Abstract
Fragment-based design was used to guide derivatization of a lead series of β-lactamase inhibitors that had heretofore resisted optimization for in vivo activity. X-ray structures of fragments overlaid with the lead suggested new, unanticipated functionality and points of attachment. Synthesis of three derivatives improved affinity over 20-fold and improved efficacy in cell culture. Crystal structures were consistent with the fragment-based design, enabling further optimization to a Ki of 50 pM, a 500-fold improvement that required the synthesis of only six derivatives. One of these, compound 5, was tested in mice. Whereas cefotaxime alone failed to cure mice infected with β-lactamase-expressing Escherichia coli, 65% were cleared of infection when treated with a cefotaxime:5 combination. Fragment complexes offer a path around design hurdles, even for advanced molecules; the series described here may provide leads to overcome β-lactamase-based resistance, a key clinical challenge.
Keywords: antibiotic resistance, antimicrobial, drug discovery, structure-based, boronic acid
Lead optimization in chemical biology and drug discovery is a multifactorial problem and frequently stalls on the way to tool molecules or clinical candidates. Confronted with an otherwise attractive compound series for which affinity or efficacy has leveled off, for instance, one seeks an efficient derivatization strategy in the face of many possible chemotypes and points of derivatization. Multiple paths may be considered, from combinatorial derivatization among accessible side chains, to structure-based placement of specific groups. Neither of these strategies can fully promise to overcome the challenge of knowing exactly where and how to derivatize a lead.
We faced such a challenge with a series of boronic acid β-lactamase inhibitors. β-lactamases are the most common cause of resistance to β-lactam antibiotics, such as penicillins and cephalosporins; they threaten what remains the most widely used class of antibiotics and have attracted much recent attention (1–5). Third- and fourth-generation cephalosporins were partly introduced to overcome these enzymes, as were inhibitors like clavulanic acid, but resistance arose rapidly to these agents, which are all themselves β-lactams and thus potential substrates of β-lactamases. Boronic acids, as non-lactam transition-state analogs, are impervious to such mutants and are often transparent to other β-lactam resistance mechanisms, such as β-lactamase upregulation. Using β-lactam functional groups on a boronic acid scaffold, we rapidly optimized an initial class of boronic acids to mid-nanomolar affinity (6, 7), and with further derivatization to 1 nM affinity against the class C β-lactamase AmpC (8). Notwithstanding their high affinity, these compounds had relatively modest activity in bacterial cell culture. Though they certainly reduced minimum inhibitory concentrations (MICs) of third-generation cephalosporins, they were unable to break through the empirical resistance threshold—typically 2 μg/mL—for most pathogens (6–9). Subsequent efforts failed to further improve affinity (9). Also, these molecules used functionality well explored among β-lactams themselves to achieve their affinity—utilizing, for instance, the R1 side chains of cephalothin and ceftazidime, and the C3(4)’ carboxylate of penicillins and cephalosporins—and so they were subject to pre-evolved resistant mutant enzymes, such as the inhibitor-resistant TEM30 (10).
To escape this cul-de-sac, we thought to replace the boronic acid R1 carboxamide, which mimics the analogous group ubiquitous among β-lactam drugs, with a sulfonamide, as this would not only change the electronic character of a key interaction but also the geometry of the inhibitor (11). This switch scrambled the structure-activity relationship previously observed with the carboxamides: Boronic acids with a carboxylate analogous to the C3(4)’ group of penicillins and β-lactams lost 10- to 100-fold activity, relative to the carboxamide series; conversely, small derivatives on the R1 side, such as compounds 1 and 2 (Table 1), achieved Ki values as low as 70 and 25 nM, 8- to 20-fold better than observed for analogous carboxamides. Unfortunately, further derivatization failed to improve affinity; indeed, even the addition of a carboxylate to 1, leading to 2, improved activity only threefold, whereas a similar derivative in an earlier series had improved affinity by over 20-fold (12). We were stuck again.
Table 1.
Ki values of leads and fragment-optimized compounds against AmpC β-lactamase
*Compounds 1, 2, and 9 are lead compounds that began this study (11).
Fragment-based discovery has anchored early lead discovery for targets that have resisted traditional methods. Fragments can optimally explore receptor pockets (13–18) and better cover chemical space (19–22), and we wondered if they could guide a late-stage optimization that had thus far floundered. In a previous fragment-based docking screen against AmpC (22) and in a defragmentation study of a known AmpC inhibitor (23), crystal structures of three anionic fragments were determined: a thiophene carboxylic acid (F1), a benzoic acid (F2), and an aryl tetrazole (F3), all of which bind in the same distal region of the active site where the aryl carboxylate of compound 2 was placed (Fig. 1 A, B, and D). The poses adopted by these fragments overlapped one another, and though in roughly the same spot as the carboxylate of compound 2, they differed in their angle of attack from the larger inhibitor (Fig. 1B). Consequently, in their crystallographic complexes with AmpC they hydrogen-bonded with the backbone amides of Ser212 and Gly320, rather than with Ser212 alone as did the 2 carboxylate. Reasoning that the fragments bound in geometries unperturbed by the restraints of the larger molecule, we thought that boronic acids capturing this geometry might have improved affinity and antibiotic efficacy. Here we describe an optimization campaign guided by these fragments, and follow these new derivatives into ever more biologically relevant investigations. A strategy for a fragment-based later-stage lead optimization is considered.
