Abstract
In vitro studies on HIV (HIV-1) replication and neutralization are usually performed in human cell cultures supplemented with FBS instead of human serum (HS). Here we show that in contrast to FBS, addition of increasing amounts of human serum from noninfected donors to the cell culture directly correlates with an increase in HIV-1 replication in vitro. This effect is independent of cell line, virus strain, or batch of pooled human serum used. We found that human serum affects viral transcription in a dose-dependent manner by activating the activator protein-1 (AP-1) member proteins c-FOS, JunD, and JunB in TZM-bl cells. Analysis of the human serum component responsible for this effect indicates that it is a protein having a molecular mass between 250 and 300 kDa. This serum protein, HIV-1 enhancing serum protein (HESP), might promote viral transcription in vivo and consequently play a role in disease progression.
Keywords: LTR, PMA, neutralization assays
The need for fast and reliable results has driven the development of numerous methods to study HIV-1 infection in vitro. However, little is known about the impact that the artificial conditions in which the virus is expanded (e.g., cell background, genetic manipulation, growth media) might have on the final outcome of the infection, or if important effects on primary cells might be masked by the in vitro conditions.
In most assays, growth media is supplemented with FBS instead of human serum (HS). In a previous study, we supplemented our culture media with HS from uninfected donors as the source of natural antibodies, to show that such antibodies could be redirected toward HIV-1 and participate in the clearance of the viral infection by activation of the innate immune system (1). During the selection and optimization process of the neutralization assays used in these studies, we found that increasing concentrations of HS in the culture media consistently correlated with a higher readout of viral infectivity. Moreover, the effect was independent of virus strain, cell type, or batch of HS used.
An increase in HIV-1 infectivity promoted by HS has been previously described and it has been mainly attributed to anti–HIV-1 antibodies (antibody-dependent enhancement, ADE) and/or to the human complement activated by both the alternative and classical pathways. Anti–HIV-1 antibodies can coat virus particles and bind to fragment crystallizable (Fc) receptors on the cell surface, thereby facilitating virus anchoring and contact with the HIV-1 CD4 receptor and coreceptors (2, 3). In addition, proteins of the complement can recognize such antibodies and mediate immune adherence to complement receptor (CR) bearing cells (C-ADE) (4–6). The complement factors can also bind to virus particles or infected cells directly, i.e., independent of antibodies, and facilitate infection in a similar fashion (7, 8).
As our experiments on the redirection of natural antibodies were based on HS from uninfected individuals and because many of the cells used in our experiments did not express CR receptors, it was unlikely that ADE or the complement alone were responsible for our observations of augmentation of HIV-1 infectivity.
HIV-1 has also been described to interact with other proteins of the plasma such as mannose binding protein, fibronectin, apoliprotein H, and chondroitin sulfate, but the binding to these molecules is either deleterious for viral infectivity or there is lack of evidence for their in vitro effect (9, 10). Human plasmin, on the other hand, may have a positive effect on viral infectivity as it has been shown to cleave the HIV-1 precursor envelope glycoprotein gp160 into gp120/gp41 (11).
The aims of the present study were to confirm that human serum indeed increases HIV-1 infectivity in vitro, to dissect which step in the viral replication cycle is affected, and whether the effect is virus and/or cell specific, as well as to characterize the factor(s) responsible for this phenomenon.
Results
Human Serum Enhances HIV-1 Infectivity.
HIV-1 infectivity can be measured by different means, and one of the most accepted systems, the TZM-bl assay, provides fast viral readouts based on the expression of luciferase that is driven by the HIV-1 long terminal repeat (LTR) promoter. We found that infection of the cells with HIV-1IIIB in the presence of increasing concentrations of heat inactivated human serum (HIHS) from normal donors was directly correlated to higher relative luminescence units (RLUs). In fact up to eight times higher values were observed in the presence of 40% (vol/vol) HIHS compared with controls cultured in advanced DMEM + 2% (vol/vol) FBS (Fig. 1A). The addition of indinavir to the culture of TZM-bl cells ensured that only one cycle of replication was monitored and because the readout was luciferase activity, the effects detected in this system did not include late steps in the viral cell cycle such as virion assembly, release, and maturation.
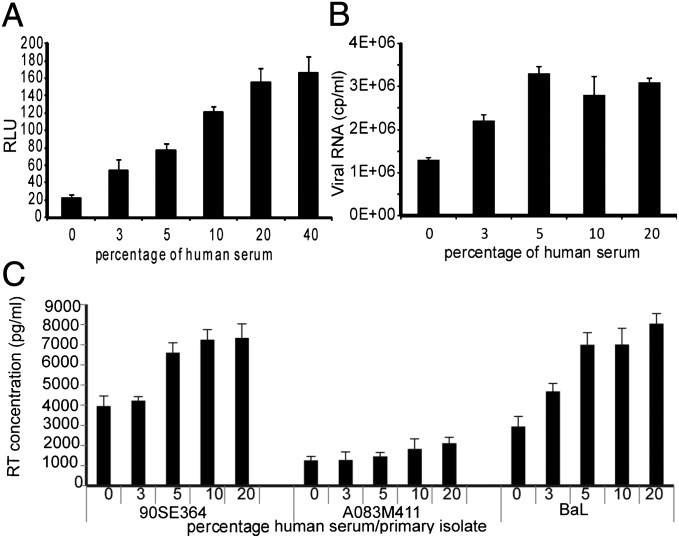
Fig. 1.
Effect of human serum on HIV-1 infectivity in different cell lines. Virus expression was measured in cell cultures supplemented with pooled HIHS from normal donors. Infectious titers are presented as (A) relative luminescence units in TZM-bl at 48 h postinfection with HIVIIIB, (B) copies of viral RNA in culture supernatant of PMA-stimulated ACH-2 cells after 48 h incubation, (C) RT activity of PBMCs infected with three primary isolates in autologous plasma at day 11 postinfection. 0 represents controls of infected cells cultured in maintenance medium. In all cases the differences between the samples cultured at the highest serum concentration and controls were statistically significant (P < 0.01). All values shown are the mean values ± the SD from four replicates.
