Abstract
The Antarctic and Arctic regions offer a unique opportunity to test factors shaping biogeography of marine microbial communities because these regions are geographically far apart, yet share similar selection pressures. Here, we report a comprehensive comparison of bacterioplankton diversity between polar oceans, using standardized methods for pyrosequencing the V6 region of the small subunit ribosomal (SSU) rRNA gene. Bacterial communities from lower latitude oceans were included, providing a global perspective. A clear difference between Southern and Arctic Ocean surface communities was evident, with 78% of operational taxonomic units (OTUs) unique to the Southern Ocean and 70% unique to the Arctic Ocean. Although polar ocean bacterial communities were more similar to each other than to lower latitude pelagic communities, analyses of depths, seasons, and coastal vs. open waters, the Southern and Arctic Ocean bacterioplankton communities consistently clustered separately from each other. Coastal surface Southern and Arctic Ocean communities were more dissimilar from their respective open ocean communities. In contrast, deep ocean communities differed less between poles and lower latitude deep waters and displayed different diversity patterns compared with the surface. In addition, estimated diversity (Chao1) for surface and deep communities did not correlate significantly with latitude or temperature. Our results suggest differences in environmental conditions at the poles and different selection mechanisms controlling surface and deep ocean community structure and diversity. Surface bacterioplankton may be subjected to more short-term, variable conditions, whereas deep communities appear to be structured by longer water-mass residence time and connectivity through ocean circulation.
Keywords: bipolar, biodiversity, next-generation sequencing, microbial ecology
Global studies of how microbial communities vary in space and along environmental gradients have highlighted key questions about the factors that control the distribution of microbes on earth (1, 2). Polar environments remain poorly studied even though they could help to identify patterns of bacterial biogeography and clarify the mechanisms responsible for them. Both of Earth’s polar ocean systems have been essentially isolated for millennia by physical barriers that limit water exchange with the other oceans (the Arctic Ocean by land masses since >60 Ma and the Southern Ocean by a strong current system since ∼20–40 Ma) (3). However, both oceans are subject to parallel extreme physical forces, such as solar irradiance that spans from 24 h of sun in the polar summer to 24 h of darkness during the polar winter. At the onset of the polar winter, low temperatures result in sea ice formation, whereas during the polar spring, ice algal blooms followed by ice melt and phytoplankton production support higher food webs at both poles (4, 5).
Despite similar climate drivers acting on the resident biota, the two polar oceans are dissimilar in several important aspects. Most notably are differences in freshwater supply to these systems. Although glacial meltwaters flow into the Southern Ocean and, to a lesser extent, the Arctic, several large river systems with large continental drainage basins profoundly influence the hydrology of the Arctic Ocean. Furthermore, the waters of the Southern Ocean completely surround the continent of Antarctica and are driven by the largest and strongest current system in the World Ocean, the Antarctic Circumpolar Current (ACC). Conversely, the Arctic Ocean is surrounded by Eurasian and North American land masses, with its basin perennially covered by ice and divided by a midbasin ridge (6).
The few direct comparisons of microbial life between the polar oceans have focused on specific taxa such as the haptophyte Phaeocystis (7), the foraminiferan Neogloboquadrina pachyderma (8), and bacteria originally isolated from ice such as Polaribacter irgensii (9) and Shewanella frigidimarina (10). Community-wide comparisons using small subunit (SSU) rRNA gene surveys of planktonic Archaea reported sequences that are 99% identical from the two poles (11). However, a latitudinal transect of the Pacific Ocean using a community fingerprinting method indicated differences in the bacterial and eukaryal communities from the two poles as well as from tropical and temperate regions (12). Likewise, sea ice microbial communities at the two poles harbor closely related organisms although differ significantly in the representation of some groups (9, 13). Finally, in silico comparison of available SSU rRNA gene sequences from marine planktonic bacteria indicate bipolar distributions of some ribotypes (14). In summary, despite the increasingly widespread application of molecular biological approaches to planktonic communities over the last 20 y, thorough comparisons of microbial communities at the two poles are inconclusive because of a lack of comparable datasets and poor coverage. This limitation highlights the lack of a standardized approach and reflects the challenges of sampling over a range of locations and depths in both polar zones.
Through a collaborative effort initially motivated by the International Census of Marine Microbes (15) and opportunities provided by the International Polar Year, we tested the hypothesis that bacterioplankton communities are the same at both poles. We used a suite of 20 Southern Ocean and 24 Arctic Ocean samples from both surface and deep waters. Samples were processed and analyzed using an identical approach based on pyrosequencing of the V6 region of the SSU rRNA gene (16). We specifically compared samples from coastal and open oceans and between winter and summer, to test whether or how environmental conditions and dispersal shape communities in the polar oceans. Finally, we analyzed an additional 48 samples from lower latitudes (Fig. 1) to investigate the polar signal in global marine bacterial biogeography.
Fig. 1.
Polar oriented maps of sample locations in the Southern (A) and Northern (B) hemispheres, including Southern Ocean (pink), Arctic (blue), and lower latitudes (gray).
Results
Similarity Among Communities.