Fig. 1.
Fragment-guided modeling of new boronic acids. (A) X-ray structures of fragments F1 (cyan) and F2 (magenta) bound to distal site defined by Ser212 and Gly320. (B) Superposition of X-ray structure of 2 (yellow) on F2. (C) Superposition of model of 3 (cyan) on F2. (D) X-ray structure of fragment F3 (magenta) bound to distal site. (E) Superposition of model of 4 (cyan) on F3. (F) Superposition of model of 5 (cyan) on F3.
Results
Fragment-Guided Design.
Superposition of the X-ray structure complex of the 25 nM compound 2 with the X-ray complexes of fragments F1 and F2 suggested an initial optimization path (Fig. 1 A and B). All three molecules contain an aryl carboxylate and all occupy the same overall site in AmpC, but they adopt different orientations (Fig. 1B). The two fragments orient their carboxylates to hydrogen-bond with the backbone amides of Ser212 and Gly320, while the carboxylate of the larger lead compound 2 hydrogen-bonds only with Ser212, leaving its second oxygen solvent exposed; the aromatic ring plane also differs. We therefore modeled analogs of 2 that could position a carboxylate or a tetrazole to superpose with the same groups in the fragments. Evaluation of several candidates suggested that a boronic acid sulfonamide bearing a benzyl side chain with a meta-carboxylate would do so (i.e., by shifting the existing carboxylate in the lead 2 by one ring position, Fig. 1C), whereas models derived from fragment F3 bearing a tetrazole (Fig. 1D) favored a para-phenyl derivative (i.e., by replacing the carboxylate in the lead 2 by a tetrazole, Fig. 1E). The pyrazolone ring in fragment F3 suggested that a pyridine would also be favorable, potentially interacting either with the Gly320 backbone or a protonated tetrazole (i.e., a more involved perturbation, adding a tetrazole and a ring nitrogen, Fig. 1F).
Activity and Structures of the First Derivatives.
Ki values of the new boronic acids 3, 4, and 5 were determined from IC50 curves, and competition with the AmpC substrate CENTA was confirmed by full Lineweaver-Burk analysis, as described (SI Appendix) (24). All were competitive inhibitors with Ki values ranging from 0.8 to 1.3 nM, an improvement of 20-to 30-fold relative to the lead 2 (Table 1). Whereas 2 had no measurable time dependence to its activity, all three of the new inhibitors exhibited an incubation effect owing to slow off-rates, as suggested by enzyme incubation and dilution experiments (SI Appendix, Fig. S3) (25).
To understand the affinity increase at atomic resolution, we determined the X-ray structures of compounds 3, 4, and 5 in complex with AmpC β-lactamase, to between 1.43 and 1.80 Å resolution (SI Appendix, Table S1). Initial Fo-Fc electron density maps allowed all three new inhibitors to be modeled unambiguously (SI Appendix, Fig. S4). As expected, the Oγ of the catalytic Ser64 forms a dative covalent bond to the boron atoms of the inhibitors, with the boronic acid adopting a tetrahedral geometry. One boronic oxygen hydrogen-bonds with the AmpC oxyanion hole, defined by backbone amides of Ser64 and Ala318, while the other oxygen hydrogen-bonds with Tyr150 and the conserved water 402 (Wat402), as observed in earlier boronic acid structures (24, 26, 27). Similarly, the key hydrogen bond between a sulfonamide oxygen and Asn152 is conserved (Fig. 2 A–F). In the AmpC/3 complex an additional hydrogen bond is formed between the sulfonamide nitrogen and the backbone carbonyl of Ala318. The new benzyl ring makes parallel π–π stacking interactions with Tyr221 while the carboxylate hydrogen bonds with backbone amides of Ser212 and Gly320, as intended by design (Fig. 2A). Meanwhile, the conserved moieties of the tetrazole 4 interact largely as observed in the AmpC/3 complex, and though the sulfonamide has shifted, it makes the same crucial hydrogen bond to Asn152 (Fig. 2B). Unlike the benzyl of 3, the phenyl ring of 4 makes edge-to-face π–π stacking interactions with Tyr221 at an angle of 55°. The tetrazole ring is almost coplanar with the phenyl ring (angle: 13°) and two nitrogen atoms of the tetrazole ring hydrogen-bond with Ser212 and Gly320 backbone amides, also foreseen by design. Compound 5 differs from 4 only in the replacement of a phenyl by a pyridine in 5, which superimposes closely on the 4 structure; an ordered water (Wat863) is observed to interact with the pyridine nitrogen (as might, too, a protonated form of the tetrazole intramolecularly) (Fig. 2C). Overall, the modeled structures may be superposed with the crystallographic results with little deviation (Fig. 3).