We tested other cell lines and other methods for quantification of viral infectivity. As the TZM-bl cells are derived from HeLa cells (cervical cancer), we also tested cell line CEM-GFP cells and H9 cells (both with T-cell background) and glioma-derived U87 cells. In all cases, the increasing amounts of HIHS correlated with a higher viral readout, i.e., green fluorescent protein (GFP) expression, p24 levels in culture supernatant, and copies per milliliter of viral RNA in culture supernatant, respectively. In these systems, however, unlike the TZM-bl cells, several cycles of replication are needed for progeny virus to be detected. (Fig. S1).
To test whether HS had an effect on attachment and fusion, we tested whether we could observe the effect of HIHS in chronically infected cell line ACH-2 cells. These cells cannot be reinfected as they lack the CD4 receptor. In Fig. 1B we show that ACH-2 cells stimulated with phorbol 12-myristate 13-acetate (PMA) and cultured in the presence of 10 or 20% (vol/vol) HIHS produced twice as much virus particles as the control cells cultured in the presence of 10% (vol/vol) FBS. Thus, there seems to be an effect of HS on HIV-1 beyond viral entry, reverse transcription, and integration.
We then tested whether the enhancement could be reproduced in freshly isolated peripheral blood mononuclear cells (PBMCs) using three primary isolates cultured with autologous plasma. As seen in Fig. 1C, at day 11 postinfection, the reverse transcriptase (RT) activity increased in cell culture supernatants in relation to the human plasma concentration. At a 10% (vol/vol) plasma concentration the RT activity was 1.8-fold higher for virus strain 90SE364, 1.5-fold higher for A083M411, and 2.4-fold higher for BaL compared with the controls cultured with 10% (vol/vol) FBS (designated as 0 in Fig. 1C).
The difference between infectivity units at the highest HS concentration and controls cultured in the presence of FBS, as well as for the samples cultured in the same amount of either FBS or HIHS, for all cell-based assays were all statistically significant (P < 0.01). The assays mentioned above were repeated at least three times with different pools of HS batches and in all cases the enhancement of infectivity was observed.
We also tested individual serum samples from uninfected donors in TZM-bl cells infected with HIVIIIB and we found that for all subjects there was an increase in infectivity in relation to the amount of HIHS; yet, at the same serum concentration, the infectivity values varied between donors (Fig. S2).
We directly compared the effect of increasing concentrations of either HIHS or FBS on HIVIIIB infectivity in the TZM-bl cells. As seen in Fig. 2A, the presence of 30% (vol/vol) HIHS clearly enhanced viral production, with a fivefold increase in the RLU values compared with the controls cultured in 2% (vol/vol) FBS (P < 0.01). Increasing concentrations of FBS had a much lower impact on the virus infectivity. At a 30% (vol/vol) concentration, the infectious titers were 2.3-fold higher in those cells cultured in the presence of HIHS than in those cultured in the same amount of FBS (P < 0.01).
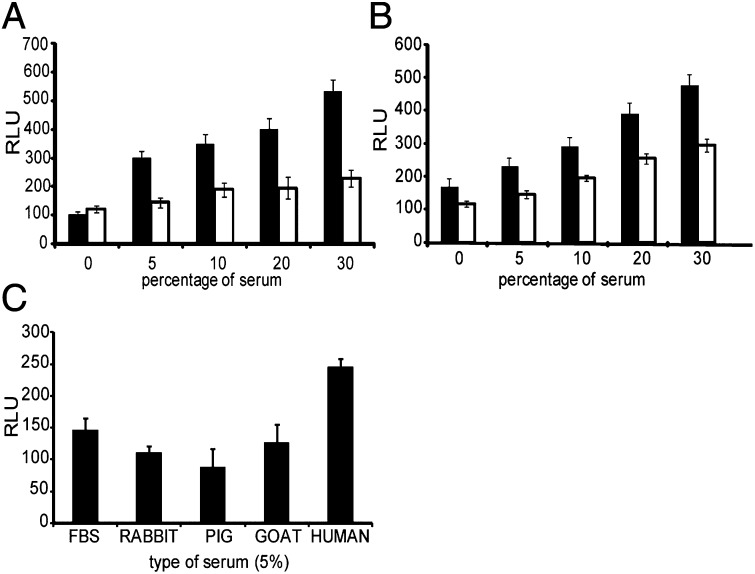
Fig. 2.
Human serum specificity. Relative luminiscence units were measured in HIVIIIB-infected TZM-bl cells after (A) supplementation of culture media with heat-inactivated FBS (white bars) or HIHS (black bars), (B) HIHS (white bars) or NHS (black bars), and (C) supplementation of culture media with 5% (vol/vol) rabbit, porcine, goat, FBS, or HS. 0 depicts controls cultured in the presence of 2% FBS. Comparisons between the highest serum concentration tested and controls were all statistically significant (P < 0.01). All values shown are the mean values ± the SD from four replicates.
As heat inactivation of HS (56 °C for 1 h) nonspecifically denatures heat-sensitive proteins, we tested the difference in effect between heat inactivated and nonheat inactivated HS (NHS). We found that the relative increase in infectivity was similar for both in relation to the controls (Fig. 2B). At 30% (vol/vol) HS a 2.6-fold increase in infectivity was observed for both NHS and HIHS in relation to the controls cultured with 2% FBS (P < 0.01). However, it was with the NHS that higher luminescence values (∼1.5 times higher at each serum concentration) were obtained. Subsequent experiments were therefore performed in the presence of NHS.
We also tested sera from three other species (rabbit, pig, and goat) in HIVIIIB-infected TZM-bl cells. As seen in Fig. 2C, none of the other sera were able to increase viral production at a 5% (vol/vol) concentration compared with HS (P < 0.01). It should be mentioned, that higher concentrations of the sera from rabbit, pig, and goat induced visible cell cytotoxicity.
We then studied the impact that increasing concentrations of NHS had on cell proliferation, cell size, cell cycle progression, as well as apoptosis on the uninfected TZM-bl cells. In Fig. S3A, we show that there was no difference in the proliferation pattern between the samples cultured in the presence of 5, 10, or 20% (vol/vol) NHS after 24 or 48 h in culture, as determined by the passive transfer to progeny cells of the intracellular dye carboxyfluorescein diacetate succinimidyl ester (CFSE). A general increase in the cell diameter was observed after 48 h in culture but there was no difference in the cell size between samples cultured with increasing amounts of NHS (Fig. S3B). To study cell cycle progression we measured the incorporation of the thymidine-nucleoside analog 5-ethynyl-2′-deoxyuridine (EdU), which is incorporated during active DNA synthesis (S phase). We found no significant differences in the percentage of EdU-positive cells in the samples cultured with 5, 10, or 20% (vol/vol) NHS after 48 h in culture (Fig. S3C). We also studied the amount of apoptotic cells and/or necrotic TZM-bl cells after treatment with NHS. We did so by staining the cells with annexin V and propidium iodide (PI), respectively. We found no significant differences in the expression of either of the markers between the samples cultured with increasing amounts of NHS (Fig. S3D).
