Abstract
Tissue-derived cultured cells exhibit a remarkable range of morphological features in vitro, depending on phenotypic expression and environmental interactions. Translation of these cellular architectures into inorganic materials would provide routes to generate hierarchical nanomaterials with stabilized structures and functions. Here, we describe the fabrication of cell/silica composites (CSCs) and their conversion to silica replicas using mammalian cells as scaffolds to direct complex structure formation. Under mildly acidic solution conditions, silica deposition is restricted to the molecularly crowded cellular template. Inter- and intracellular heterogeneity from the nano- to macroscale is captured and dimensionally preserved in CSCs following drying and subjection to extreme temperatures allowing, for instance, size and shape preserving pyrolysis of cellular architectures to form conductive carbon replicas. The structural and behavioral malleability of the starting material (cultured cells) provides opportunities to develop robust and economical biocomposites with programmed structures and functions.
Keywords: sol-gel, biomineralization, biopreservation, frustule
The synthesis of inorganic materials with controlled and complex forms has been facilitated through discoveries such as vesicle, micelle, and liquid crystalline templating of silicates (1–3), which provided inspiration to explore a range of templating strategies based on self-assembled molecular precursors (4–8), colloids (9–11), and biological templates and vessels (12–14). A driving force for these efforts is the many complex inorganic structures found in nature. An oft-cited example is the hierarchical composites built by silica condensing microorganisms such as diatoms, which have generated substantial scientific interest for over a century (15). Diatoms display complex three-dimensional (3D) architectures with great structural control over nano- to millimeter length scales. However, despite some success toward elucidating mechanisms of diatom biomineralization, the in vitro synthesis of 3D diatom-like forms has remained elusive. Diatom silica has found numerous applications including as a chemical stabilizer, absorbent, filter medium, and fine abrasive, and the lack of synthetic analogues has facilitated recent investigations to employ diatom frustules as starting materials for shape-preserving chemical transformations into functional nanomaterials (16–18). Given the potential of this biosilica, it would be desirable to be able to wield control over the silica structure to achieve broader applicability (19); however, strategies to direct diatom morphology using chemical (20) and genetic approaches (21) has proven challenging. Therefore, an ability to generate cell frustules from more malleable templates such as mammalian cells would provide greater access to natural and engineered cell heterogeneity—both structure and function—to be exploited in the design of complex materials.
To these ends, biomineralization by silica condensing microorganisms offers key lessons. The discovery of biogenic peptides that catalyze silica condensation (22–24) subsequently has motivated the extensive investigation of the interaction of natural and synthetically derived peptides and proteins with silica and its precursors (5, 22, 23, 25–30). Identification of silica-associated biomolecules such as long-chain polyamines (22) and the silaffin peptides (23) has led to a general understanding of the tenets by which macromolecules control polymerization of silica precursors into silica assemblies (31). However, silica morphogenesis at the meso- and microscales must involve both transport of soluble silica precursors and their directed deposition by biomolecular templating or structural elements (32–34). Likely, these larger scale molecular assemblies direct the assembly of silica building blocks, formed in the silica deposition vesicle (SDV), into complex structures. This reasoning led us to question whether synthetic 3D protein scaffolds could direct/template silica deposition provided the appropriate silica precursors and chemical conditions. Indeed, we recently showed that microfabricated protein hydrogels could template silica volumetrically into mechanically stable, nano to microscale biocomposites with user-defined 3D features identical in size and shape to those of the template. These features were preserved following removal of the organic component to form a porous silica replica (35). Importantly, proteins of diverse properties (e.g., isoelectric point; pI) directed silica condensation under identical solution conditions (100 mM silicic acid, pH 3), which is somewhat contrary to the generally held understanding that cationic species (e.g., proteins with pI > 7) are required for biogenic silica deposition (27). These protein hydrogels are highly concentrated (> 40% protein by wt vol-1), producing a locally crowded 3D molecular environment, which acts to capture and concentrate silica precursors (mono-, oligosilicic acid, and nanoparticles) via hydrogen bonding and other noncovalent interactions, promoting their further condensation and conversion to covalently bonded siloxane replicas. Based on these observations, we hypothesized that naturally crowded molecular environments, such as cells, would also direct silica condensation under similar conditions.
Results and Discussion
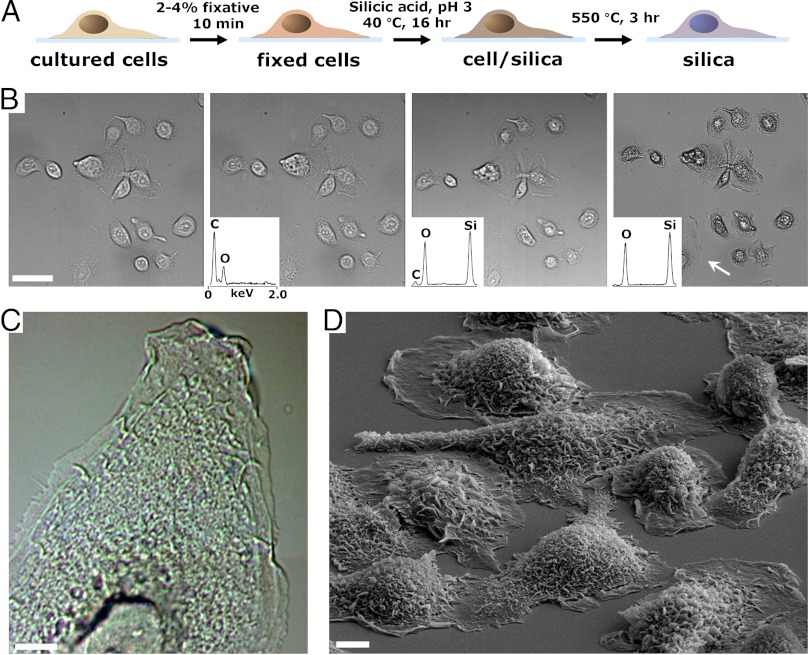
To address this question, we incubated chemically fixed mammalian cells in dilute, silicic acid solutions as illustrated schematically in Fig. 1A. In a typical experiment, cells plated onto glass substrates were fixed using 2–4% fixative (formaldehyde and glutaraldehyde produced qualitatively similar results) for at least 10 min. Cells were rinsed and immersed overnight (approximately 16 h) in a solution of 100 mM silicic acid at pH 3 and approximately 40 °C resulting in a composite comprising primarily silicon, oxygen, and carbon [cell/silica composites (CSCs)]. Fig. 1B shows brightfield images of the identical grouping of differentiated AsPC-1 pancreatic carcinoma cells: Live, after fixation, silicification and drying, and calcination at 550 °C. We observe structural features and dimensions at each stage of the process to be nearly identical to those of the parent (cell) templates albeit with some minor cracking observed, from SEM images of substrate bound, calcined CSCs (Fig. 1D). Additionally, features of hydrated living cells that were virtually transparent under brightfield microscopy appeared sharply resolved in calcined CSCs (e.g., the calcined sample imaged in Fig. 1C) due to the increase in refractive index contrast.