Fig. 2.
X-ray structures of the new sulfonamide boronic acids bound to AmpC. AmpC carbon atoms depicted in grey, ligand carbon atoms in yellow, oxygens in red, nitrogens in blue, sulfurs in yellow, borons in amaranth pink, chlorides in green, fluorides in light blue. Red dashes represent hydrogen bonds, water molecules are represented by red spheres. (A) AmpC/3. (B) AmpC/4. (C) AmpC/5. (D) AmpC/6. (E) AmpC/7. (F) AmpC/8.
Fig. 3.
X-ray structures of the new compounds superposed on the fragments and on the initial models. Carbon atoms of new ligands depicted in yellow, carbons of fragments in magenta, carbons of models in cyan, carbons of AmpC residues in grey. Red dashes represent hydrogen bonds. (A) Compound 3 and F2. (B) Compound 3 and model of 3. (C) Compound 4 and F3. (D) Compound 4 and model of 4. (E) Compound 5 and F3. (F) Compound 5 and model of 5.
Further Optimization.
Encouraged by the high affinity of these compounds, we sought derivatives with even further improved affinity. Aware that the tetrazole derivatives might have better cell penetrance than the carboxylates (28), owing to a higher pKa value, we chose to derivatize 4. We sought compounds that might improve steric complementarity with the enzyme without disrupting other interactions. Modeling suggested that chloro derivatives ortho- and meta- to the tetrazole were easily accommodated by the site, as was a trifluoromethyl group in the meta-position. Compounds 6, 7, and 8 were thus synthesized and tested; all three were competitive, slow off-rate inhibitors of AmpC. While the 3-chloro derivative 6 had a worse Ki of 3 nM, the 2-chloro and 2-trifluoromethyl derivatives 7 and 8 showed 6-and 24-fold improved affinities compared to 4 with Ki values of 200 and 50 picomolar, respectively. X-ray crystal structures of AmpC complexes with 6, 7, and 8, determined at 1.43, 1.44, and 1.49 Å, respectively, confirm that the new substitutions point away from Tyr221 and towards Ala318 as anticipated (Fig. 2 D–F). In 7 and 8, the derivatives correspond closely to the precursor 4, while in 6 the plane of the phenyl is rotated by 20° to avoid a clash between the 3-chloro and backbone atoms of Gly320. In 7, the 2-chloro is in van der Waals contact with the Cβ and backbone carbonyl atoms of Ala318 (3.8 and 3.6 Å, respectively). Similarly, in 8 the 2-trifluoromethyl makes comparable interactions and also packs with the backbone of Thr319 (3.2–3.7 Å). The fluorines appear to also interact with the sulfonamide amide hydrogen (F-N distance: 2.8 Å), forming orthogonal multipolar interactions with the sulfonamide oxygen (F-O distance: 2.8 Å). To our knowledge, the 50 pM activity of 8 makes it the most potent reversible β-lactamase inhibitor described to date, and ranks it among the most potent inhibitors described for most enzymes, having 200-fold better affinity than most drugs for their main targets (29).
Selectivity.
To assess the selectivity of the new molecules we determined Ki values against three common serine proteases—Trypsin, Elastase, and α-Chymotrypsin—as well as that of a class A β-lactamase, CTX-M-9 (SI Appendix, Table S2) (30–33). Affinity for AmpC was typically 105-to 106-fold better than for the serine proteases (SI Appendix, Fig. S5). Notwithstanding the boronic acid warhead shared by these inhibitors, the compounds show clear specificity for their target over protease off-targets. Affinity was also substantially better for AmpC than CTX-M-9, which, though speaking to specificity, may portend difficulties for clinical relevance, as one would ideally prefer a compound active against both class C and class A enzymes. Still, several of the analogs retained substantial affinity for CTX-M-9, especially 3, which was a 45 nM inhibitor of CTX-M-9.
Microbiology.
The anti-resistance activity of inhibitors was investigated by the determination of the minimum inhibitory concentrations (MICs) of the β-lactam/inhibitor combination necessary to inhibit the growth of clinically isolated bacteria resistant to third-generation cephalosporins via expression of class A or class C β-lactamases (Table 2). Used by themselves, the antibiotics cefotaxime and ceftazidime had high MIC values, often greater than 64 μg/mL, certainly much higher than the break point for empirical resistance levels ≥2 μg/mL (34). Conversely, in combination with the new inhibitors, the MIC values of these third-generation cephalosporins improved substantially, typically by 64-fold or more. For 75% of the clinical isolates measured, MICs dropped into the susceptible range (MICs ≤ 1 μg/mL) with compounds 3 and 5, for 50% with compounds 7 and 8, and for 25% of those treated with the cephalosporins and 4 or 6 (SI Appendix, Table S3). For many clinical isolates, MIC values for ceftazidime and cefotaxime combined with 5 and 7 dropped to 0.5 μg/mL and below (SI Appendix, Fig. S6), which represents an 8-to 16-fold improvement of MIC values compared to previously tested boronic acids (6, 8, 11).