Effect of Human Serum on HIV-1 Replication Cycle.
We showed in Fig. 1B that HS increases viral production in chronically infected ACH-2 cells. To confirm that NHS does not affect virus binding and fusion, we preinfected TZM-bl cells with HIV-1IIIB virus for 2.5 h and washed off unbound virus before the addition of NHS. In Fig. 3A we show that compared with those samples in which the virus was present in the culture during the whole assay (48 h), the samples that were preinfected showed lower infectivity values. However, the luminescence values increased under both conditions compared with controls with FBS and the ratio between FBS controls and NHS-treated samples was similar for preinfected and nonpreinfected, respectively. At the highest serum concentration the difference between preinfected samples and controls was 4.5-fold, and in the samples where serum and virus were added simultaneously it was sixfold (P < 0.01).
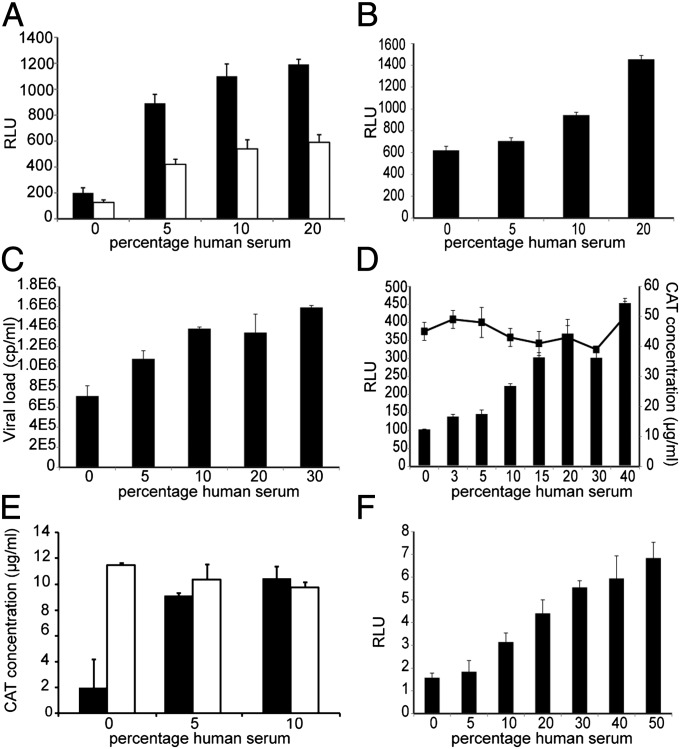
Fig. 3.
Effect of the serum on the HIV-1 replication cycle. (A) Preinfection of TZM-bl cells with HIVIIIB for 2.5 h followed by wash and addition of NHS (white bars) vs. culture of the cells in the presence of both HIVIIIB and NHS for 48 h (black bars). (B) Transfection of TZM-bl cells with infectious plasmid pNL4.3 followed by addition of NHS. (C) Copies of viral RNA in culture supernatant at day 9 postinfection of SupT1 cells infected with a Δtat IIIB virus. (D) Cotransfection of TZM-bl cells with CMV-CAT and CMV-tat. Represented are the CAT concentration values (line, Right axis) or RLU values reflecting tat activity (bars, Left axis) in cell lysate after 48 h of incubation with NHS. (E) CAT concentration at 48 h posttransfection of 293T cells with LTR-CAT (black bars) or CMV-CAT (white bars). (F) Luciferase expression in uninfected TZM-bl cells after treatment with NHS (RLU scale is 1/100 of those of infected cells). Comparison between serum-treated cultures and controls cultured in maintenance medium (depicted as 0) were all statistically significant (P < 0.01). All values shown are the mean values ± the SD from three replicates.
We also performed a transfection of the TZM-bl with the HIV-1 infectious plasmid pNL4-3 to circumvent the fusion step. As seen in Fig. 3B at a 20% (vol/vol) concentration of NHS, the RLU values were more than doubled compared with the controls cultured in the presence of FBS (P < 0.01).
To test whether the results with higher amounts of HS were an effect of better nutritious conditions, we cotransfected TZM-bl cells with two plasmids: one that expresses the Tat protein of HIV-1 and the other chloramphenicol acetyltransferase (CAT), both driven by the CMV promoter. The Tat produced would act on the transactivating response element from the LTR promoter of the luciferase reporter gene and so its activity was measured as luminescence units, whereas CAT was quantified by ELISA. Because both Tat and CAT had the same promoter, it was assumed that they would have similar levels of expression. However, we found that the CAT values were stable throughout the different serum dilutions (even in those cells cultured with 40% NHS), whereas the RLU values driven by Tat were dose dependent (Fig. 3D). This indicated a possible role for Tat or LTR in the HS enhancement of infectivity.
As Tat plays a key role in orchestrating the transcription of HIV-1, we evaluated the contribution of NHS in a Tat-free environment, by infecting SupT1 cells with a Δtat IIIB virus. As seen in Fig. 3C, the amount of virus particles produced by the cells was almost twofold higher already at a 10% (vol/vol) NHS concentration compared with the controls cultured in 10% FBS (P < 0.01).
We then checked the specificity for LTR by transfecting 293T cells with either LTR-CAT or CMV-CAT plasmids and cultured them in the presence of 10% FBS, 5% (vol/vol) NHS, or 10% NHS. In Fig. 3E we show that, whereas the CAT produced in the CMV-CAT transfected cells remained constant regardless of the amount of serum added to the cells, the CAT expression in those cells transfected with LTR-CAT was augmented in the presence of NHS. We found a fivefold difference in CAT expression between cells cultured with 10% NHS and 10% FBS (P < 0.01).
To confirm the effect of NHS on the LTR, we measured the luciferase expression in uninfected TZM-bl cells. As shown in Fig. 3F, we observed that indeed the luciferase protein was produced in higher amounts in the presence of NHS (P < 0.01), albeit at much lower levels than when infected with HIV-1.