All Southern Ocean (20), Arctic Ocean (24), and lower latitude (48) datasets (Fig. 1 and Table S1) were randomly resampled to ensure the number of sequence tags from each sample were equal to the number of tags from the sample with the fewest sequences (9,107). This resulted in 837,844 sequence tags (400,708 from the polar samples) falling into 26,902 OTUs (0.03 distance threshold) for all samples, with 11,441 from the polar datasets. Hierarchical clustering based on Bray–Curtis dissimilarities revealed that the global pattern of pelagic bacterial diversity was driven by two main factors: vertical distribution and oceanic subregion; analysis of similarity (ANOSIM) showed both factors to be highly significant (UniFrac and P test significance, both P < 0.001) (Fig. 2). Additionally, the influence of environmental parameters on global changes in bacterial community structure was statistically analyzed by canonical correspondence analysis (CCA). However, few variables were available across all datasets and the best model using temperature, depth, and latitude explained only 17.2% of the variance (P < 0.05). We also inspected the relationship between latitude and the Chao1 richness estimator and between temperature and Chao1 for the surface (defined as 0–40 m) and deep (defined as samples >200 m) datasets (Fig. S1). The trends for both the surface and the deep samples generally decreased with latitude although were not significant (Spearman’s R = −0.19, P = 0.20, n = 44; and Spearman’s R = −0.17, P = 0.34, n = 33, respectively). The relationships with temperature were slightly positive, although not significant [surface dataset (n = 44): R = 0.12, P = 0.43; deep dataset (n = 33): R = 0.14, P = 0.45].
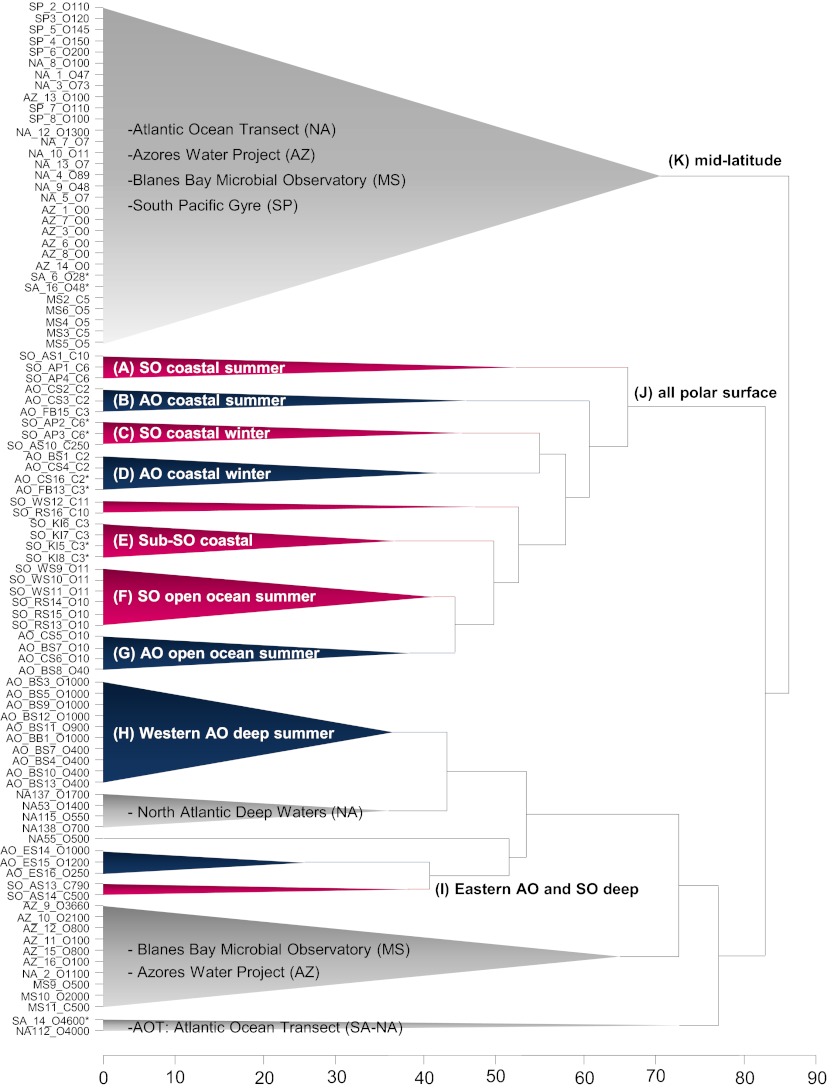
Fig. 2.
Unweighted pair group method with arithmetic mean (UPGMA) dendrogram based on Bray–Curtis dissimilarities of V6 16 rRNA tags from pelagic samples taken in Arctic (blue), Southern Ocean (purple), and in lower latitudes (gray). The x axis represents Bray–Curtis dissimilarity. Ecosystem clusters are represented with labels and letters A through I. Sample names at left are encoded as ocean region, VAMPS code number, open (O) or coastal (C) sites, and water depth in meters. Polar ocean regions in the Southern Ocean (SO) are Amundsen Sea (AS), Antarctic Peninsula (AP), Kerguelen Islands (KI), Ross Sea (RS), and Weddell Sea (WS). In the Arctic Ocean (AO), regions sampled are Baffin Bay (BB), Beaufort Sea (BS), Chukchi Sea (CS), East Siberian Sea (ES), and Franklin Bay (FB). A sample name followed by an asterisk refers to winter sampling.