Table 2.
Minimum inhibitory concentrations (MICs) of third-generation cephalosporins alone and in combination with the inhibitors 3–8 (dosed at a cephalosporin:inhibitor ratio of 1∶4) for clinical bacteria exhibiting a high level of resistance
| Bacterial strains |
MICs (μg/mL) of ceftazidime alone or in combination |
MICs (μg/mL) of cefotaxime alone or in combination |
||||||||||||
| alone | 3 | 4 | 5 | 6 | 7 | 8 | alone | 3 | 4 | 5 | 6 | 7 | 8 | |
| Escherichia coli 1† | 128 | 1 | 4 | 0.5 | 1 | 0.5 | 1 | 8 | 0.5 | 2 | 0.25 | 1 | 0.5 | 1 |
| Escherichia coli 2† | 128 | 2 | 4 | 1 | 2 | 1 | 1 | 16 | 0.5 | 2 | 0.5 | 2 | 0.5 | 1 |
| Escherichia coli 3† | 64 | 1 | 2 | 0.5 | 0.5 | 0.5 | 1 | 8 | 0.5 | 1 | 0.5 | 0.5 | 0.25 | 0.5 |
| Escherichia coli 4† | 64 | 1 | 2 | 1 | 2 | 1 | 1 | 16 | 1 | 2 | 0.5 | 1 | 0.5 | 0.5 |
| Escherichia coli 5† | 32 | 1 | 1 | 0.5 | 1 | 0.5 | 1 | 4 | 0.5 | 1 | 0.5 | 2 | 0.5 | 1 |
| Escherichia coli 6† | 8 | 0.5 | 1 | 0.5 | 0.5 | 0.5 | 1 | 4 | 0.25 | 1 | 0.5 | 0.5 | 0.25 | 1 |
| Enterobacter cloacae 1† | 64 | 1 | 2 | 0.5 | 4 | 1 | 2 | 64 | 1 | 4 | 1 | 2 | 1 | 1 |
| Enterobacter cloacae 2† | 128 | 4 | 4 | 4 | 2 | 1 | 2 | 128 | 4 | 4 | 4 | 4 | 2 | 2 |
| Citrobacter freundii 1† | 64 | 1 | 2 | 0.5 | 1 | 1 | 2 | 16 | 0.5 | 1 | 0.5 | 2 | 1 | 1 |
| Citrobacter freundii 2† | 128 | 1 | 4 | 2 | 2 | 2 | 2 | 64 | 1 | 2 | 1 | 4 | 2 | 2 |
| Pseudomonas aeruginosa 1† | 32 | 2 | 2 | 2 | 4 | 2 | 2 | > 128 | 8 | 16 | 8 | 16 | 16 | 16 |
| Klebsiella pneumonia 1† | 32 | 1 | 2 | 0.5 | 4 | 1 | 2 | 8 | 1 | 2 | 0.5 | 4 | 4 | 8 |
| Escherichia coli 7‡ | 2 | 0.25 | 2 | 1 | 4 | 2 | 4 | 256 | 4 | 16 | 32 | 8 | 8 | 16 |
†AmpC-overproducing bacteria.
‡Escherichia coli–producing CTX-M-14.
Intriguingly, substantial decreases in MIC values were observed for a strain producing the plasmid-mediated class A β-lactamase CTX-M-14 (8-to 64-fold), especially for compounds 3 and 7, which had the broadest spectrums of activity. This offers preliminary evidence that the sulfonamide boronic acids may inhibit both class C and class A β-lactamases, consistent with their in vitro activity against this class of enzymes.
Efficacy in a Mouse Model of Infection.