Effect of Human Serum on HIV-1 Transcription.
Several transcription factors can act on the LTR and the precise mapping of the binding sites for different molecules has already been described (12–14). Factors like nuclear factor kappa B (NFκB), the activator protein-1 (AP-1) family, and specificity protein 1 (SP-1) have been shown to play important roles on HIV-1 transcription.
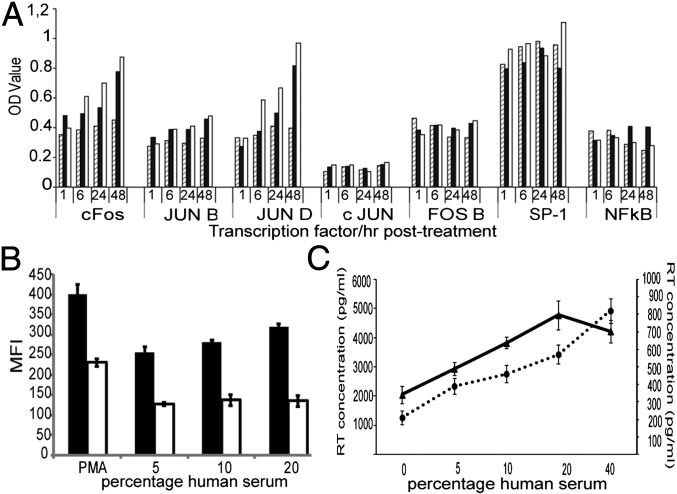
To test whether HS would have an effect on the levels of these proteins we cultured TZM-bl cells in the presence of 5% FBS, 5% NHS, or 10% (vol/vol) NHS and the nuclear extracts were collected at 1, 6, 24, and 48 h. In Fig. 4A we show that the AP-1 member proteins c-FOS, JunD, and JunB exhibited a time and dose-dependent response to the HS, whereas c-JUN, FOS B, SP-1, and NFκB were not altered by the treatment. Similar results were obtained with HIVIIIB-infected TZM-bl cells cultured in the presence of NHS (Fig S4A). We proceeded to confirm these results by transfecting TZM-bl cells with a plasmid encoding the complete LTR sequence of pNL4-3, which was serving as a promoter of enhanced green fluorescent protein (EGFP) expression (wtLTR-EGFP) and another plasmid containing in total seven point mutations in the AP-1 binding sites of LTR (ΔAP-1LTR-EGFP) that have been described to hinder binding of the respective transcription factors (15). In Fig. 4B we show that the cells transfected with the mutant vector had a twofold lower expression of EGFP compared with those of the wtLTR vector (P < 0.01) and there was no difference across serum concentrations. As a control of functional LTR we used PMA. This compound potently activates signal transduction and promotes viral transcription by activation mainly of NFκB but an effect on c-FOS, JunD, and JunB has also been described (16). In Fig. 4B we show that there was a 1.7-fold difference in EGFP expression in the cells transfected with the mutant LTR, thus, underlining the importance that AP-1 factors have on the PMA-induced transcription initiation. We then measured the actual input of NHS on viral transcription by culturing ACH-2 cells in the presence or absence of PMA. In Fig. 4C we show that the RT activity increased in relation to the concentration of NHS in the culture both in the presence or absence of PMA, but the viral production was 7–10 times lower in those samples cultured with NHS alone. Nevertheless, in the absence of PMA there was a dose-dependent response to NHS. The viral production at 10% (vol/vol) NHS was twice as high as in the control wells cultured in the same concentration of FBS (P < 0.01).
Fig. 4.
Effect of the serum on HIV-1 transcription. (A) TMZ-bl cells were cultured with 5% (vol/vol) FBS (diagonal-line bars), 5% NHS (black bars), or 10% NHS (white bars). Represented are the expression levels over time of the AP-1 family, SP-1, and NFκB transcription factors. (B) TZM-bl cells were transfected with a vector expressing EGFP driven by either wild-type LTR (wtLTR-EGFP; black bars) or an LTR with mutations in each of the three AP-1 binding sites (ΔAP-1LTR-EGFP; white bars) and cultured in the presence of NHS for 48 h. Represented is the mean fluorescence intensity of three replicates of EGFP+ cells. Difference between the mean fluorescence intensity of WT and mutant-expressing cells were all statistically significant (P < 0.01). (C) ACH-2 cells were cultured in the presence (solid line, Left axis) or absence of PMA (dotted line, Right axis) plus NHS. All values shown are the mean values ± the SD from three replicates.
We also tested the impact that U0126, a potent MEK1/2 inhibitor, would have on the NHS-induced AP-1 activation by pretreating the cells with 20 μM of the drug before the addition of NHS. We found that the drug reduced by over twofold the NHS-induced cFOS up-regulation and it had also a lower impact on JunB and JunD expression (Fig. S4B).
Test of HS Components and Fractionation.
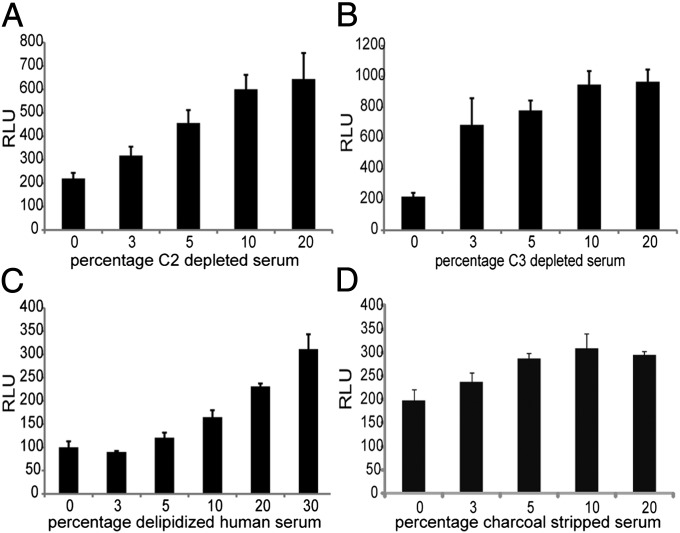
In Fig. 2B we showed that heat inactivation only partially compromised the effect of HS. However, as proteins of the complement (particularly C2 and C3) have been implicated in the enhancement of HIV-1 infectivity, we tested sera depleted from C3 and C2 in the TZM-bl system. Increasing luminescence values were also observed with the C2- and C3-depleted HS giving three- and fourfold higher RLU values, respectively, compared with the HIV-1–infected cells cultured with 2% FBS (P < 0.01) (Fig. 5 A and B). Also, the HS enhancement was still observed using serum depleted of either its lipid fraction or hormones (charcoal stripped). In Fig. 5 C and D, respectively, we observe that at a 20% (vol/vol) concentration, there was a threefold and a 1.5-fold increase in infectivity in relation to the controls (P < 0.01).