Within the polar surface group (cluster J; Fig. 2), Southern and Arctic Ocean bacterioplankton communities always clustered apart from each other. Southern and Arctic Ocean summer coastal communities were 65% dissimilar (clusters A and B), winter coastal communities were 55% dissimilar (clusters C and D), and open ocean communities were 46% dissimilar (clusters F and G). Surface layer polar communities were highly dissimilar (86%) to those from lower latitudes (clusters J and K; Fig. 2).
Bacterial communities in polar deep waters were also different (72% dissimilar) from those in temperate zones. Water-mass connectivity was also evident; for example, the deep Western Arctic communities (cluster H) clustered closely with bacteria (44% dissimilar) from the North Atlantic Ocean sites, which were influenced by Labrador Sea water masses. Furthermore, bacterial communities from the deep Southern Ocean cluster with communities retrieved from the Eurasian Arctic (cluster I), indicating that there were also fewer differences between poles for deep samples. However, both communities clustered apart from each other (41% dissimilar; Fig. 2).
Phylogenetic Diversity Accumulation in Surface and Deep Communities.
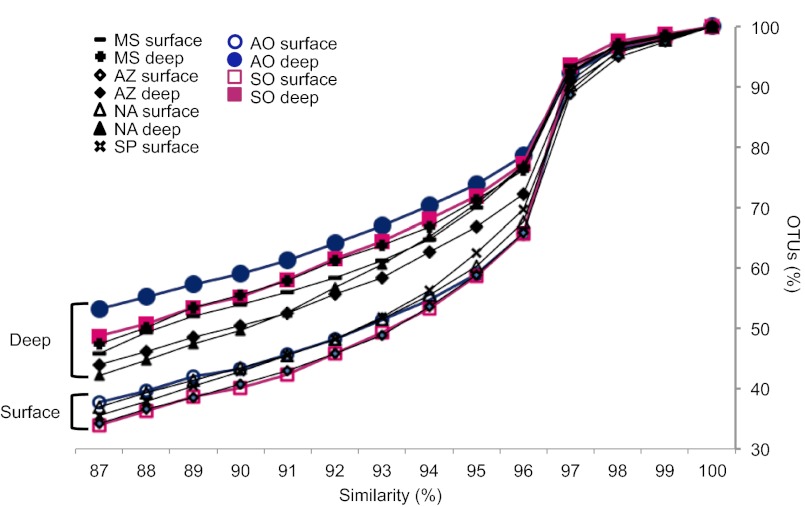
To further test factors that could influence the biogeography of marine bacterial communities, we calculated phylogenetic diversity accumulation curves (Fig. 3) in which the number of OTUs vs. the sequence similarity threshold used to define the OTUs are plotted. The curve shape indicates the degree of phylogenetic relatedness (or distance) among taxa within each community. If taxa in a community have low SSU rRNA gene sequence divergence, shown by shorter branch lengths in the phylogenetic tree, the diversity accumulation curve is expected to indicate a higher percentage of OTUs at high similarity thresholds and fewer OTUs at the lower similarity thresholds than for communities containing more distantly related taxa. Interestingly, differences in the diversity accumulation curves were largest between surface and deep communities (ANOSIM R = 0.36; P < 0.0001; n = 92), with significantly fewer OTUs at low SSU rRNA gene sequence similarity in the surface compared with deep waters (Fig. 3). In the surface samples, a rapid increase in the number of OTUs appeared at the sequence similarity threshold ≥97% identity (Fig. 3, break in slopes). This indicates lower divergence in the SSU rDNA gene sequences in surface compared with deep waters. Deep communities were more variable than surface communities in their diversity accumulation curve shapes, with a greater percentage of OTUs remaining at low sequence similarity thresholds. The polar deep samples showed the highest degree of diversity accumulation.
Fig. 3.
Diversity accumulation in deep vs. surface communities represented by the percentage of OTUs in a dataset when the sequence similarity threshold used to define OTUs increases. Datasets included Mediterranean Sea (MS), Azores (AZ), North Atlantic (NA), South Pacific (SP), Arctic Ocean (AO) and Southern Ocean (SO).
Unique and Shared OTUs Between Polar Microbial Communities.
When we compared the surface polar oceans to each other (at a 0.03 distance threshold), 78% of the Southern Ocean and 70% of the Arctic Ocean OTUs were exclusive to each pole. For the deep samples, 45% of the OTUs were unique to the Southern Ocean, whereas 85% were unique to the Arctic Ocean where more deep samples were surveyed (Fig. S2). When the entire dataset including lower latitude stations was considered, 70% of surface Southern Ocean OTUs and 39% of deep Southern Ocean OTUs were never found in the Arctic or the temperate oceans. Similarly for the Arctic Ocean, 61% of surface and 60% of deep were only found in the Arctic (Fig. S3). Furthermore, 59% of the shared polar surface and 26% of the shared polar deep OTUs were not present in the lower latitude samples. Only 2% of OTUs were found in all surface waters and 4% of OTUs were found in all deep waters.