Fig. 1.
Silicification of mammalian cells cultured on flat substrates. (A) Schematic describing the process of cell silicification. (B) Image field of AsPC-1 cells throughout the steps (noted above) of silicification. Images in (i) and (ii) show hydrated cells and (iii) and (iv) show dehydrated composites and silica replicas. Insets show representative Energy Dispersive Spectroscopy spectra of cells at the various stages. (C) Close-up differential interference contrast (DIC) image of the cell noted by the white arrow in B, Right. (D) Representative SEM image of AsPC-1 templated cell silica following calcination. (Scale bars, B: 40 μm ; C and D: 5 μm.)
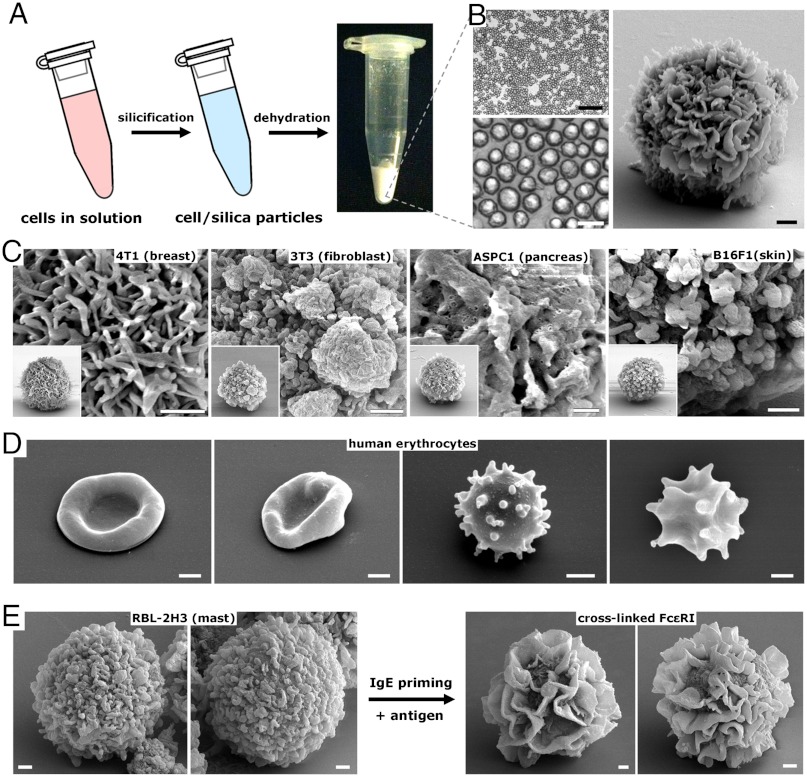
Cellular and subcellular morphology is dependent on genetic and environmental factors and therefore can be highly malleable and responsive to, for instance, physical interactions with a substrate. Initial experiments showed that the morphology of cells differentiated on a substrate can be faithfully captured in silica (Fig. 1). We wished to further explore the procedure under conditions that give rise to more physically homogenous CSCs with high throughput. Therefore, we fixed and silicified cells under suspension conditions that resulted in a population of essentially monodisperse composite microparticles (e.g., average diameter of 4T1-derived CSCs in Fig. 2B = 8.9 μm ± 1.4) with complex surface features (Fig. 2). For fast growing CHO cells (doubling time approximately 12 h) a standard 225 cm2 flask of adherent cells at 80% confluency (approximately 2.0 × 107 cells) yielded approximately 10–20 mgs dry weight of CSCs, indicating a means to rapidly produce gram scale quantities from cell lines such as CHO using large capacity bioreactors (36, 37). We tested this procedure on cultured cells derived from a variety of tissues and observed similar particle sizes within a given clonal cell line but widely differing surface morphologies both within and across the cell lines examined (Fig. 2C). Membrane ruffles, filaments, blebs, clusters, and smooth surfaces—common features of cell membrane dynamics—are captured in CSCs and calcined CSCs with high fidelity. Importantly, surface features of silica replicas could be directly modified by inducing cell behaviors such as apoptosis (Fig. S1) and surface ruffling prior to silicification. Fig. 2E shows RBL-2H3-templated CSCs following calcinations, which display the predicted grainy to ruffled membrane surface transformation accompanying surface receptor cross-linking (38).
Fig. 2.
CSC particles derived from cell suspensions. (A) Schematic representing the formation of CSC particles. Following silicification as for adherent cells, dehydration results in a dry powder comprised of monodisperse CSC particles (B; Right shows a closeup SEM of a 4T1-templated CSC displaying a ruffled external surface). (C) Close-up SEMs of CSC particles derived from a variety of tissues. Insets show the whole particle. (D) Calcined CSCs templated from human erythrocytes showing normal to increasingly abnormal/crenate morphology resulting from increasing levels of osmotic stress (Left to Right). (E) Calcined CSCs derived from RBL-2H3 cells before (Left) and after (Right) IgE cross-linking. (Scale bars, C–E: 1 μm.)