Fig. 5.
Test of human serum components. (A) C2-depleted serum, (B) C3-depleted serum, (C) delipidized HS, and (D) charcoal stripped serum. Comparison between serum treated and controls were all statistically significant (P < 0.01). All values shown are the mean values ± the SD from four replicates.
We also tested purified human plasmin for its potential effect on increasing gp120 availability in the viral surface, as well as, albumin, IgG, and IL-2, which have been described to aid the infectivity of other pathogens (17–21). Contrary to what is known, plasmin dramatically reduced infectivity as did IgG. Albumin and IL-2 did not show a dose-dependent increase of infectivity (Fig. S5).
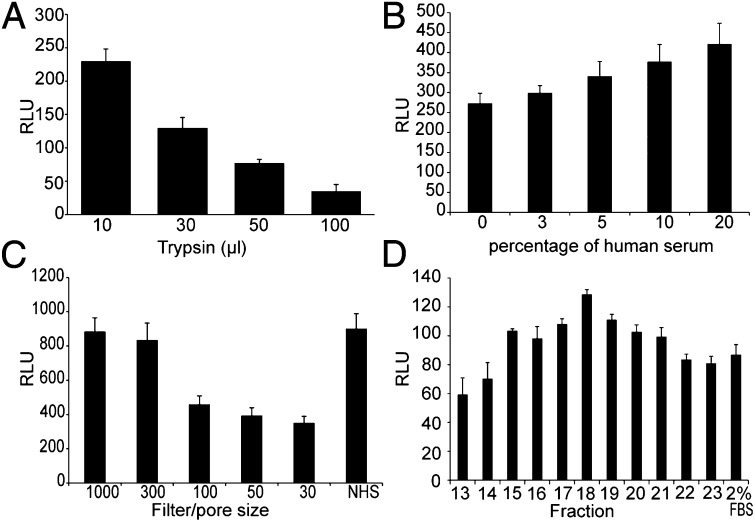
Because the depletion of the lipid and hormone fractions did not seem to affect the enhancing ability of HS in a major way, the factor(s) responsible is most likely a protein. To test this, we treated NHS with increasing amounts of trypsin. Following digestion of the HS, the trypsin was inactivated and the serum was tested in the TZM-bl assay. In Fig. 6A we show that after trypsin digestion the HS did not retain its enhancing activity (P < 0.01).
Fig. 6.
Fractionation and analysis of NHS. (A) Tryptic digestion of NHS followed by trypsin inactivation. (B) Treatment of NHS with 2.5% CHAPSO followed by centrifugation on a 100-kDa filter. The difference between serum treated and control of 2% (vol/vol) FBS (depicted as 0) was statistically significant (P < 0.01). The NHS was fractionated by (C) subsequent centrifugation with filters of different pore size. (D) Protein fractionation by size exclusion chromatography. All values shown are the mean values ± the SD from four replicates.
To test whether the enhancement was due to one protein or to a protein complex, we added increasing concentrations of a mild detergent (CHAPSO) to disaggregate-possible complexes. Before adding the treated NHS to the TZM-bl cells, it was centrifuged on a 100-kDa filter to rid it of the detergent and also of possible smaller complex subunits. As seen in Fig. 6B, this treatment did not affect the enhancement in infectivity, indicating that a single protein and not a complex was responsible for the effects on HIV-1 infectivity by HS. The difference between samples cultured with 20% (vol/vol) of detergent-pretreated NHS and controls was statistically significant (P < 0.01).
We then fractionated the HS proteins by size. We fractionated NHS by centrifugation through filters of different pore size. First we added the NHS to a 1,000-kDa filter and after centrifugation the flow-through was added to a 300-kDa filter and subsequently to 100-, 50-, and 30-kDa filters. An aliquot from each flow-through was collected and tested in the TZM-bl assay. Similar enhancement in infectivity, as observed with unfiltered NHS, was seen with those aliquots that passed through the 1,000 kDa and the 300-kDa cutoff filters, but was markedly reduced when collected after passing filters with cutoffs of 100 kDa or less (Fig. 6C). Dialysis of NHS also reproduced an increased infectivity in the fraction above 100 kDa (Fig. S6A).
We also carried out size exclusion chromatography using Superdex 200 (effective range 10–600 kDa). We found the increase in infectivity with the pooled fractions 15–21 (Fig. S6B) and with the split pool particularly in fraction 18 (Fig. 6D), indicating a molecular mass of 250–300 kDa. We name this HS protein: HIV-1 enhancing serum protein (HESP).
Discussion
In the present study we could show that HS from uninfected individuals increases the in vitro production of HIV-1. This effect was reproducible in five different cell lines infected with the HIV-1IIIB virus, as well as in PBMC infected with three primary isolates, the latter when cultured in syngenic plasma. Moreover, an enhancement of HIV-1 infectivity was seen independent of batch of pooled serum used and was present in all tested individual serum samples, although we observed variations in the infectious levels reached by the different donors. We also showed that the effect was specific to HS at least compared with bovine, porcine, rabbit, or goat serum and that it was not dependent on complement factors C2 or C3.
To dissect which step within the viral replication cycle was being affected by the NHS, we used two different single virus replication cycle cell-based assay systems, the ACH-2 and TZM-bl cells. We were able to show that neither the early steps (fusion, reverse transcription, and integration) nor the late steps (virion assembly, release, and maturation) seem to be affected by HS in these two cell lines. Wu et al. (22) previously reported on a possible effect of HS on HIV adsorption to PBMCs but they only noted this effect at concentrations of HS of 40% or higher, i.e., much higher concentrations than what was used in the present study. Instead we showed that HS triggers the activation of the LTR promoter, at least in epithelial-derived cell lines, thereby facilitating virus replication and infectivity. This finding is also supported by transfection of TZM-bl and 293T cells with LTR reporter genes. Further analysis of the transcription factors possibly involved, showed that three members of the AP-1 family, c-Fos, JunD, and JunB, increased in a time- and dose-dependent manner in relation to the amount of NHS present in the culture of TZM-bl cells, but this was not the case with the other AP-1 members c-JUN and FOS B, nor with SP-1 or NFκB. The significance of this family of transcription factors was also confirmed by a decrease in the expression of EGFP, which was driven by an LTR promoter with point mutations in each of the three AP-1 binding sites. Also, the LTR-CAT plasmid used contains not only the 3′ AP-1 sites but also the downstream AP-1 binding sites (23) of which the +160 down street element (relative to the transcription initiation site) has been shown to bind c-Fos and JunD (15, 24, 25).
