This observation led us to investigate whether MITD1 binds to membranes. Both sedimentation assays (Fig. 4C) and protein-lipid FRET assays (Fig. 4D), using PIP2-containing lipid vesicles, showed robust binding of MITD1 to lipid membranes (Kd 0.75 ± 0.07 μM). Mutation of three basic residues in the positively charged surface patch, R168E/R220E/R231E (MUT RRR) (Fig. 4B), greatly decreased binding of MITD1 to liposomes (Fig. 4 C and D), showing a background binding similar to WT MITD1 binding to liposomes lacking PIP2 (Fig. 4D). The same RRR mutation did not alter MITD1 binding to ESCRT-III or dimerization, as shown by yeast two-hybrid assays (Fig. S8A). MITD1 shows a broad preference for phosphoinositides (Fig. 4C). Similar broad phosphoinositide preference has been shown for ESCRT-II and ESCRT-III subunits (32–34).
Fig. 4.
MITD1 binds membranes. (A) A surface representation of the MITD1 MIT_C domain dimer, colored according to electrostatic surface potential, shows a distinct basic patch. The electrostatic potential was calculated by using the APBS plugin in PyMOL and is plotted between ±7 kT/e. (B) Model of the full-length MITD1 dimer on membranes. Three basic residues whose mutation decreases membrane binding are highlighted as ball-and-stick. (C) Lipid sedimentation assays showing MITD1 binding to lipid vesicles. S and P indicate supernatant and pellet after ultracentrifugation and analysis by SDS/PAGE and Coomassie staining. (D) A FRET-based lipid binding assay of wild-type (WT) or mutant (RRR) MITD1. Excitation of intrinsic Trp residues served as the FRET donor and liposomes containing 10% (wt/vol) Dansyl-PS served as the FRET acceptor. The “no PIP2” lipids contain an additional 5% (wt/vol) PS instead of PIP2. (E) Rescue of the premature abscission induced by MITD1 depletion. Cells stably expressing mCherry-Tubulin and YFP (as a control) or siRNA-resistant YFP-MITD1 proteins (YFP-MITD1r, YFP-MITD1r E73A, or YFP-MITD1r RRR) were treated with the indicated siRNA and imaged live. The graphs show the abscission times for cells that divided without becoming multinucleated. The number of cells (n) for each group is indicated per condition and is representative of two independent experiments.
Lastly, the role of membrane binding in midbody recruitment was determined by scoring 100 midbodies from cells, either in telophase or in cytokinesis, using confocal microscopy of HeLa cells stably expressing WT or mutant HA-MITD1. As shown in Fig. S8B, the RRR mutation ablated midbody localization compared with WT MITD1. A similar inhibition of midbody localization was observed in MITD1 mutants that are defective for ESCRT-III binding (E73A) and dimerization (YFY), suggesting that these functions are important for MITD1’s function in cytokinesis. Accordingly, the premature abscission time observed in MITD1 depleted cells is rescued by stably expressing YFP-tagged, siRNA-resistant MITD1 (YFP-MITD1r), whereas replacement of endogenous MITD1 with YFP-MITD1r E73A or YFP-MITD1r RRR mutants is not (Fig. 4E). Surprisingly, the dimerization defective mutant (YFP-MITD1r YFY) rescued MITD1 knockdown, suggesting that the mutant retains sufficient membrane binding to enable the protein function or, alternatively, that YFP is mediating MITD1 dimerization (Fig. S8C).
Discussion
We have shown that MITD1 interacts with the core ESCRT machinery through an N-terminal MIT domain that binds MIMs in a subset of ESCRT-III subunits. At a structural level, this interaction shows close conservation with other MIT-containing proteins despite poor sequence identity. MITD1 has a role in cytokinesis by facilitating ESCRT-III–dependent events in cell division, whereas it is dispensable for other ESCRT-mediated functions.