Most OTUs in both polar oceans belonged to Gammaproteobacteria, Alphaproteobacteria, and Flavobacteria. However, there were some differences for other groups. For example, Betaproteobacteria, Actinobacteria, and Acidobacteria were more common in the Arctic (3–4% each; Fig. S2) than in the Southern Ocean (less than 2% each). Compared with surface, deep waters from both polar regions had a lower proportion of Flavobacteria (8% at deep and 14% in surface) and a higher proportion of Deltaproteobacteria (9% at deep and 3% in surface).
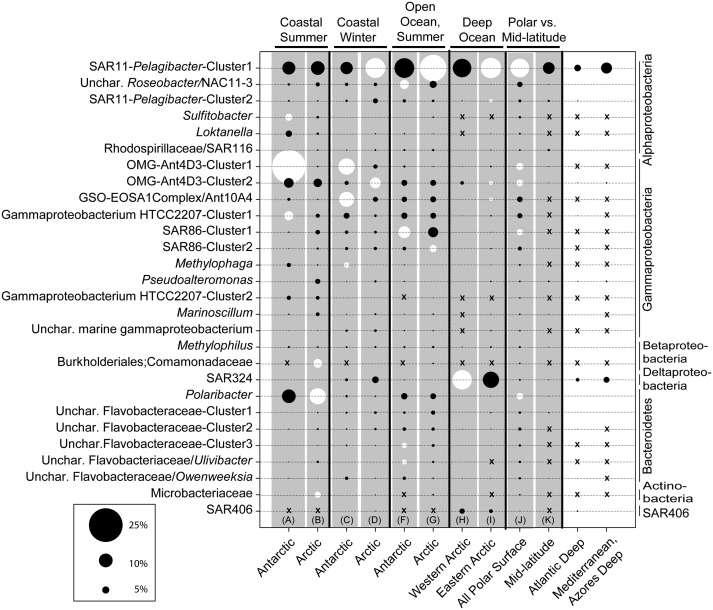
Samples were then grouped at the level of major ecosystems derived from the clusters in Fig. 2. Using the polar data only, we identified the most frequent OTUs (28 in total) that accounted for ≥5% of the sequences in each of the polar ecosystem clusters: Coastal Summer, Coastal Winter, Open Ocean Summer, and Deep Arctic Ocean Surface (Fig. 4). Independently, we identified the OTUs primarily responsible for dissimilarities between pairs of these clusters (e.g., Southern Ocean vs. Arctic coastal summer) as well as between polar and mid latitude samples (Fig. 4) using similarity percentage analysis (SIMPER). In most cases, these OTUs were a subset of the 28 most represented OTUs (Fig. 4). Among those top 28 OTUs, 14 were unique to polar areas and not found in the lower latitude samples; see the comparison of all polar surface with midlatitude clusters J vs. K (Fig. 4). The most important polar bacteria OTUs according to SIMPER that influenced the dissimilarity between ecosystem clusters were SAR11-Pelagibacter-Cluster1, OMG-Ant4D3-Cluster1 and -2, and SAR86-Cluster1, and Polaribacter. Further inspection revealed differences between poles for different environments (coastal and open ocean) and seasons (summer and winter). In summer coastal waters, OMG-Ant4D3-Cluster1, Gammaproteobacterium HTCC2207-Cluster 1, and Sulfitobacter were more abundant in the Southern Ocean, while Polaribacter, Burkholderiales, and Microbacteriaceae OTUs were more abundant in the Arctic (Fig. 4). In winter, there were also large differences between the poles, with OMG-Ant4D3-Cluster1, GSO-EOSA1Complex/Ant10A4, and Methylophaga explaining the majority of the difference for the Southern Ocean, whereas SAR11-Pelagibacter-Cluster1 and OMG-Ant4D3-Cluster2 were more abundant in the Arctic Ocean. Polar winter coastal communities had a higher frequency of SAR324 and uncultivated Gammaproteobacteria OTUs compared with summer samples from the same regions.
Fig. 4.
Bacterial OTUs associated with the polar ecosystem clusters identified in Fig. 2 (clusters A to H). SSU rRNA gene tags (distance of 0.03) were averaged across each ecosystem cluster (3–29 samples per cluster) and summed across the eight polar ecosystems. Shown are OTUs representing 5% or greater in each compared ecosystem cluster (totaling 28 OTUs), where the circle size corresponds to the relative average abundance the OTU in each cluster. OTUs assigned to the highest taxonomic level possible using a Bayesian classification tool (RDP) BLAST and sequence alignments to polar SSU rRNA gene clone libraries, are grouped phylogenetically on the vertical dimension. OTUs with average relative abundances < 0.01 (rare OTUs in a fraction of the samples in an ecosystem cluster) are indicated with an x, whereas very low values (0.01–0.5) appear as a continuation of dashes. Pairwise comparisons of ecosystem clusters (delimited with vertical black lines and gray background) were conducted using SIMPER to determine OTUs contributing to dissimilarity between the clusters. Dominant tags (top five to six OTUs) that contributed to dissimilarity between two clusters are in white. For example, the six frequent OTUs explaining the dissimilarity between the coastal surface summer samples in the Antarctic and the Arctic included three Antarctic OTUs: Sulfitobacter, OMGAnt4D3-cluster1, Gammaproteobacterium HTTC 2207-Cluster1, and three Arctic OTUs: Burkholderiales, Polaribacter, and Microbacteriaceae); the discerning OTUs are often distributed between the two clusters being compared.