In this context it should be noted that the effect of transcription factors and other transcription regulatory proteins on LTR might vary between different cell types (26, 27). However, we observed the enhancing effect of HS not only in epithelial-derived cells but also in H9 cells, U87 cells, CEM-GFP cells, and PBMC. These latter in vitro systems require several cycles of replication in order for progeny virus to be detected. It is therefore possible that other effects on the HIV-1 replication cycle may be operative in these cells. Of note, Colin et al. (16) found that all three intragenic enhancer AP-1 binding sites in the HIV-1 pol gene recruit and bind c-Fos, JunB, and JunD, the very AP-1 proteins up-regulated in the presence of HS in our study upon PMA stimulation, and that mutations of these intragenic AP-1 binding sites negatively affected HIV-1 expression in TZM-bl cells, as well as HIV-1 replication in Jurkat T cells, promonocytic U937 cells, and primary monocyte-derived macrophages.
The AP-1 family of transcription factors mediates gene regulation in response to cytokines and growth factors (generally below 50 kDa). However, preliminary characterization of the HS factor(s) responsible for the HS-induced stimulation of HIV-1 replication in the present study indicated that the factor is a single HIV-1 HESP with a molecular mass between 250 and 300 kDa. Therefore, cytokines or growth factors do not seem to be responsible for the enhancement of infectivity described here. Further studies to try to define and characterize HESP using inter alia mass spectrometry are under way.
Because viral load is correlated to disease progression in infected individuals (28, 29), it would be interesting to evaluate possible differences in the HIV-1 enhancing effect of sera from rapid progressors to those from elite controllers. The paper also highlights the importance of having proper controls when, e.g., studying HIV neutralizing antibodies in serum from patients in clinical vaccination studies.
In summary, we present here results indicating that a protein in HS (HESP) increases the transcription of the proviral HIV-1 genome, possibly by signal transduction and activation of AP-1.
Materials and Methods
Cells, Virus, and Serum.
The following cell lines were obtained from the National Institutes of Health (NIH) AIDS Research and Reference Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), NIH: H9, ACH-2, CEM-GFP, SupT1, TZM-bl, and U87.CXCR4 cells. The 293T cells were purchased from the Interlab Cell Line Collection. Maintenance medium for suspension cells was RPMI + 10% (vol/vol) FBS and for adherent cells was advanced DMEM + 2% FBS. The PBMCs were obtained as buffy coats from the blood bank at Karolinska University Hospital and isolated by Ficoll-Paque plus gradient (GE Healthcare) according to the manufacturer’s protocol.
The viruses used in this study, i.e., HIV-1IIIB; primary isolates 90SE364, A083M411, and BaL; and the HIV-1 MC99IIIBΔTat-Rev (referred as Δtat IIIB virus in the text) were all obtained from the NIH AIDS Research and Reference Program, Division of AIDS, NIAID, NIH.
The HS was of type AB from a pool of uninfected individuals and several batches were tested from different sources: Cambrex, Innovative Research, and 3H Biomedicals. Individual AB serum samples were obtained from 3H Biomedicals. For the studies on PBMCs, the plasma collected in citrate buffer from the same anonymous donor was purchased from the blood bank of Karolinska Hospital. The C3- and C2-depleted sera were obtained from Sigma-Aldrich, whereas the charcoal stripped and delipidized sera were obtained from Innovative Research. Rabbit, porcine, and goat sera were purchased from Invitrogen. Complement inactivation of the sera was done by heat inactivation at 56 °C in a water bath for 1 h.
Cell-Based Assays.
i) TZM-bl cells: performed as previously specified (30) and infected with 150 50% tissue culture infective dose (TCID50) of the HIV-1IIIB virus in advanced DMEM (Gibco) containing 1 mM indinavir. Each serum dilution/concentration was tested in quadruplicates in the same plate and in at least five independent experiments. When testing the effect of HS on uninfected TZM-bl cells, the cells were seeded in six-well plates at a concentration of 400,000 cells per well and total volume of 1 mL.
ii) ACH-2 cells: 500,000 cells per well were incubated with HS in the absence or presence of 100 nM PMA for 48 h. Each concentration was run in triplicate in the same plate and tested in at least four separate experiments. The supernatant tested for quantification of viral RNA using the COBAS AmpliPrep/COBAS TaqMan HIV-1 test (Roche) or by RT quantification with RT activity kit (Cavidi).
iii) H9 cells: Assays were performed as previously specified (1). The cultures were infected with 150 TCID50 of the HIV-1IIIB plus HIHS. At day 7, the supernatants were evaluated for p24 levels with an in-house ELISA (31). All conditions were run in quadruplicates in the same plate and on three separate occasions.
iv) U87.CXCR4 cells: Assays were performed as previously specified (1). The cultures were infected with 150 TCID50 of HIV-1IIIB plus HIHS in advanced DMEM. Each virus/serum dilution was tested in quadruplicates in the same plate. The viral supernatant was collected and tested for quantification of viral RNA using the COBAS AmpliPrep/COBAS TaqMan HIV-1 test.
v) CEM-GFP cells: 500,000 cells were infected with 150 TCID50 of HIV-1IIIB plus HIHS at a final volume of 2 mL/well. Each HS concentration was run in triplicate and in three separate experiments. GFP expression was measured at day 7 postinfection in a FACSCalibur flow cytometer (BD Biosciences).
vi) PBMC: 200,000 freshly isolated, phytohemagglutinine-stimulated PBMCs were infected with 200 TCID50 of the primary isolate and cultured with human plasma and 200 IU interleukin-2. Control wells were cultured in 10% FBS. All samples were measured in quadruplicates in the same plate and in three different plates. RT was quantified at days 7 and 11 postinfection using the RT activity kit (Cavidi).
vii) SupT1 cells: The cells were infected with MC99IIIBΔTat-Rev virus in a volume of 1 mL for 3 h followed by addition of NHS or 10% FBS in triplicates. The viral RNA was measured in the supernatant at days 5, 7, and 9 postinfection with COBAS AmpliPrep/COBAS TaqMan HIV-1 test.