Our experiments reveal that MITD1 depletion results in cytokinesis failure due to both midbody instability and abscission arrest. Therefore, it is possible that MITD1 coordinates abscission with earlier events in cytokinesis. As cytokinesis progresses, the cytoskeleton needs to undergo a dramatic reorganization before abscission takes place. In particular, actin filaments within the cleavage furrow need to be depolymerized, and microtubules spanning the midbody need to be removed (35, 36). Simultaneously, ESCRT-III forms helical contractile filaments at the intercellular bridge, which enables full constriction at the abscission site (11, 12). Through its ability to bind membranes, MITD1 could stabilize the cortex flanking the furrow after its ingression. Accordingly, loss of MITD1 results in cortical instability, as revealed by increased blebbing and premature abscission in the dividing cells. In this context, MITD1 could coordinate actin removal from the midbody and abscission by ESCRT-III. Interestingly, MITD1 may interact with a guanine nucleotide exchange factor for Rho (MCF2) (37), suggesting a potential link with actin remodelling activities. Other connections between the ESCRT machinery and actomyosin are suggested by a reported ALIX/actin interaction and the remodelling of actin filaments observed in TSG101 knockout cells (38, 39).
The late cytokinesis phenotype observed in MITD1-depleted cells resembles the phenotypes observed upon depletion of other ESCRT proteins. This result, together with the fact that the interaction with ESCRT-III is needed for MITD1’s localization at the midbody, suggests that MITD1 acts in concert with the ESCRT machinery to facilitate abscission. Our structural and biochemical data are consistent with a model whereby MITD1 dimers bind multiple ESCRT-III subunits and lipids simultaneously (Fig. 4B), suggesting that MITD1 could stabilize ESCRT-III filaments and anchor them to the midbody during the late stages of cytokinesis.
The C-terminal domain of MITD1 has an intriguing and unanticipated PLD-like fold. In contrast to a dimeric PLD active site as found in bacterial Nuc (31), dimer formation by MITD1 in solution does not place the putative catalytic histidine residues near each other. Instead, these residues are on opposing ends of MITD1 (separated by approximately 60 Å) (Fig. 4A and Fig. S7A), forming an “inside out” type dimer compared with Nuc (Fig. S7 A and C), suggesting that the observed MITD1 dimer cannot operate by the canonical PLD mechanism. Accordingly, we have been unable to show that MITD1 functions as a PLD enzyme. Interestingly, the MITD1 dimer that we observe exposes the HxR motif as part of a highly conserved surface patch (Fig. S7D). This conserved surface could interact with another as yet unidentified protein to complement the MITD1 active site and yield a catalytically active heterodimer (Fig. S7E). Complementation could also be mediated by MITD1 itself through oligomerisation of the crystallographic dimers on membranes. Alternatively, MITD1 might have evolved from a PLD-type enzyme, but then lost catalytic activity so that it has a purely structural role in regulating ESCRT-III assemblies on membranes.
Materials and Methods
Additional experimental details are provided in SI Materials and Methods, including summary of plasmids (Table S2), siRNA oligos (Table S3), and antibodies (Table S4).
Plasmid Construction.
Human MITD1 was amplified by PCR from IMAGE clone 4509694. All deletions and point mutant derivatives were generated by PCR in a similar way.
Protein Purification.
CHMP1A184–196, full-length MITD1, MITD19–85, or MITD191–242 were cloned in pOPTH. The expression and purification was performed as described in SI Materials and Methods.
Crystallization, Data Collection, Structure Solution, and Refinement.
Full-length human MITD1, MITD19–85 in complex with CHMP1A184–196, and MITD191–242 were crystallized and the structures were solved as described in the SI Materials and Methods.
Imaging.
HeLa cells were seeded onto glass coverslips, fixed, permeabilized, stained with the relevant antibody and imaged as described in SI Materials and Methods.
Time Lapse.
HeLa cells stably expressing mCherry-Tubulin and transfected with siRNA were imaged on a Nikon Ti-Eclipse widefield inverted microscope and analyzed as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the staff of Diamond beamlines I03 and I04 for help with X-ray data collection; and Chris Johnson for help with MALLS. Small angle X-ray scattering at the SIBYLS beamline BL12.3.1 of Advanced Light Source, Lawrence Berkeley National Laboratory, was supported in part by the Department of Energy program Integrated Diffraction Analysis Technologies. R.L.W. was funded by Wellcome Trust Programme Grant 083639/Z/07/Z and Medical Research Council Grant U105184308. J.M.-S. was funded by the Lister Institute for Preventative Medicine. J.M.-S. and M.A. were funded by Wellcome Trust Grant WT093056MA. We also thank the UK National Institute for Health Research Comprehensive Biomedical Research Centre at Guy’s and St. Thomas’s National Health Service Foundation Trust and King’s College London for an equipment grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206839109/-/DCSupplemental.