Polar summer coastal communities were easily distinguished from summer open ocean communities [Figs. 2 and 4; similarity profile testing (SIMPROF) analysis]. For example, the coastal communities were dominated by the OMG-Ant4D3-Cluster1, along with fewer Polaribacter-, Sulfitobacter-, and Loktanella-affiliated OTUs, whereas the open ocean samples contained a higher proportion of SAR11, uncharacterized Roseobacter NAC11-3, and SAR86-Cluster1 and -2. Summer polar surface open ocean samples (Fig. 4, columns F and G) could be distinguished by the uncharacterized Roseobacter/NAC11-3 OTU, SAR86-Cluster1, uncharacterized Flavobacteriaceae-Cluster3, and an Ulvibacter-related OTU, which were dominant in the Southern Ocean, whereas the SAR11-Pelagibacter-Cluster1 and SAR86-Cluster2 were more abundant in the Arctic. The deep polar ocean clusters (Fig. 4, columns H and I) differed the most because of the SAR324 OTU being more abundant in the Western Arctic than in the Eastern Arctic and Southern Ocean. The SAR11-Pelagibacter-Cluster1 and -Cluster2, OMG-Ant4D3-Cluster2, and GSO-EOSAA1Complex/Ant10A4 OTUs also contributed to the dissimilarity between the clusters. The typical deep water clade SAR406 was equally represented at both poles.
Discussion
The two polar oceans share similar extreme environmental conditions that are very different from other oceans. Moreover, polar ocean circulation patterns and cold seawater temperatures have been present for up to 25 million years (3). Few studies have compared bacterioplankton communities at the poles; one transect along the axis in the Pacific Ocean from the Arctic to the Southern Ocean used a DNA fingerprinting technique, which indicated differences between the two poles, as well as mid latitudes (12). That study detected, but did not identify, ∼11 taxa per sample and covered only a small area of each polar zone. Because seasonal, regional, and ecological differences also influence bacterial communities, the extent of differences between the poles required deeper, more extensive, sampling coverage to include the less abundant and rare taxa that make up a substantial portion of bacterial communities in the oceans. Our analysis, based on over 830,000 V6 rRNA gene sequences from 92 samples distributed among multiple polar and lower latitudes areas at different times of the year and including surface and deep waters, showed that there were profound differences between Southern and Arctic Ocean bacterioplankton communities.
Limited dispersal capacity could account for differences between polar areas. If bacteria do not remain viable during long-range transport, communities would differentiate from each other according to a classical allopatric model. However, gene flow between the poles has been suggested for other microorganisms and benthic foraminifera (17) and very similar (≥99%) SSU rRNA gene sequences belonging to an ice dinoflagellate (18), Planctomycetales (9), other bacteria (14), and marine group I Crenarchaeota (11) are suggestive of bipolar distributions. We found that about 15% of the OTUs were common to both poles, including representatives of the dominant Alphaproteobacteria, Gammaproteobacteria, and Flavobacteria groups that are cosmopolitan in marine waters. Several OTUs matched cultivated species from sea ice, including the psychrophile Polaribacter sp. and the psychrophilic gas-vacuolate bacterium Octadecabacter sp. (19). The potential bipolar distribution of a subset of the bacterioplankton is intriguing, and our results point toward lineages that warrant further investigation. At the same time, we found that the majority (85%) of the OTUs appeared to have pole-specific distributions, suggesting incomplete dispersal between the poles; geographical isolation, physiological characteristics, and ecological traits could work together inhibit widespread bacterioplankton dispersal.
Environmental filters may act strongly at the two poles with greater community differences occurring where selection is greatest. One difference between the poles is the terrestrial influence on coastal areas. Arctic surface waters are modified by large rivers that bring in significant sediment loads and dissolved organic carbon, whereas freshwater flows into the Southern Ocean are smaller and mostly from glaciers. The different qualities and quantities of freshwater drive differences in nutrient regimes. For example, we would expect greater differences in coastal than in the open ocean when comparing the two poles. Accordingly, although coastal surface communities of the Arctic and Southern Oceans were more similar to each other compared with the open oceans, they were distinct (65% Bray–Curtis dissimilarity in summer) from each other. Indeed, there was greater representation of some freshwater bacteria including Betaproteobacteria and Actinobacteria (20) in the surface Arctic Ocean. We found fewer differences between the two polar coastal regions in winter (55% Bray–Curtis dissimilarity), consistent with the lack of river runoff in the Arctic during winter, and presumed environmental convergence of polar winter conditions. Our results support the notion that environmental conditions structure bacterial communities of surface polar waters strongly, in particular in the coastal areas.