Cell proliferation, cell cycle, and apoptosis were measured in the TZM-bl cells at 0 and 48 h postculture with NHS, following the same culture conditions as previously specified. They were measured using CellTrace CFSE cell proliferation kit, Click-iT EdU kit, Alexa Fluor 488, and Dead Cell Apoptosis kit with Annexin Alexa Fluor 488 and PI, respectively, following manufacturer protocol (Invitrogen) and analyzed by flow cytometry in FACScalibur (BD Biosciences). The experiment was performed in triplicates in two separate occasions.
Plasmids and Transfections.
The following plasmids were used in this study: pNL4-3 (NIH AIDS reagent facility), pCMVCAT (Invitrogen), pCMVtat (provided by S. Schwartz, Lund University, Lund, Sweden), pNLCATW (referred as LTR-CAT in the text) (23). The latter contains LTR nucleotides from the first nucleotide in U3 through the nucleotide at position 743. The complete LTR sequence derived from pNL4.3 (789 bp) and one including seven point mutations as previously specified (15) were synthesized and cloned into the pEGFP-N1 vector (Clontech) by Genscript (wtLTR-EGFP and ΔAP-1LTR-EGFP, respectively). The LTR sequence was located upstream of EGFP in replacement of the CMV promoter and multiple cloning site.
For the transfections, 400,000 TZM-bl per well were preseeded in six-well plates and transfected with 1 μg/well of the plasmids + 8 μL Fugene HD (Roche) and incubated for 7 h at 37 °C. Then the cells were washed twice and incubated in advanced DMEM with the appropriate concentration of NHS for 48 h at 37 °C. Each dilution was run in triplicate. The cells were lysed with Glo Lysis buffer (300 μL/dish) and the lysate was either tested for luminescence as mentioned above or tested for CAT concentration with CAT ELISA kit (Roche) as per manufacturer protocol. EGFP-expressing cells were detached from the plate, washed, and analyzed by flow cytometry using FACScalibur (BD Biosciences).
A similar set up was followed for the transfection of 293T cells with pCMVCAT and pNLCATB.
Transcription Factors Expression.
A total of 400,000 TZM-bl cells per well were cultured in advanced DMEM with either 5% FBS or 5–10% NHS in triplicates. The nuclear fraction was isolated at specified time points using a nuclear extract kit (Active Motif) following manufacturer protocol. Transcription factor levels were measured using 2 μg of nuclear extracts and following manufacturer protocol for TransAM AP-1 family, TransAM Sp1, and TransAM NFκB (Active Motif) ELISAs. Control samples were pretreated with 20 μM U0126 (Sigma), 30 min before the addition of 20% NHS.
Analysis of HS.
Filtration NHS was diluted 1:2 in PBS and centrifuged in Vivaspin 6 filters (Sartorius) with pore size of 1,000 kDa. The flow-through was further centrifuged through a 300-kDa filter followed by subsequent centrifugation through 100-, 50-, and 30-kDa filters. An aliquot from each flow-through was collected and tested at 20% concentration on the TZM-bl cells.
Size exclusion chromatography.
Size exclusion chromatography was run at 4 °C on an XK16/100 column using Superdex 200 prep grade (GE Healthcare) following manufacturer protocol. The fractions were collected over an 8- to 12-h period at a flow rate of 0.5 mL/min using PBS as running buffer. Samples were concentrated by centrifugation at 3,200 × g in Vivaspin 20 (Sartorius) before test in the TZM-bl cells.
Tryptic digestion.
Tryptic digestion was performed using Trypsin gold (Promega) at serum:trypsin ratios of 30:1, 10:1, 5:1, and 1:1 followed by overnight incubation at 37 °C on gentle agitation. To stop digestion, soybean protease inhibitor (Sigma-Aldrich) was used at an inhibitor:trypsin ratio of 2:1.
Detergent treatment.
HS was diluted 1:2 in PBS supplemented with 2.5% CHAPSO and incubated for 2 h on a rocking platform. The serum was then filtered by centrifugation through a 100-kDa filter (Sartorius).
Dialysis.
A total of 4 mL NHS was dialyzed overnight using a 5-mL volume Flot-A-Lyzer (SpectrumLabs) with molecular weight (MW) cutoff of 8, 20, 50, and 100 kDa as recommended by the manufacturer.
The following individual serum components were tested on the TZM-bl assay as described above: albumin fraction V high purity, human plasmin, and total human IgG (Millipore).
Statistical Analysis.
A Student t test was used to assess statistical differences between two sets of data. In most cases, it was used to compare samples cultured at the highest concentration of HS and controls cultured in maintenance medium.
Supplementary Material
Acknowledgments
This study was supported by the Swedish Medical Foundation (Grant K2000-06X-09501-10B).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206893109/-/DCSupplemental.