References
- 1.Williams RL, Urbé S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8(5):355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 2.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: It’s all in the neck. Nat Rev Mol Cell Biol. 2010;11(8):556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21(1):77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science. 2007;316(5833):1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 5.Morita E, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26(19):4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindås AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci USA. 2008;105(48):18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322(5908):1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caballe A, Martin-Serrano J. ESCRT machinery and cytokinesis: The road to daughter cell separation. Traffic. 2011;12(10):1318–1326. doi: 10.1111/j.1600-0854.2011.01244.x. [DOI] [PubMed] [Google Scholar]
- 9.Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322(5901):576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlton JG, Agromayor M, Martin-Serrano J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc Natl Acad Sci USA. 2008;105(30):10541–10546. doi: 10.1073/pnas.0802008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci USA. 2011;108(12):4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guizetti J, et al. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331(6024):1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- 13.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464(7290):864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458(7235):172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morita E, et al. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Natl Acad Sci USA. 2010;107(29):12889–12894. doi: 10.1073/pnas.1005938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlton JG, Caballe A, Agromayor M, Kloc M, Martin-Serrano J. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science. 2012;336(6078):220–225. doi: 10.1126/science.1217180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obita T, et al. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449(7163):735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 18.Stuchell-Brereton MD, et al. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449(7163):740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 19.Lata S, et al. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321(5894):1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connell JW, Lindon C, Luzio JP, Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10(1):42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang D, et al. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat Struct Mol Biol. 2008;15(12):1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lind GE, et al. SPG20, a novel biomarker for early detection of colorectal cancer, encodes a regulator of cytokinesis. Oncogene. 2011;30(37):3967–3978. doi: 10.1038/onc.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renvoisé B, et al. SPG20 protein spartin is recruited to midbodies by ESCRT-III protein Ist1 and participates in cytokinesis. Mol Biol Cell. 2010;21(19):3293–3303. doi: 10.1091/mbc.E09-10-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agromayor M, et al. Essential role of hIST1 in cytokinesis. Mol Biol Cell. 2009;20(5):1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsang HT, et al. A systematic analysis of human CHMP protein interactions: Additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 2006;88(3):333–346. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Kieffer C, et al. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev Cell. 2008;15(1):62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 27.Bajorek M, et al. Biochemical analyses of human IST1 and its function in cytokinesis. Mol Biol Cell. 2009;20(5):1360–1373. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hewitt EW, et al. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 2002;21(10):2418–2429. doi: 10.1093/emboj/21.10.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agromayor M, et al. The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain. Structure. 2012;20(3):414–428. doi: 10.1016/j.str.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gottlin EB, Rudolph AE, Zhao Y, Matthews HR, Dixon JE. Catalytic mechanism of the phospholipase D superfamily proceeds via a covalent phosphohistidine intermediate. Proc Natl Acad Sci USA. 1998;95(16):9202–9207. doi: 10.1073/pnas.95.16.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stuckey JA, Dixon JE. Crystal structure of a phospholipase D family member. Nat Struct Biol. 1999;6(3):278–284. doi: 10.1038/6716. [DOI] [PubMed] [Google Scholar]
- 32.Im YJ, Hurley JH. Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev Cell. 2008;14(6):902–913. doi: 10.1016/j.devcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teo H, et al. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125(1):99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 34.Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem. 2005;280(13):12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- 35.Dambournet D, et al. Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nat Cell Biol. 2011;13(8):981–988. doi: 10.1038/ncb2279. [DOI] [PubMed] [Google Scholar]
- 36.Guizetti J, Gerlich DW. Cytokinetic abscission in animal cells. Semin Cell Dev Biol. 2010;21(9):909–916. doi: 10.1016/j.semcdb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Papin J, Subramaniam S. Bioinformatics and cellular signaling. Curr Opin Biotechnol. 2004;15(1):78–81. doi: 10.1016/j.copbio.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 38.Morris CR, Stanton MJ, Manthey KC, Oh KB, Wagner KU. A knockout of the Tsg101 gene leads to decreased expression of ErbB receptor tyrosine kinases and induction of autophagy prior to cell death. PLoS ONE. 2012;7(3):e34308. doi: 10.1371/journal.pone.0034308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan S, et al. Involvement of the conserved adaptor protein Alix in actin cytoskeleton assembly. J Biol Chem. 2006;281(45):34640–34650. doi: 10.1074/jbc.M602263200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.