Although our results are consentient with the emerging global view of horizontal differences in bacterial communities across oceans (e.g., refs. 2 and 21), vertical structure is more striking and evident in essentially all other oceans including the Pacific and Atlantic Oceans (22, 23) and the Southern Ocean (24). In the absence of light and more quiescent conditions, environmental drivers may be weaker compared with those in surface waters. In support of this, we found that surface communities differed more than deep communities (Fig. 2): a finding consistent with environmental rather than geographical isolation being the main determinant of the bacterial community composition in surface waters. In addition, when we addressed the question of whether there was a relationship between community richness and latitude, as has been reported by others (e.g., refs. 14 and 25) with lower levels of resolution or depth, we found little support for a relationship. Admittedly, the coverage, in even this large study, is not adequate to address this intriguing question sufficiently. The data analyzed here offer a window into significant variation within latitude (e.g., 40° and 64°) and between seasons, in particular, at the high latitudes, where winter samples were more diverse than their summer counterparts.
An intriguing aspect of deep water bacterioplankton in this study was the geographic patterns of similarity among communities. Deep bacterial communities from the East Siberian Sea clustered with deep Southern Ocean communities from the Amundsen Sea but not with other deep Arctic communities (cluster I in Fig. 2). The deep community connection between poles could be explained by the export of deep water formed in the Eurasian Arctic Basin (Eastern Arctic), which exits the Arctic Ocean east of Greenland through the 2,600-m-deep Fram Strait. This dense water moves relatively fast through the Atlantic Ocean along the deep conveyor belt, reaching the Southern Ocean on the order of hundreds of years (26). Because environmental conditions in this water mass would be relatively stable, the deep Arctic bacterial communities could be maintained.
It is also noteworthy that even with this larger dataset, the eastern and western Arctic samples did not cluster together (27). These deep basins are separated by the Lomonosov Ridge, which rises to 650 m below the surface of the ocean (6), thereby separating the two deep Arctic Ocean Basins. The Canadian Arctic Ocean bacterial communities from the deep Beaufort Sea and deep Baffin Bay clustered together along with North Atlantic Deep Water that is derived from Labrador Current waters (cluster H in Fig. 2; Western Arctic Deep Summer). The deep bacterial communities from the Canadian Arctic were from the Beaufort Gyre, the cold, dense waters of which eventually exit the Arctic Ocean through Baffin Bay. These waters are then exported from Baffin Bay to the North Atlantic Water Mass sampled here (28). This global connectivity implies that the biogeography of deep-water communities may be largely controlled by ocean circulation. Deep polar communities were dominated by OTUs in the SAR324 cluster of Deltaproteobacteria, a typical deep ocean phylotype (e.g., ref. 29). Notably, this OTU and, to a much smaller degree, SAR406 (average 1.8–4.7% compared with 0.1–0.3%) was also recovered in surface winter samples from both poles, suggesting at least some microbes can persist in cold dark waters irrespective of depth.
Surface and deep communities had different phylogenetic diversity patterns, as defined by diversity accumulation curves (30) based on the slowly evolving SSU rRNA gene. The curves provide a general framework for understanding patterns of phylogenetic diversity. Despite the relatively short length of the V6 region of the SSU rRNA gene, we found that levels of sequence divergence differ substantially between the surface and deep pelagic environments, with less divergent SSU rRNA gene pools in surface communities. This is consistent with previous reports of high phylogenetic turnover of surface communities at the global scale (31). Such a depth difference argues for different speciation patterns and perhaps different evolutionary pressures shaping community structure in surface vs. the deep waters, because phylogenetic diversity should be lower in a region with recent and more rapid speciation events and higher in a region with slower speciation rates (32). We propose that the differences in phylogenetic diversity between surface and deep polar communities are attributable to (i) rapid speciation at the surface as consequence of dynamic surface condition, in contrast to (ii) spatially and temporally constant environmental conditions over longer timescales in the deep sea. These longer scales would allow more time to select for phenotypes adapted to the specific conditions, with concomitant loss of less adapted phylotypes. Phylogenetically distant populations would be expected to co-occur if most speciation followed allopatric speciation, whereas co-occurrences of phylogenetically closely related populations would be controlled by sympatric speciation (33). The more divergent OTUs found in deep waters suggest communities shaped by allopatry, whereas the surface communities, with less divergence between OTUs (maximum differences at the poles), are more likely to be sympatric.
Our bipolar comparisons highlighted global patterns of biogeography and diversity in pelagic bacterioplankton. A notable lack of correlation between latitude and richness warrants additional attention, and further research is needed to address the ecological and evolutionary processes underlying these patterns. Our comprehensive global dataset, encompassing diverse oceanic regions, suggests that surface communities are driven by environmental selection, whereas deep communities are more constrained by historical events and connected through oceanic circulation, providing evidence for biogeographically defined communities in the global ocean.