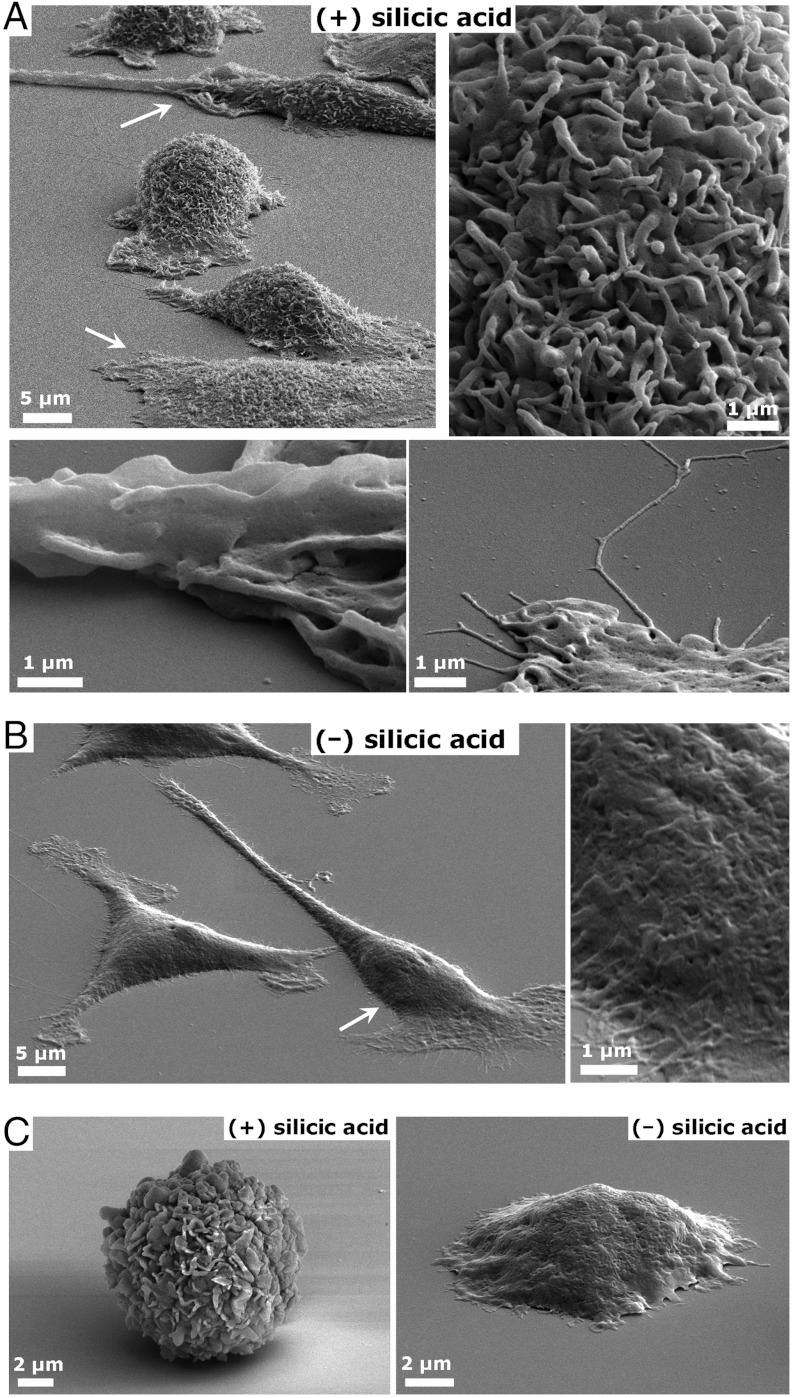
External features of CSCs (Fig. 3) show more defined and detailed surface structures compared to the identical cell line prepared using the well-established benchtop electron microscopy preparation procedures (i.e., no supercritical drying or rapid freezing) of fixation followed by careful dehydration in increasing concentrations of ethanol and drying from hexamethyldisilazane (HMDS) (Fig. 3B), a procedure shown to provide identical feature preservation as critical point drying (39, 40). Suspension cells silicified in solution showed particularly dramatic differences compared to nonsilicified cells. CSC particles dried in contact with a substrate (and even calcined) were resistant to deformation, remaining stiff and spherical, whereas the parent fixed cells deformed significantly with loss of surface features during drying (Fig. 3C) and of course were completely obliterated upon calcination (Fig. S2). Thus, silicification acts to mechanically stabilize the cellular architecture during drying and particularly during calcination, by forming a continuous, mechanically connected interpenetrating network throughout the cell hydrogel, analogous to our results from protein-templated silica hydrogels (35). This approach therefore may provide a simple alternative to common methods for specimen preparation/preservation that does not require extensive optimization, expertise, or specialized equipment (e.g., critical point dryer), and particularly when tolerance to extreme environments (e.g., temperature) is required. We note that silicification can alter the size of nanoscale cellular features in comparison to drying from HMDS. SEM comparisons of substrate-bound differentiated AsPC-1 cells indicates an increase in the size of nanoscale cellular features throughout the procedure (approximately 10 nm increase in width of CSC filopodia outgrowths versus nonsilicified cells), which is attributed to silica deposition (Fig. S3).
Fig. 3.
SEM comparison of AsPC-1-templated CSC features (a) versus cells fixed and dehydrated using standard procedures (b). Magnified features are indicated by arrows in A and B. (C) SEM images of SK-OV-3 suspension cultured cells dried against a substrate with and without silicic acid treatment.