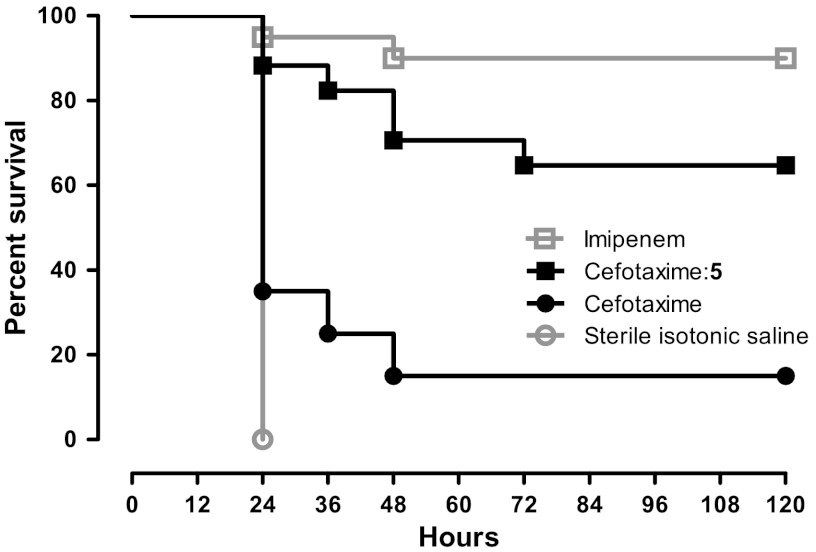
We had not observed such substantial reversal of bacterial resistance to β-lactams, across such a broad spectrum of clinical isolates, for previous series of boronic acid inhibitors of β-lactamase; indeed, this lack of efficacy had motivated this study. These new cell-culture MIC values inspired us to investigate the efficacy of one of these compounds in a mouse model of bacteremia and sepsis. Oncins France 1 mice were infected with a hospital-derived strain of Escherichia coli that overproduces AmpC and is highly resistant to cefotaxime (MIC: 32 μg/mL). Mice were treated with cefotaxime alone, cefotaxime combined with compound 5, with sterile isotonic saline, and with imipenem as a reference treatment of systemic infections by cephalosporin-resistant enterobacteriaceae (Fig. 4); 20 mice were used in each clade. The animals became severely sick 5 h after infection and all untreated animals (sterile isotonic saline) died within 12–24 h. With a clinical dose of 50 mg/kg, imipenem was almost fully active (90% survival at 120 h postinfection). Only 15% of mice treated with cefotaxime alone survived by at 120 h post infection. Cefotaxime:5 treatment, conversely, rescued 65% of animals at the 120-h postinfection timepoint, and those mice that did die did so later than with cefotaxime alone. Statistical analysis confirms a significant increase in the percent survival for the combination of 5 with cefotaxime (p ≤ 0.0005 versus cefotaxime alone). No significant difference was observed with imipenem treatment (p≥0.1148 for the comparison cefotaxime:5 versus imipenem). Consistent with the expectation that the cefotaxime:5 treatment has a direct effect on bacteria, the colony forming unit (CFU) counts of imipenem- and cefotaxime:5-treated mice showed reductions in all organs and blood compared to treatment with cefotaxime alone and to untreated controls (SI Appendix, Fig. S7 and Table S4).
Fig. 4.
Percentage of survival of mice infected with AmpC–overproducing Escherichia coli over the course of 5 days (120 h). Mice were inoculated by intraperitoneal injection with AmpC–overproducing Escherichia coli strain 4 (1 ± 0.5 109 colony-forming units) and treated at 0.5, 3.5, and 6.5 h after infection by intraperitoneal injection of 50 mg/kg imipenem (open square, n = 20), 50∶200 mg/kg cefotaxime:5 combination (black filled square, n = 17), 50 mg/kg cefotaxime (black filled circle, n = 20), or sterile isotonic saline (open circle, n = 20).
Discussion
The use of fragments in hit-to-lead development has become popular in drug discovery, especially for difficult drug targets. Fragments benefit from binding to pockets and surfaces unperturbed by restraints found in larger molecules, and often do so with high ligand efficiency. By merging, linking, or growing fragments, high-affinity leads may be obtained. A second advantage of fragments is that they cover much more chemical space than lead-like molecules (13–15, 19–21).
Here we used both virtues to optimize a series already exhibiting decent affinity but insufficient biological activity. First, we exploited the geometric information contained in aryl-carboxylate fragments. The lead compound 2 had a Ki of 25 nM against AmpC β-lactamase and lowered MIC values eightfold on average. Modeling suggested that 3 could pick up the interactions observed in fragments F1 and F2. In fact, only a different orientation for the benzoic acid substructure contained in the lead 2 seemed necessary, which could be obtained by moving the carboxylate from para- to meta-. This improved affinity of compound 3 almost 20-fold over 2, while the affinity 3 is 54-fold better than 1 (ΔΔG = 1.9 kcal/mol), which can be attributed to the carboxylate in a preferred environment. Indeed, the placement of the distal carboxylate of 3 between Ser212 and Gly320 superposes well with that observed in F2 (Fig. 3A), recapitulating the designed structure with a RMSD of 0.3 Å (Fig. 3B). The improved affinity also improved antimicrobial activity: MIC values dropped 64-fold on average and the median MIC for 3 was 1 μg/mL against 12 highly resistant strains (SI Appendix, Table S3), below the empirical break point for hospital infections. Fortuitously, compound 3 also inhibits class A β-lactamases efficiently, with a Ki of 45 nM against CTX-M-9. Correspondingly, it lowers MIC values for bacteria expressing this enzyme, in combination with ceftazidime and cefotaxime, by 8-and 64-fold respectively, making it the compound with best broad-spectrum activity within this series.