Materials and Methods
All samples (Fig. 1) were part of the International Census of Marine Microbes (ICoMM), which developed sample and metadata submission, sample processing, and data analysis pipelines, ensuring similar treatment for all samples. The ICoMM hypervariable V6 region SSU rRNA pyrotag dataset and geospatial parameters are available at http://icomm.mbl.edu/microbis and are reported in Table S1. All Southern Ocean samples except the Amundsen Sea vertical profile were coordinated by the Census of Antarctic Marine Life (CAML) with the goal of representing spatial, temporal, and latitudinal variability across four subregions of the Southern Ocean including the Weddell and Ross Seas, the Antarctic Peninsula, and Kerguelen Islands. Samples collected over the annual cycle in the Antarctic Peninsula and Kerguelen Islands have been described recently (34). The Amundsen Sea profile was collected on the same cruise as described recently (35). We classified samples based on whether they were from the Southern or the Arctic Ocean, coastal or open ocean (limit of 200 km from the coast), surface (<40 m) or intermediate and deep waters (>200 m), summer or winter. The Arctic Ocean samples included in this study have been reported previously (30, 36, 37).
Details concerning the methodological approach and data analysis pipeline are described in SI Materials and Methods. In brief, the approach included data resampling, and a pipeline using Mothur (38) including multiple alignment, preclustering, distance matrix calculation, and clustering to define OTUs at similarity thresholds ranging from 87% to 100% sequence identity. Details of the statistical approaches used are also described in the supplementary information and included hierarchical agglomerative clustering of Bray–Curtis similarities, SIMPROF, one-way ANOSIM, similarity percentage (SIMPER), UniFrac, canonical correspondence analysis (CCA), and Spearman rank correlations. All sequence tags used are publicly available at http://vamps.mbl.edu as trimmed FASTA sequences (used in our analysis) or as ICoMM processed datasets.
Supplementary Material
Acknowledgments
We thank the members of field teams, shipboard crews, and logistics support personnel from all national polar programs involved in sample collection, without whom this study would not have been possible. The Census of Antarctic Marine Life, funded by the Sloan Foundation, facilitated this collaboration. Pyrosequencing was provided by the International Census of Marine Microbes (ICoMM) with financial support from a W. M. Keck Foundation award to the Marine Biological Laboratory in Woods Hole. Funding to support sample collection was provided by the Institut Français pour la Recherche et la Technologie Polaires (J.-F.G.); the Spanish Ministry of Education and Science (C.P.-A.); the New Zealand International Polar Year-Census of Antarctic Marine Life Project [Phases 1 (So001IPY) and 2 (IPY2007-01); to E.W.M.); the Natural Sciences and Engineering Council (NSERC) of Canada (C.L.); National Science Foundation Grants OPP-0124733 (to D.L.K.), ANT-0632389 (to A.E.M.), and ANT-0741409 (to P.L.Y.); and the Swedish Polar Research Secretariat (S.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: FASTA and processed DNA sequence data have been deposited in the the Visualization and Analysis of Microbial Population Structures (VAMPS) database, http://vamps.mbl.edu.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208160109/-/DCSupplemental.
References
- 1.Nemergut DR, et al. Global patterns in the biogeography of bacterial taxa. Environ Microbiol. 2011;13:135–144. doi: 10.1111/j.1462-2920.2010.02315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zinger L, et al. Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS ONE. 2011;6:e24570. doi: 10.1371/journal.pone.0024570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegert MJ, et al. Recent advances in understanding Antarctic climate evolution. Antarct Sci. 2008;4:313–325. [Google Scholar]
- 4.Karl DM. In: Antarctic Microbiology. Friedmann EI, editor. New York: Wiley; 1993. pp. 1–63. [Google Scholar]
- 5.Wheeler PA, et al. Active cycling of organic carbon in the central Arctic Ocean. Nature. 1996;380:697–699. [Google Scholar]
- 6.Cochran JR, Edwards MH, Coakley BJ. Morphology and structure of the Lomonosov Ridge, Arctic Ocean. Geochem Geophys Geosyst. 2006;7:Q05019. [Google Scholar]
- 7.Verity PG, et al. Current understanding of Phaeocystis ecology and biogeochemistry, and perspectives for future research. Biogeochemistry. 2007;83:311–330. [Google Scholar]
- 8.Darling KF, Kucera M, Wade CM. Global molecular phylogeography reveals persistent Arctic circumpolar isolation in a marine planktonic protist. Proc Natl Acad Sci USA. 2007;104:5002–5007. doi: 10.1073/pnas.0700520104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Staley JT, Gosink JJ. Poles apart: Biodiversity and biogeography of sea ice bacteria. Annu Rev Microbiol. 1999;53:189–215. doi: 10.1146/annurev.micro.53.1.189. [DOI] [PubMed] [Google Scholar]