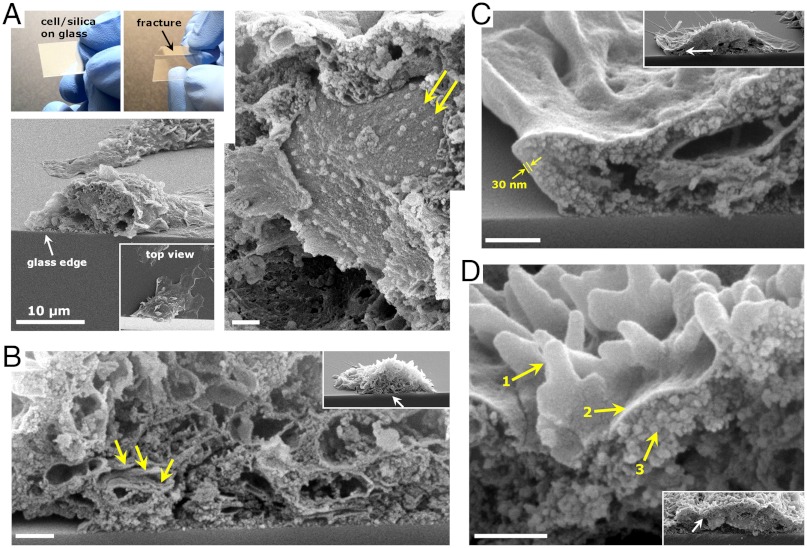
To examine the internal features of CSCs in greater detail, AsPC-1 cells were plated onto glass substrates, silicified, and dried. Glass substrates were scored on the surface opposite the cells and fractured (Fig. 4A). Because of the brittle fracture characteristics of the CSCs, cells lying across the fracture edge were often cleanly sectioned, allowing cross-sectional analysis using SEM. Fig. 4A shows a sectioned cell revealing intracellular structures such as the nuclear membrane, indicated by 100 nm diameter ring-like features (presumably nuclear pore complexes). Comparison of fractured CSCs (e.g., Fig. 4 A and B) and fractured calcined CSCs (Fig. 4 C and D) showed no obvious difference in size or shape of internal features after calcination. Examining calcined structures such as those in Fig. 4 C and D as well as transmission electron microscopy (TEM) cross-sectional images of CSC particles (Fig. S4) a conformal silica coating of ca. 30-nm thick is apparent, elaborated around filapodia-like features (Fig. 4D) and encasing the intracellular-templated structures and void spaces. In a eukaryotic cell, the membrane is defined by the phospholipid bilayer anchored to the cell cortex via membrane-bound proteins. The cortex is composed of fibrous proteins such as spectrin and actin, forming a meshwork that provides mechanical strength to the membrane. High-resolution atomic force microscopy (AFM) imaging of relatively flat regions of calcined external surfaces were featureless at approximately 2 nm resolution indicting the absence of a primary feature or particle size (Fig. S5). Similar observations were made in AFM studies of select diatom cell surfaces (41). In comparison, silica templated by single component protein hydrogel scaffolds was observed to be granular with a primary feature size of approximately 16 nm (35).
Fig. 4.
Cross-sectional imaging of CSCs enabled by simple fracture technique. (A) Fracture of CSCs on a coverslip provides clean sectioning to reveal intracellular features using SEM. (Right) A close-up view of the sectioned cell. Arrows indicate nuclear pore complexes. (B) SEM section of a CSC shows multilayer, endoplasmic reticulum-like structures (arrows). (C) Calcined CSC sectioned on glass shows a 30 nm membrane templated silica structure. (D) Filopodia-templated upright protrusions (1) are encased in a smooth silica membrane (2) overlying roughened, particle-based features (3) in a calcined and sectioned CSC. Arrows in B–D insets point to the area of magnification. (Scale bars, 500 nm.)
In an effort to understand the mechanism of cell silicification we conducted a series of experiments. First, cells subjected to silicification conditions without fixation were observed to swell significantly, as a result of hypotonic stress, but nonetheless formed CSCs [albeit with drastic differences in morphology due to membrane swelling and other stresses incurred during silicic acid incubation (Fig. S6)]. Erythrocytes are particularly sensitive to osmolarity and were found to lyse in the silicic acid solution when fixed for short timescales (Fig. S6). Through a modified fixation process and use of an osmotically balanced silicic acid solution (addition of 0.9% NaCl), CSCs and calcined erythrocytes silica replicas were achieved that faithfully replicated the parent cell morphology (Fig. 2D).
Complete solubilization of the membrane of fixed cells using a mild detergent (0.5% Triton X-100) prior to silicification resulted in CSCs with deformed features, most likely incurred as a result of settling against the reaction tube surface (Fig. S7). However, staining of the outer lipid membrane and intracellular proteins followed by silicification showed some delocalization of lipid following incubation in the silicic acid solution (also, confirmed by poststaining CSCs using a lipid-associating dye) while the protein dye remained stationary (Fig. S8). Triton X-100 is not expected to disrupt the cortical layer or other cytoskeletal constituents, or denature most proteins at this concentration. Taken together, these results indicate that the whole membrane complex (lipid bilayer + cortex) is necessary to maintain the mechanical integrity of CSC surfaces, but that a portion of the lipid component is gradually displaced during silica deposition.
Indeed, time-lapse imaging of a lipid membrane dye (Fig. S9) indicates that the presence of dilute methanol (hydrolysis product of tetramethyl orthosilicate (TMOS) in the silicification solution provides relatively slow and mild permeabilization of cell membranes (compared to methods used for immunostaining such as Triton and 100% methanol) that enables silica precursors to penetrate into the cell while maintaining the mechanical integrity of external cell features during silica deposition. Additionally, CSCs derived from Escherichia coli do not retain cellular structure following calcination (Fig. S10), which indicates incomplete silica templating, most likely as a consequence of inhibited intracellular penetration of silica precursors past the prokaryotic cell envelope.