In the tetrazole series, we were guided by the geometric information contained in fragment F3. The tetrazoles of the designed molecules 4 and 5 superpose well with that of F3, and the initial models agree well with the subsequent crystal structures, with RMSD values of 0.9 Å and 0.7 Å, respectively (Fig. 3 C–F). Comparison with the Ki of molecule 9 (Ki 210 nM) suggests that the tetrazole added about 3 kcal/mol of affinity, improving the Ki 170-fold in compound 4 and 250-fold in the pyridine derivative 5. Tetrazoles are common bioisosteres of carboxylates and often have better bioavailability (28, 35). Although they are not unprecedented in β-lactam antibiotics (e.g., cefazolin), boronic acid inhibitors of β-lactamases have not yet exploited this chemotype in this region of the active site. While previous generations of boronic acids focused on mimicking β-lactam substrates, these fragment-derived boronic acids exhibit greater novelty and may be more robust against pre-evolved mutant enzymes that overcome boronic acids more closely resembling β-lactams (10).
Whereas fragments have been used previously for new chemotype discovery (13–22) and merging, their use in late stage optimization has remained largely unexplored. Of course nothing prevented this, and indeed such an idea is implicit in the fragment approach and anticipated by computational design methods like LUDI, HOOK, GrowMol, and MCSS (36–39). Still, late-stage optimization with fragments seems underdeveloped; it can reveal derivatization strategies, both in geometry and in chemotype, that may otherwise remain unknown without an industrial-scale hit-to-lead campaign.
Certain caveats deserve attention. This approach to optimizing leads with fragments is restricted to targets where proximal binding sites can be detected and for which fragment orientations can be accurately determined. It also requires a decomposable lead series where substantial inhibition remains with only a core chemotype, which is not always the case (23, 40). Whereas these compounds did turn out to be additive in affinity gained—compared to the naked sulfonamide they added over 2-logs, while the Ki values of the fragments were between 3 and 40 mM—this too will not always hold. Indeed, in another series of analogs that also tried to exploit the fragment placement, no improvement in affinity was achieved. There are also cautions to the mouse experiments—in the cephalosporin/inhibitor combination clade, we preserved the cefotaxime/inhibitor ratio of 1∶4 of the MIC experiments. This resulted in a final concentration of 200 mg/kg for inhibitor 5, which is very high. Future studies may focus on the analysis and improvement of pharmacokinetic properties and proper evaluation of toxicity and activity against a larger panel of bacteria. Fortunately, because the molecules remain small (molecular weights range from 270 to 350 Da), there is room for further optimization.
These caveats should not obscure the central observation of this study—two series of fragments, bound in a particular pocket revealed an opportunity to derivatize a relatively advanced series in a direction, and with chemotypes, that had not been previously explored or imagined. This overcame what had been an unsurmounted barrier in efficacy. Not only was the resulting series potent, with sub-nanomolar to mid-picomolar affinities, but also it had clinically relevant MIC values and activity in a mouse model of bacterial infection. In short, beginning with a structural study and guided synthesis, we ended up with molecules active in vivo in a mammalian system. In themselves, these inhibitors hold promise as leads to overcome a pervasive and growing threat to public health. More generally, whereas fragments are widely used to nucleate early discovery (13–22), this study suggests that they also may be used to guide late-stage optimization into chemotypes and geometries that would be hard to systematically sample by other methods.
Methods
Modeling of Distal Site Binders.
Boronic acids were modeled manually and subsequently minimized using PLOP (41) (SI Appendix, Supporting Methods).
Synthesis.
Sulfonamidomethaneboronic acids (1–10) were obtained from functionalized sulfonyl chlorides. Microwave assisted cycloaddition yielded tetrazoles 4–8 (SI Appendix, Supporting Methods).
Enzymology.
Enzyme inhibition was measured by the method of initial rates (SI Appendix, Supporting Methods).
Crystallography.
All AmpC/inhibitor X-ray structures were obtained by co-crystallization and determined by molecular replacement (SI Appendix, Supporting Methods). The atomic coordinates and structure factors for AmpC with compounds 3–8 and 10 have been deposited in the Protein Data Bank (PDB), www.rcsb.org (PDB ID codes 4E3I, 4E3J, 4E3K, 4E3L, 4E3M, 4E3N, 4E3O).
Microbiology.
Susceptibility testing followed the guidelines of CLSI (34). Each MIC value reported reflects the average of three independent experiments (SI Appendix, Supporting Methods).
In Vivo Efficacy Studies.
The experiments were approved by the Animal Care Committee of Auvergne University, Clermont-Ferrand, France (SI Appendix, Supporting Methods).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Prof. Y. Chen and A. Doak for CTX-M-9 and AmpC and Dr. A. O’Donoghue for serine proteases and substrates. We thank Drs. K. Ziebart and M. Merski for assisting with the slow binding kinetics and I. Fish and S. Pierre for structure refinements. We thank Dr. H.T.T. Nguyen for technical assistance Microbes, Intestin, Inflammation et Susceptibilité de l'Hôte and Dr. A. Alloui for animal care. We thank Dr. M. Fischer and A. Doak for reading this manuscript, and the Centro Interdipartimentale Grandi Strumenti of Modena for NMR and MS spectra. This study was supported by National Institutes of Health Grant GM63815 and by Institut National de la Santé et de la Recherche Médicale and Institut National de la Recherche Agronomique.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4E3I, 4E3J, 4E3K, 4E3L, 4E3M, 4E3N, and 4E3O).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208337109/-/DCSupplemental.