- 10.Zeng Y, Zheng T, Yu Y, Chen B, He J. Relationships between Arctic and Antarctic Shewanella strains evaluated by a poly- phasic taxonomic approach. Polar Biol. 2010;33:531–541. [Google Scholar]
- 11.Bano N, Ruffin S, Ransom B, Hollibaugh JT. Phylogenetic composition of Arctic Ocean archaeal assemblages and comparison with Antarctic assemblages. Appl Environ Microbiol. 2004;70:781–789. doi: 10.1128/AEM.70.2.781-789.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baldwin AJ, et al. Microbial diversity in a Pacific Ocean transect from the Arctic to Antarctic circles. Aquat Microb Ecol. 2005;41:91–102. [Google Scholar]
- 13.Brinkmeyer R, et al. Diversity and structure of bacterial communities in Arctic versus Antarctic pack ice. Appl Environ Microbiol. 2003;69:6610–6619. doi: 10.1128/AEM.69.11.6610-6619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pommier T, Pinhassi J, Hagström Å. Biogeographic analysis of ribosomal RNA clusters from marine bacterioplankton. Aquat Microb Ecol. 2005;41:79–89. [Google Scholar]
- 15.Amaral-Zettler L, et al. In: Life in the World’s Oceans: Diversity, Distribution, and Abundance. McIntyre AD, editor. Oxford: Wiley-Blackwell; 2010. pp. 223–245. [Google Scholar]
- 16.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pawlowski J, et al. Bipolar gene flow in deep-sea benthic foraminifera. Mol Ecol. 2007;16:4089–4096. doi: 10.1111/j.1365-294X.2007.03465.x. [DOI] [PubMed] [Google Scholar]
- 18.Montresor M, Lovejoy C, Orsini L, Procaccini G, Roy S. Bipolar distribution of the cyst-forming dinoflagellate Polarella glacialis. Polar Biol. 2003;26:186–194. [Google Scholar]
- 19.Gosink JJ, Herwig RP, Staley JT. Octadecabacter arcticus gen. nov., sp. nov., nonpigmented, psychrophilic gas vacuolate bacteria from polar sea ice and water. Syst Appl Microbiol. 1997;20:356–365. [Google Scholar]
- 20.Crump BC, et al. Circumpolar synchrony in big river bacterioplankton. Proc Natl Acad Sci USA. 2009;106:21208–21212. doi: 10.1073/pnas.0906149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghiglione JF, Larcher M, Lebaron P. Spatial and temporal scales of variation in bacterioplankton community structure in the NW Mediterranean Sea. Aquat Microb Ecol. 2005;40:229–240. [Google Scholar]
- 22.Lee SH, Fuhrman JA. Spatial and temporal variation of natural bacterioplankton assemblages studied by total genomic DNA cross-hybridization. Limnol Oceanogr. 1991;36:1277–1287. [Google Scholar]
- 23.Field KG, et al. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray AE, et al. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuhrman JA, et al. A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA. 2008;105:7774–7778. doi: 10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aagaard K, Swift JH, Carmack EC. Thermohaline circulation in the Arctic Mediterranean Seas. J Geophys Res. 1985;90:4833–4846. [Google Scholar]
- 27.Galand PE, Potvin M, Casamayor EO, Lovejoy C. Hydrography shapes bacterial biogeography of the deep Arctic Ocean. ISME J. 2010;4:564–576. doi: 10.1038/ismej.2009.134. [DOI] [PubMed] [Google Scholar]
- 28.Beszczynska-Möller A, Woodgate RA, Lee C, Melling H, Karcher M. A synthesis of exchanges through the main Oceanic gateways to the Arctic Ocean. Oceanography (Wash DC) 2011;24:82–99. [Google Scholar]
- 29.Gordon DA, Giovannoni SJ. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific oceans. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barberán A, Fernández-Guerra A, Auguet JC, Galand PE, Casamayor EO. Phylogenetic ecology of widespread uncultured clades of the Kingdom Euryarchaeota. Mol Ecol. 2011;20:1988–1996. doi: 10.1111/j.1365-294X.2011.05057.x. [DOI] [PubMed] [Google Scholar]
- 31.Pommier T, Douzery EJ, Mouillot D. Environment drives high phylogenetic turnover among oceanic bacterial communities. Biol Lett. 2012;8:562–566. doi: 10.1098/rsbl.2011.0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson MTJ, Stinchcombe JR. An emerging synthesis between community ecology and evolutionary biology. Trends Ecol Evol. 2007;22:250–257. doi: 10.1016/j.tree.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 33.Whitaker RJ. Allopatric origins of microbial species. Philos Trans R Soc Lond B Biol Sci. 2006;361:1975–1984. doi: 10.1098/rstb.2006.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghiglione JF, Murray AE. Pronounced summer to winter differences and higher wintertime richness in coastal Antarctic marine bacterioplankton. Environ Microbiol. 2012;14:617–629. doi: 10.1111/j.1462-2920.2011.02601.x. [DOI] [PubMed] [Google Scholar]
- 35.Alonso-Sáez L, Andersson A, Heinrich F, Bertilsson S. High archaeal diversity in Antarctic circumpolar deep waters. Environ Microbiol. 2011;3:689–697. doi: 10.1111/j.1758-2229.2011.00282.x. [DOI] [PubMed] [Google Scholar]
- 36.Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C. Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J. 2009;3:860–869. doi: 10.1038/ismej.2009.23. [DOI] [PubMed] [Google Scholar]
- 37.Kirchman DL, Cottrell MT, Lovejoy C. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol. 2010;12:1132–1143. doi: 10.1111/j.1462-2920.2010.02154.x. [DOI] [PubMed] [Google Scholar]
- 38.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.