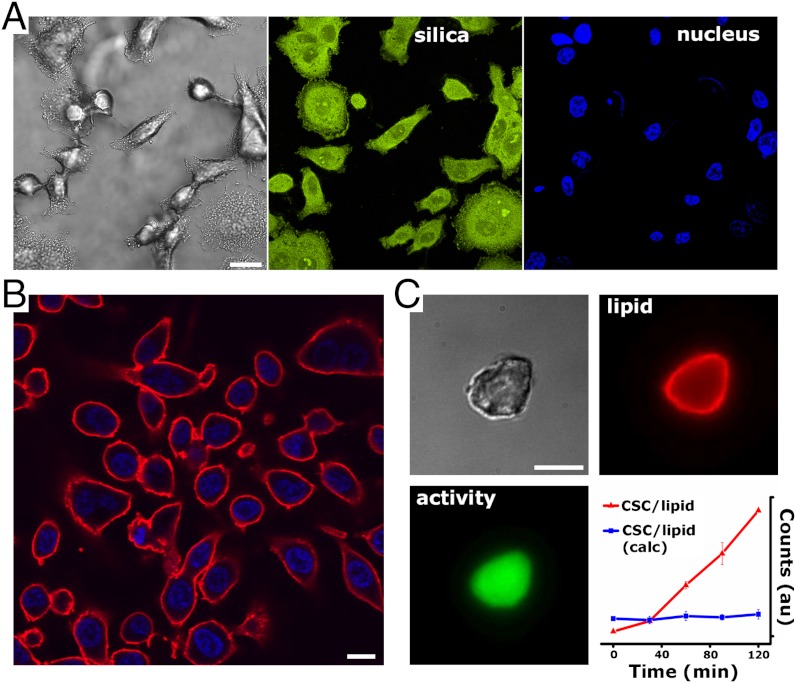
Silica localization throughout the CSC was observed during silicification using PDMPO ([2-(4-pyridyl)-5-((4-(2-dimethylaminoethylamino- carbamoyl)methoxy)phenyl)oxazole]), which has been shown to incorporate with silica as it condenses (32). Staining was observed throughout the entirety of the cytoplasm and nucleus following incubation for 16 h (Fig. 5A). This observation indicates that although silica condensation is likely to occur over variable timescales at the (macro)molecular scale, it eventually infiltrates all discernable subcellular structures and organelles—with the notable exception of large, fluid filled vacuoles (Fig. S11). Further, the nuclear stain DAPI is shown to localize exclusively within the nuclear region of CSCs with little background signal (Fig. 5 A and B), indicating that when CSCs are incubated in an aqueous solution of the dye molecule, the DNA helical structure remains intact and molecularly accessible within the nucleus—despite silicification throughout the nuclear region. N2 sorption isotherms (Fig. S12) obtained from CHO-templated silica particles (representing a silica imprint of the internal and external cellular structure) indicated a Brunauer–Emmett–Teller surface area of approximately 365 m2/g and a broad range of pore dimensions, although with no appreciable microporosity. Hysteresis in the desorption branch indicated two populations of mesopore restrictions, which we interpret as the presence of large interstitial pores defined by the volume between cellular structures connected through two subsets of smaller pores.
Fig. 5.
Distribution of silica in CSCs, nuclear staining, and lipid membrane reconstitution. (A) DIC and confocal fluorescence images of AsPC-1-templated CSCs show that silica is continuous throughout the cytoplasm and nucleus as indicated by PMPDO staining (Middle). (Right) Localization of DAPI nuclear stain. (B) Confocal fluorescence image slice of substrate grown AsPC-1 CSCs showing surface localization of lipid (red) and internal location of the nucleus (blue). (c) CSC particles supporting lipid layers show accumulation of esterase fluorogenic products (red line). The blue line shows activity of calcined CSCs supporting lipid bilayers. Error bars describe the standard deviation (n≥5) of the maximum intensity value. (Scale bars, A and B: 10 μm; 5 μm C.)
The results from the above series of experiments indicate that the silica deposition process occurs throughout the complete volume of the cell to produce a faithful replica of the exterior and interior cellular structures. Based on the featurelessness of silica deposits in select areas, we conclude that deposition at pH 3 involves weakly charged monomeric or small oligomeric silicic acid species that interact noncovalently with the crowded biomolecular components comprising the cell. The high fidelity replication and self-limiting characteristics suggest a mechanism where silicic acid is distributed uniformly over and throughout the cell scaffold, where it undergoes acid or base catalyzed condensation promoted by the spectrum of proximal functional groups such as protein surface residues. In this manner, the process is inherently self-limiting to form a continuous silica replica throughout the cell. Remarkable is that the silicified cell, although nanostructured, withstands drying and sintering to 550 °C with minimal shrinkage (Fig. S3). Generally, drying (capillary) and sintering stresses would result in enormous volumetric changes (42). The absence of appreciable shrinkage speaks to the mechanical integrity of the cell-catalyzed silica replica. The absence of primary particles and microporosity reduces greatly both drying and sintering stresses, which scale roughly inversely with particle or pore size. One mechanistic hypothesis consistent with these observations is that at pH 3, where silicic acid monomers and oligomers are uncharged (26, 43), silicic acid incorporates within the continuous hydrogen bonded water network encompassing cellular surfaces where it becomes locally concentrated and subsequently condensed amphoterically via surface moieties (e.g., acidic and basic protein residues).
In essence, the structural complexity of cells is captured via self-limiting nanoscale replication in a hard material, providing a platform in which to preserve and reconstitute cellular functions. For example, amphiphilic lipid bilayers introduced as liposomes localize (selectively as compared to on the adjoining substrate) on the outer surfaces of CSCs demonstrating that the membrane lipid component could, in principle, be reconstituted. Subsequent, incubation with a lipid diffusible fluorogenic stain used to assess cellular viability indicated retention of some level of enzyme activity; sequestration of the dye (based on esterase cleavage to form a lipid insoluble fluorophore) was observed in CSCs supporting lipid membranes versus calcined CSCs (Fig. 5C). These initial results provide an avenue to begin to explore CSCs as an alternative route to biocatalyst stabilization where the current state-of-the-art employs prefabricated (mesoporous) silicas for subsequent enzyme loading (44–47). By using this general approach as a starting point, more complex and specific biocatalyst stabilization can be targeted, by stabilizing enzymes and enzyme complexes in their optimized, crowded in vivo configurations.