References
- 1.Perdomo MF, Levi M, Sällberg M, Vahlne A. Neutralization of HIV-1 by redirection of natural antibodies. Proc Natl Acad Sci USA. 2008;105(34):12515–12520. doi: 10.1073/pnas.0805777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell WM, Ding L, Gabriel J. Inactivation of a common epitope responsible for the induction of antibody-dependent enhancement of HIV. AIDS. 1998;12(2):147–156. doi: 10.1097/00002030-199802000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Füst G. Enhancing antibodies in HIV infection. Parasitology. 1997;115(Suppl):S127–S140. doi: 10.1017/s0031182097001819. [DOI] [PubMed] [Google Scholar]
- 4.Soelder BM, et al. Complement receptors: Another port of entry for HIV. Lancet. 1989;2(8657):271–272. doi: 10.1016/s0140-6736(89)90452-2. [DOI] [PubMed] [Google Scholar]
- 5.Prohászka Z, et al. Two parallel routes of the complement-mediated antibody-dependent enhancement of HIV-1 infection. AIDS. 1997;11(8):949–958. doi: 10.1097/00002030-199708000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Robinson WE, Jr, Montefiori DC, Gillespie DH, Mitchell WM. Complement-mediated, antibody-dependent enhancement of HIV-1 infection in vitro is characterized by increased protein and RNA syntheses and infectious virus release. J Acquir Immune Defic Syndr. 1989;2(1):33–42. [PubMed] [Google Scholar]
- 7.Boyer V, Desgranges C, Trabaud MA, Fischer E, Kazatchkine MD. Complement mediates human immunodeficiency virus type 1 infection of a human T cell line in a CD4- and antibody-independent fashion. J Exp Med. 1991;173(5):1151–1158. doi: 10.1084/jem.173.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dierich MP, et al. HIV and human complement: Mechanisms of interaction and biological implication. Immunol Today. 1993;14(9):435–440. doi: 10.1016/0167-5699(93)90246-H. [DOI] [PubMed] [Google Scholar]
- 9.Spear GT. Interaction of non-antibody factors with HIV in plasma. AIDS. 1993;7(9):1149–1157. doi: 10.1097/00002030-199309000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Stefas E, et al. Human plasmatic apolipoprotein H binds human immunodeficiency virus type 1 and type 2 proteins. AIDS Res Hum Retroviruses. 1997;13(1):97–104. doi: 10.1089/aid.1997.13.97. [DOI] [PubMed] [Google Scholar]
- 11.Okumura Y, et al. The extracellular processing of HIV-1 envelope glycoprotein gp160 by human plasmin. FEBS Lett. 1999;442(1):39–42. doi: 10.1016/s0014-5793(98)01612-3. [DOI] [PubMed] [Google Scholar]
- 12.Gaynor R. Cellular transcription factors involved in the regulation of HIV-1 gene expression. AIDS. 1992;6(4):347–363. doi: 10.1097/00002030-199204000-00001. [DOI] [PubMed] [Google Scholar]
- 13.Pereira LA, Bentley K, Peeters A, Churchill MJ, Deacon NJ. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28(3):663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilareski EM, Shah S, Nonnemacher MR, Wigdahl B. Regulation of HIV-1 transcription in cells of the monocyte-macrophage lineage. Retrovirology. 2009;6:118. doi: 10.1186/1742-4690-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Lint C, et al. Transcription factor binding sites downstream of the human immunodeficiency virus type 1 transcription start site are important for virus infectivity. J Virol. 1997;71(8):6113–6127. doi: 10.1128/jvi.71.8.6113-6127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colin L, et al. The AP-1 binding sites located in the pol gene intragenic regulatory region of HIV-1 are important for viral replication. PLoS ONE. 2011;6(4):e19084. doi: 10.1371/journal.pone.0019084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Somner EA, Black J, Pasvol G. Multiple human serum components act as bridging molecules in rosette formation by Plasmodium falciparum-infected erythrocytes. Blood. 2000;95(2):674–682. [PubMed] [Google Scholar]
- 18.Mayumi M, Imai M, Miyakawa Y. HBsAg particles with a receptor for polymerized albumin. Gastroenterology. 1979;77(1):203. [PubMed] [Google Scholar]
- 19.Asahi H, Kanazawa T, Hirayama N, Kajihara Y. Investigating serum factors promoting erythrocytic growth of Plasmodium falciparum. Exp Parasitol. 2005;109(1):7–15. doi: 10.1016/j.exppara.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Imai M, Yanase Y, Nojiri T, Miyakawa Y, Mayumi M. A receptor for polymerized human and chimpanzee albumins on hepatitis B virus particles co-occurring with HBeAg. Gastroenterology. 1979;76(2):242–247. [PubMed] [Google Scholar]
- 21.Lavillette D, et al. Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J Virol. 2005;79(10):6023–6034. doi: 10.1128/JVI.79.10.6023-6034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu SC, et al. Human plasma enhances the infectivity of primary human immunodeficiency virus type 1 isolates in peripheral blood mononuclear cells and monocyte-derived macrophages. J Virol. 1995;69(10):6054–6062. doi: 10.1128/jvi.69.10.6054-6062.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan W, Felber BK, Zolotukhin AS, Pavlakis GN, Schwartz S. Efficient expression of the human papillomavirus type 16 L1 protein in epithelial cells by using Rev and the Rev-responsive element of human immunodeficiency virus or the cis-acting transactivation element of simian retrovirus type 1. J Virol. 1995;69(9):5607–5620. doi: 10.1128/jvi.69.9.5607-5620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabbi MF, Saifuddin M, Gu DS, Kagnoff MF, Roebuck KA. U5 region of the human immunodeficiency virus type 1 long terminal repeat contains TRE-like cAMP-responsive elements that bind both AP-1 and CREB/ATF proteins. Virology. 1997;233(1):235–245. doi: 10.1006/viro.1997.8602. [DOI] [PubMed] [Google Scholar]
- 25.Roebuck KA, Brenner DA, Kagnoff MF. Identification of c-fos-responsive elements downstream of TAR in the long terminal repeat of human immunodeficiency virus type-1. J Clin Invest. 1993;92(3):1336–1348. doi: 10.1172/JCI116707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canonne-Hergaux F, Aunis D, Schaeffer E. Interactions of the transcription factor AP-1 with the long terminal repeat of different human immunodeficiency virus type 1 strains in Jurkat, glial, and neuronal cells. J Virol. 1995;69(11):6634–6642. doi: 10.1128/jvi.69.11.6634-6642.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naghavi MH, et al. Intracellular high mobility group B1 protein (HMGB1) represses HIV-1 LTR-directed transcription in a promoter- and cell-specific manner. Virology. 2003;314(1):179–189. doi: 10.1016/s0042-6822(03)00453-7. [DOI] [PubMed] [Google Scholar]
- 28.Mellors JW, et al. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272(5265):1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 29.Tetali S, et al. Plasma virus load evaluation in relation to disease progression in HIV-infected children. AIDS Res Hum Retroviruses. 1998;14(7):571–577. doi: 10.1089/aid.1998.14.571. [DOI] [PubMed] [Google Scholar]
- 30.Montefiori DC. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol. 2009;485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- 31.Abdurahman S, Höglund S, Goobar-Larsson L, Vahlne A. Selected amino acid substitutions in the C-terminal region of human immunodeficiency virus type 1 capsid protein affect virus assembly and release. J Gen Virol. 2004;85(Pt 10):2903–2913. doi: 10.1099/vir.0.80137-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.