References
- 1.Frere JM. Beta-lactamases and bacterial resistance to antibiotics. Mol Microbiol. 1995;16:385–395. doi: 10.1111/j.1365-2958.1995.tb02404.x. [DOI] [PubMed] [Google Scholar]
- 2.Bush K. Beta-lactamases of increasing clinical importance. Curr Pharm Des. 1999;5:839–845. [PubMed] [Google Scholar]
- 3.Nukaga M, Kumar S, Nukaga K, Pratt RF, Knox JR. Hydrolysis of third-generation cephalosporins by class C beta-lactamases: Structures of a transition state analog of cefotaxime in wild-type and extended spectrum enzymes. J Biol Chem. 2004;279:9344–9352. doi: 10.1074/jbc.M312356200. [DOI] [PubMed] [Google Scholar]
- 4.Fisher JF, Meroueh SO, Mobashery S. Bacterial resistance to beta-lactam antibiotics: Compelling opportunism, compelling opportunity. Chem Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 5.Drawz SM, Bonomo RA. Three decades of beta-lactamase inhibitors. Clin Microbiol Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caselli E, et al. Energetic, structural, and antimicrobial analyses of beta-lactam side chain recognition by beta-lactamases. Chem Biol. 2001;8:17–31. doi: 10.1016/s1074-5521(00)00052-1. [DOI] [PubMed] [Google Scholar]
- 7.Powers RA, Caselli E, Focia PJ, Prati F, Shoichet BK. Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: Implications for resistance mutations and inhibitor design. Biochemistry. 2001;40:9207–9214. doi: 10.1021/bi0109358. [DOI] [PubMed] [Google Scholar]
- 8.Morandi F, et al. Nanomolar inhibitors of AmpC beta-lactamase. J Am Chem Soc. 2003;125:685–695. doi: 10.1021/ja0288338. [DOI] [PubMed] [Google Scholar]
- 9.Morandi S, Morandi F, Caselli E, Shoichet BK, Prati F. Structure-based optimization of cephalothin-analogue boronic acids as beta-lactamase inhibitors. Bioorg Med Chem. 2008;16:1195–1205. doi: 10.1016/j.bmc.2007.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, et al. Recognition and resistance in TEM beta-lactamase. Biochemistry. 2003;42:8434–8444. doi: 10.1021/bi034242y. [DOI] [PubMed] [Google Scholar]
- 11.Eidam O, et al. Design, synthesis, crystal structures, and antimicrobial activity of sulfonamide boronic acids as beta-lactamase inhibitors. J Med Chem. 2010;53:7852–7863. doi: 10.1021/jm101015z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tondi D, Morandi F, Bonnet R, Costi MP, Shoichet BK. Structure-based optimization of a non-beta-lactam lead results in inhibitors that do not up-regulate beta-lactamase expression in cell culture. J Am Chem Soc. 2005;127:4632–4639. doi: 10.1021/ja042984o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rees DC, Congreve M, Murray CW, Carr R. Fragment-based lead discovery. Nat Rev Drug Discovery. 2004;3:660–672. doi: 10.1038/nrd1467. [DOI] [PubMed] [Google Scholar]
- 14.Murray CW, Blundell TL. Structural biology in fragment-based drug design. Curr Opin Struct Biol. 2010;20:497–507. doi: 10.1016/j.sbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Fischer M, Hubbard RE. Fragment-based ligand discovery. Mol Interventions. 2009;9:22–30. doi: 10.1124/mi.9.1.7. [DOI] [PubMed] [Google Scholar]
- 16.Ciulli A, Williams G, Smith AG, Blundell TL, Abell C. Probing hot spots at protein-ligand binding sites: A fragment-based approach using biophysical methods. J Med Chem. 2006;49:4992–5000. doi: 10.1021/jm060490r. [DOI] [PubMed] [Google Scholar]
- 17.Allen KN, et al. An experimental approach to mapping the binding surfaces of crystalline proteins. J Phys Chem. 1996;100:2605–2611. [Google Scholar]
- 18.Landon MR, et al. Detection of ligand binding hot spots on protein surfaces via fragment-based methods: Application to DJ-1 and glucocerebrosidase. J Comput Aided Mol Des. 2009;23:491–500. doi: 10.1007/s10822-009-9283-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hann MM, Leach AR, Harper G. Molecular complexity and its impact on the probability of finding leads for drug discovery. J Chem Inf Comput Sci. 2001;41:856–864. doi: 10.1021/ci000403i. [DOI] [PubMed] [Google Scholar]
- 20.Fink T, Bruggesser H, Reymond JL. Virtual exploration of the small-molecule chemical universe below 160 daltons. Angew Chem Int Ed Engl. 2005;44:1504–1508. doi: 10.1002/anie.200462457. [DOI] [PubMed] [Google Scholar]
- 21.Fink T, Reymond JL. Virtual exploration of the chemical universe up to 11 atoms of C, N, O, F: Assembly of 26.4 million structures (110.9 million stereoisomers) and analysis for new ring systems, stereochemistry, physicochemical properties, compound classes, and drug discovery. J Chem Inf Model. 2007;47:342–353. doi: 10.1021/ci600423u. [DOI] [PubMed] [Google Scholar]