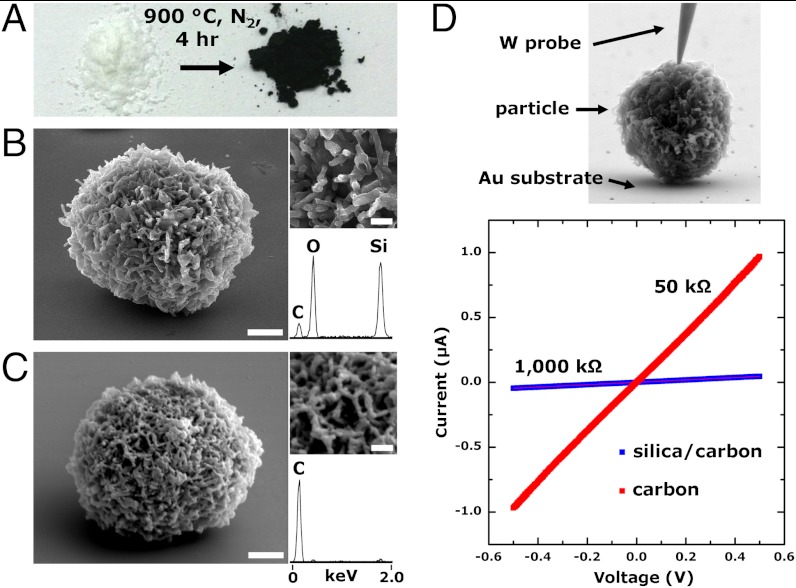
Finally, the ability to replicate both surface and intracellular molecular architectures with silica provides opportunities to investigate shape-preserving chemical transformations of CSCs to other materials, for instance, using approaches analogous to those developed for diatom silica (16–18). To begin to explore these properties, we investigated the ability of CSCs to render porous carbon structures, a class of materials with substantial utility in fuel cell, decontamination, and sensor technologies. We subjected CSC particles to high-temperature pyrolysis conditions (900 °C, 4 h, under N2 atmosphere), which resulted in an opaque powder (Fig. 6A), with individual particles [carbonized-cell/silica composites (c-CSCs)] displaying similar morphologies to that of the starting material (Fig. 6B). Subsequent dissolution of the silica support [6 M potassium hydroxide (KOH), 4 d] resulted in free-standing carbon particles retaining cellular morphologies (Fig. 6C). In situ SEM electrical characterization (Fig. 6D) showed ohmic conductivity through the particles. Representative current–voltage (IV) curves for c-CSCs and carbon replicas are shown in Fig. 6D, Lower. Note that removal of the insulative silica support decreased particle resistance approximately 20-fold. These results indicate that the wide heterogeneity of in vitro soft cellular architectures can now be considered for use as a feedstock for most materials processing procedures, including those requiring high temperature and pressure.
Fig. 6.
Shape-preserving carbonization of 4T1 CSCs. (A) Pyrolysis of CSCs produces an opaque powder comprised of particles that have retained cellular structure (C). Etching of the silica produces a carbon rich replica. (D) In situ electrical characterization of carbonized particles shows a 20-fold decrease in electrical resistance across a particle following silica etching. (Scale bars, B and C: 2 μm; Insets, 500 nm).
We have described a simple approach to derive functional biomorphic composites, silica frustules, and carbon replicas from mammalian cells, which should allow straightforward customization of structure and function via chemical and genetic engineering. This procedure does not require preinfiltration of templating molecules [e.g., cationic polymers; (48)] or multistep layer by layer assembly and is distinct from other inorganic biotemplating strategies that simply coat external surfaces to produce hollow shells or low fidelity inverse structures following calcination (12, 29). In contrast to the majority of studies describing cell encapsulation in silica (49) where the primary goal of maintaining cell viability necessitates reaction conditions near neutral pH and cells become physically entrapped within (nonconformal) gels, here the charge of silicic acid is essentially neutral (pH 3) (26, 43) and thus hydrogen bonding and other noncovalent silica/molecular interactions govern deposition (35, 43). To date, individual cellular/biomolecular components, peptides, proteins, lipid vesicles, polysaccharides, cytoskeletal filaments, etc. have all been shown to interact with, and often template silica in vitro but with no control over 3D structure (12, 27, 29, 43). Presented on and within a cell, these collective silica/molecular interactions are exploited here under molecularly crowded environments using stable sols (e.g., limited homopolymerization, no gel formation, etc.) such that deposition is targeted to cell structures, resulting in a process that is inherently conformal and self-limiting due to slow solution silica polymerization kinetics (43). The apparent generalizability of this process should allow for the synthetic production of complex and durable composites and minerals with structural diversity approaching that of natural biomineralizing microorganisms.
Methods
Cell Silicification and Carbonization.
Cells were incubated in closed containers of 100 mM TMOS solution in 1 mM HCl at approximately 40 °C for 16–18 h. CSCs were dehydrated by sequential soaking in deionized water, 1∶1 DI water∶methanol, and 100% methanol (2X) for 10 min in each solution, followed by drying in air. Calcination was performed in air at 550 to 600 °C for 3–4 h, which eliminated the majority of organics. Fig. S13 shows a representative thermogravimetric analysis (TGA) curve acquired from CHO-derived CSCs. Pyrolysis and etching were performed using a previously described method (50). Briefly, pyrolysis was carried out at 900 °C in a tube furnace under flowing nitrogen for 4 h and silica was etched in 6 M KOH for 4 d.
Cell Culture, Detailed Silicification, Imaging, and Other Analyses.
See SI Methods for details.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Constantine Y. Khripin, Wendy M. Patterson, and Diane S. Lidke for useful discussions and Eric Coker and Steven Jett for technical assistance. We acknowledge support from US Department of Energy, Office of Science, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering. This work was performed, in part, at the Center for Integrated Nanotechnologies, a US Department of Energy, Office of Basic Energy Sciences user facility. Sandia National Laboratories is a multiprogram laboratory managed and operated by Sandia Corporation, a wholly owned subsidiary of Lockheed Martin Corporation, for the US Department of Energy’s National Nuclear Security Administration under contract DE-AC04-94AL85000. Some images in this paper were generated in the University of New Mexico, Cancer Center Fluorescence Microscopy Facility, supported by National Center for Research Resources, National Science Foundation, and National Cancer Institute. J.L.T. acknowledges support from Air Force Office of Scientific Research grant, FA 9550-10-1-0054 and the New Mexico Cancer Nanotech Training Center Postdoctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205816109/-/DCSupplemental.