- 22.Teotico DG, et al. Docking for fragment inhibitors of AmpC beta-lactamase. Proc Natl Acad Sci USA. 2009;106:7455–7460. doi: 10.1073/pnas.0813029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babaoglu K, Shoichet BK. Deconstructing fragment-based inhibitor discovery. Nat Chem Biol. 2006;2:720–723. doi: 10.1038/nchembio831. [DOI] [PubMed] [Google Scholar]
- 24.Weston GS, Blazquez J, Baquero F, Shoichet BK. Structure-based enhancement of boronic acid-based inhibitors of AmpC beta-lactamase. J Med Chem. 1998;41:4577–4586. doi: 10.1021/jm980343w. [DOI] [PubMed] [Google Scholar]
- 25.Morrison JF, Walsh CT. The behavior and significance of slow-binding enzyme inhibitors. Adv Enzymol Relat Areas Mol Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 26.Strynadka NC, Martin R, Jensen SE, Gold M, Jones JB. Structure-based design of a potent transition state analogue for TEM-1 beta-lactamase. Nat Struct Biol. 1996;3:688–695. doi: 10.1038/nsb0896-688. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Minasov G, Roth TA, Prati F, Shoichet BK. The deacylation mechanism of AmpC beta-lactamase at ultrahigh resolution. J Am Chem Soc. 2006;128:2970–2976. doi: 10.1021/ja056806m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemke TL, Williams DA. Foye’s Principles of Medicinal Chemistry. 6th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 29.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discovery. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 30.Pouvreau L, et al. Effect of pea and bovine trypsin inhibitors on wild-type and modified trypsins. FEBS Lett. 1998;423:167–172. doi: 10.1016/s0014-5793(98)00062-3. [DOI] [PubMed] [Google Scholar]
- 31.Del Mar EG, Largman C, Brodrick JW, Fassett M, Geokas MC. Substrate specificity of human pancreatic elastase 2. Biochemistry. 1980;19:468–472. doi: 10.1021/bi00544a011. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez-Martinez JA, Rivera-Rivera I, Sola RJ, Griebenow K. Enzymatic activity and thermal stability of PEG-alpha-chymotrypsin conjugates. Biotechnol Lett. 2009;31:883–887. doi: 10.1007/s10529-009-9947-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Delmas J, Sirot J, Shoichet B, Bonnet R. Atomic resolution structures of CTX-M beta-lactamases: Extended spectrum activities from increased mobility and decreased stability. J Mol Biol. 2005;348:349–362. doi: 10.1016/j.jmb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 34.Clinical and Laboratory Standards Institute. 20th Informational Supplement. Wayne, PA: 2010. Performance standards for antimicrobial susceptibility testing; pp. M100–S20. [Google Scholar]
- 35.Meanwell NA. Synopsis of some recent tactical application of bioisosteres in drug design. J Med Chem. 2011;54:2529–2591. doi: 10.1021/jm1013693. [DOI] [PubMed] [Google Scholar]
- 36.Bohm HJ. The computer program Ludi: A new method for the de novo design of enzyme inhibitors. J Comput Aided Mol Des. 1992;6:61–78. doi: 10.1007/BF00124387. [DOI] [PubMed] [Google Scholar]
- 37.Eisen MB, Wiley DC, Karplus M, Hubbard RE. HOOK: A program for finding novel molecular architectures that satisfy the chemical and steric requirements of a macromolecule binding site. Proteins. 1994;19:199–221. doi: 10.1002/prot.340190305. [DOI] [PubMed] [Google Scholar]
- 38.Bohacek RS, McMartin C. Multiple highly diverse structures complementary to enzyme binding sites: Results of extensive application of a de novo design method incorporating combinatorial growth. J Am Chem Soc. 1994;116:5560–5571. [Google Scholar]
- 39.Joseph-McCarthy D, Hogle JM, Karplus M. Use of the multiple copy simultaneous search (MCSS) method to design a new class of picornavirus capsid binding drugs. Proteins. 1997;29:32–58. doi: 10.1002/(sici)1097-0134(199709)29:1<32::aid-prot3>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 40.Barelier S, Pons J, Marcillat O, Lancelin JM, Krimm I. Fragment-based deconstruction of Bcl-x(L) inhibitors. J Med Chem. 2010;53:2577–2588. doi: 10.1021/jm100009z. [DOI] [PubMed] [Google Scholar]
- 41.Kalyanaraman C, Bernacki K, Jacobson MP. Virtual screening against highly charged active sites: Identifying substrates of alpha-beta barrel enzymes. Biochemistry. 2005;44:2059–2071. doi: 10.1021/bi0481186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.