References
- 1.Kresge CT, Leonowicz ME, Roth WJ, Vartuli JC, Beck JS. Ordered mesoporous molecular-sieves synthesized by a liquid-crystal template mechanism. Nature. 1992;359:710–712. [Google Scholar]
- 2.Beck JS, et al. A new family of mesoporous molecular-sieves prepared with liquid-crystal templates. J Am Chem Soc. 1992;114:10834–10843. [Google Scholar]
- 3.Mann S, Ozin GA. Synthesis of inorganic materials with complex form. Nature. 1996;382:313–318. [Google Scholar]
- 4.Brinker CJ, Lu YF, Sellinger A, Fan HY. Evaporation-induced self-assembly: Nanostructures made easy. Adv Mater. 1999;11:579–585. [Google Scholar]
- 5.Pouget E, et al. Hierarchical architectures by synergy between dynamical template self-assembly and biomineralization. Nat Mater. 2007;6:434–439. doi: 10.1038/nmat1912. [DOI] [PubMed] [Google Scholar]
- 6.Chen CL, Rosi NL. Peptide-based methods for the preparation of nanostructured inorganic materials. Angew Chem Int Ed. 2010;49:1924–1942. doi: 10.1002/anie.200903572. [DOI] [PubMed] [Google Scholar]
- 7.Shopsowitz KE, Qi H, Hamad WY, MacLachlan MJ. Free-standing mesoporous silica films with tunable chiral nematic structures. Nature. 2010;468:422–425. doi: 10.1038/nature09540. [DOI] [PubMed] [Google Scholar]
- 8.Boissiere C, Grosso D, Chaumonnot A, Nicole L, Sanchez C. Aerosol route to functional nanostructured inorganic and hybrid porous materials. Adv Mater. 2011;23:599–623. doi: 10.1002/adma.201001410. [DOI] [PubMed] [Google Scholar]
- 9.Holland BT, Blanford CF, Stein A. Synthesis of macroporous minerals with highly ordered three-dimensional arrays of spheroidal voids. Science. 1998;281:538–540. doi: 10.1126/science.281.5376.538. [DOI] [PubMed] [Google Scholar]
- 10.Hatton B, Mishchenko L, Davis S, Sandhage KH, Aizenberg J. Assembly of large-area, highly ordered, crack-free inverse opal films. Proc Natl Acad Sci USA. 2010;107:10354–10359. doi: 10.1073/pnas.1000954107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein A, Li F, Denny NR. Morphological control in colloidal crystal templating of inverse opals, hierarchical structures, and shaped particles. Chem Mater. 2008;20:649–666. [Google Scholar]
- 12.Paris O, Burgert I, Fratzl P. Biomimetics and biotemplating of natural materials. MRS Bull. 2010;35:219–225. [Google Scholar]
- 13.Van Opdenbosch D, Fritz-Popovski G, Paris O, Zollfrank C. Silica replication of the hierarchical structure of wood with nanometer precision. J Mater Chem. 2011;26:1193–1202. [Google Scholar]
- 14.van Bommel KJC, Friggeri A, Shinkai S. Organic templates for the generation of inorganic materials. Angew Chem Int Ed. 2003;42:980–999. doi: 10.1002/anie.200390284. [DOI] [PubMed] [Google Scholar]
- 15.Fratzl P, Weiner S. Bio inspired materials–mining the old literature for new ideas. Adv Mater. 2010;22:4547–4550. doi: 10.1002/adma.201002127. [DOI] [PubMed] [Google Scholar]
- 16.Behrens P, Baeuerlein E, editors. Handbook of Biomineralization: Biomimetic and Bioinspired Chemistry. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2007. Inorganic preforms of biological origin: Shape-preserving reactive conversion of biosilica microshells (diatoms) pp. 235–253. [Google Scholar]
- 17.Losic D, Mitchell JG, Voelcker NH. Diatomaceous lessons in nanotechnology and advanced materials. Adv Mater. 2009;21:2947–2958. [Google Scholar]
- 18.Bao Z, et al. Chemical reduction of three-dimensional silica micro-assemblies into microporous silicon replicas. Nature. 2007;446:172–175. doi: 10.1038/nature05570. [DOI] [PubMed] [Google Scholar]
- 19.Hildebrand M. Prospects of manipulating diatom silica nanostructure. J Nanosci Nanotechnol. 2005;5:146–157. doi: 10.1166/jnn.2005.013. [DOI] [PubMed] [Google Scholar]
- 20.Townley HE, Woon KL, Payne FP, White-Cooper H, Parker AR. Modification of the physical and optical properties of the frustule of the diatom Coscinodiscus wailesii by nickel sulfate. Nanotechnology. 2007;18:295101–295106. [Google Scholar]
- 21.Kroger N. Prescribing diatom morphology: Toward genetic engineering of biological nanomaterials. Curr Opin Chem Biol. 2007;11:662–669. doi: 10.1016/j.cbpa.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Kroger N, Deutzmann R, Bergsdorf C, Sumper M. Species-specific polyamines from diatoms control silica morphology. Proc Natl Acad Sci USA. 2000;97:14133–14138. doi: 10.1073/pnas.260496497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroger N, Deutzmann R, Sumper M. Polycationic peptides from diatom biosilica that direct silica nanosphere formation. Science. 1999;286:1129–1132. doi: 10.1126/science.286.5442.1129. [DOI] [PubMed] [Google Scholar]
- 24.Cha JN, et al. Silicatein filaments and subunits from a marine sponge direct the polymerization of silica and silicones in vitro. Proc Natl Acad Sci USA. 1999;96:361–365. doi: 10.1073/pnas.96.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kröger N, Lorenz S, Brunner E, Sumper M. Self -assembly of highly phosphorylated silaffins and their function in biosilica morphogenesis. Science. 2002;298:584–586. doi: 10.1126/science.1076221. [DOI] [PubMed] [Google Scholar]
- 26.Coradin T, Coupé A, Livage J. Interactions of bovine serum albumin and lysozyme with sodium silicate solutions. Colloids Surf B. 2003;29:189–196. [Google Scholar]
- 27.Bassindale A, Taylor P, Abbate V, Brandstadt K. Simple and mild preparation of silica-enzyme composites from silicic acid solution. J Mater Chem. 2009;19:7606–7609. [Google Scholar]
- 28.Gautier C, et al. Biomimetic dual templating of silica by polysaccharide/protein assemblies. Colloids Surf B. 2008;65:140–145. doi: 10.1016/j.colsurfb.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 29.Dickerson M, Sandhage K, Naik R. Protein- and peptide-directed syntheses of inorganic materials. Chem Rev. 2008;108:4935–4978. doi: 10.1021/cr8002328. [DOI] [PubMed] [Google Scholar]
- 30.Patwardhan SV, Clarson SJ, Perry CC. On the role(s) of additives in bioinspired silicification. Chem Commun. 2005;9:1113–1121. doi: 10.1039/b416926c. [DOI] [PubMed] [Google Scholar]
- 31.Hildebrand M. Diatoms, biomineralization processes, and genomics. Chem Rev. 2008;108:4855–4874. doi: 10.1021/cr078253z. [DOI] [PubMed] [Google Scholar]
- 32.Tesson B, Hildebrand M. Extensive and intimate association of the cytoskeleton with forming silica in diatoms: Control over patterning on the meso- and micro-scale. PloS One. 2010;5:e14300. doi: 10.1371/journal.pone.0014300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brunner E, et al. Chitin-based organic networks: An integral part of cell wall biosilica in the diatom thalassiosira pseudonana. Angew Chem Int Ed. 2009;48:9724–9727. doi: 10.1002/anie.200905028. [DOI] [PubMed] [Google Scholar]
- 34.Scheffel A, Poulsen N, Shian S, Kroger N. Nanopatterned protein microrings from a diatom that direct silica morphogenesis. Proc Natl Acad Sci USA. 2011;108:3175–3180. doi: 10.1073/pnas.1012842108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khripin CY, Pristinski D, Dunphy DR, Brinker CJ, Kaehr B. Protein-directed assembly of arbitrary three-dimensional nanoporous silica architectures. ACS Nano. 2011;5:1401–1409. doi: 10.1021/nn1031774. [DOI] [PubMed] [Google Scholar]
- 36.Warnock JN, Al-Rubeai M. Bioreactor systems for the production of biopharmaceuticals from animal cells. Biotechnol Appl Biochem. 2006;45:1–12. doi: 10.1042/BA20050233. [DOI] [PubMed] [Google Scholar]
- 37.Xing Z, Kenty BM, Li ZJ, Lee SS. Scale-up analysis for a CHO cell culture process in large-scale bioreactors. Biotechnol Bioeng. 2009;103:733–746. doi: 10.1002/bit.22287. [DOI] [PubMed] [Google Scholar]
- 38.Wilson BS, et al. Calcium-dependent clustering of inositol 1,4,5-trisphosphate receptors. Mol Biol Cell. 1998;9:1465–1478. doi: 10.1091/mbc.9.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braet F, De Zanger R, Wisse E. Drying cells for SEM, AFM and TEM by hexamethyldisilazane: A study on hepatic endothelial cells. J Microsc. 1997;186:84–87. doi: 10.1046/j.1365-2818.1997.1940755.x. [DOI] [PubMed] [Google Scholar]
- 40.Bray DF, Bagu J, Koegler P. Comparison of hexamethyldisilazane (HMDS),Peldri II and critical-point drying methods for scanning electron microscopy of biological specimens. Microsc Res Tech. 1993;26:489–495. doi: 10.1002/jemt.1070260603. [DOI] [PubMed] [Google Scholar]
- 41.Hildebrand M, Doktycz MJ, Allison DP. Application of AFM in understanding biomineral formation in diatoms. Pflügers Arch. 2008;456:127–137. doi: 10.1007/s00424-007-0388-y. [DOI] [PubMed] [Google Scholar]
- 42.Brinker CJ, Scherer GW. Sol-Gel Science. San Diego: Academic; 1990. [Google Scholar]
- 43.Iler RK. The Chemistry of Silica: Solubility, Polymerization, Colloid and Surface Properties, and Biochemistry. New York: Wiley; 1979. [Google Scholar]
- 44.Hanefeld U, Gardossi L, Magner E. Understanding enzyme immobilization. Chem Soc Rev. 2008;38:453–468. doi: 10.1039/b711564b. [DOI] [PubMed] [Google Scholar]
- 45.Hudson S, Cooney J, Magner E. Proteins in mesoporous silicates. Angew Chem Int Ed. 2008;47:8582–8594. doi: 10.1002/anie.200705238. [DOI] [PubMed] [Google Scholar]
- 46.Betancor L, Luckarift HR. Bioinspired enzyme encapsulation for biocatalysis. Trends Biotechnol. 2008;26:566–572. doi: 10.1016/j.tibtech.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 47.Avnir D, Coradin T, Lev O, Livage J. Recent bio-applications of sol–gel materials. J Mater Chem. 2005;16:1013–1030. [Google Scholar]
- 48.Niu L, et al. Infiltration of silica inside fibrillar collagen. Angew Chem Int Ed. 2011;50:11688–11691. doi: 10.1002/anie.201105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meunier CF, Dandoy P, Su BL. Encapsulation of cells within silica matrixes: Towards a new advance in the conception of living hybrid materials. J Colloid Interface Sci. 2010;342:211–224. doi: 10.1016/j.jcis.2009.10.050. [DOI] [PubMed] [Google Scholar]
- 50.Carroll NJ, Pylypenko S, Atanassov PB, Petsev DN. Microparticles with bimodal nanoporosity derived by microemulsion templating. Langmuir. 2009;25:13540–13544. doi: 10.1021/la900988j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.