Abstract
Histophilus somni is an economically important pathogen of cattle and other ruminants and is considered one of the key components of the bovine respiratory disease (BRD) complex, the leading cause of economic loss in the livestock industry. BRD is a multifactorial syndrome, in which a triad of agents, including bacteria, viruses, and predisposing factors or “stressors,” combines to induce disease. Although vaccines against H. somni have been used for many decades, traditional bacterins have failed to demonstrate effective protection in vaccinated animals. Hence, the BRD complex continues to produce strong adverse effects on the health and well-being of stock and feeder cattle. The generation of recombinant proteins may facilitate the development of more effective vaccines against H. somni, which could confer better protection against BRD. In the present study, primers were designed to amplify, clone, express, and purify two recombinant lipoproteins from H. somni, p31 (Plp4) and p40 (LppB), which are structural proteins of the outer bacterial membrane. The results presented here demonstrate, to our knowledge for the first time, that when formulated, an experimental vaccine enriched with these two recombinant lipoproteins generates high antibody titers in rabbits and sheep and exerts a protective effect in mice against septicemia induced by H. somni bacterial challenge.
INTRODUCTION
Bovine respiratory disease (BRD) is a major cause of economic losses in the livestock industry (15, 47). Technological, biological, and pharmacological advances have facilitated the development of several products to combat BRD, including vaccines. Nevertheless, the bacterial component of the BRD complex continues to provoke important adverse effects on the health and well-being of stock and feeder cattle (14, 20, 33).
BRD is a multifactorial syndrome, in which a triad of agents, including bacteria, viruses, and predisposing factors or “stressors,” combines to induce disease (reviewed in reference 49). BRD involves complex interactions among viral and bacterial pathogens that can lead to intense pulmonary inflammation (fibrinous pleuropneumonia) (10). Histophilus somni is among the bacterial pathogens associated with this disease (formerly Haemophilus somnus) (1), an unencapsulated Gram-negative coccobacillus and member of the Pasteurellaceae family that exhibits strict ruminant host specificity (9, 23, 39). H. somni is considered one of the key components of the BRD complex (11), and it is an economically important pathogen that causes respiratory disease, septicemia, thrombotic meningoencephalitis, myocarditis, arthritis, and abortion (9, 37, 44). H. somni is an opportunistic normal habitant of the lower reproductive tract and upper respiratory tract of cattle and other ruminants (9, 39), and it acts as a pathogen under certain circumstances via mechanisms that remain poorly understood (55).
Pathogens involved in BRD have developed intricate mechanisms to thwart both the innate and adaptive immune responses of their hosts. These immune evasive strategies probably contribute to the failure of currently available vaccines to provide complete protection against these pathogens (24, 29, 48). Consequently, the combination of multiple antigens in an enriched vaccine may provide more effective protection.
Lipooligosaccharide and various outer membrane proteins (OMPs) have been proposed as potential virulence factors of H. somni, although experimental evidence is lacking (7). A comparative genomic analysis of virulence factors from two H. somni strains (2336 and 129Pt) identified genes and gene products that are putatively involved in H. somni virulence, placing particular emphasis on those that were common to both strains (44). Nonetheless, a recent detailed sequence analysis of the H. somni transcriptional map (28) suggests that other immune-active proteins may also be involved.
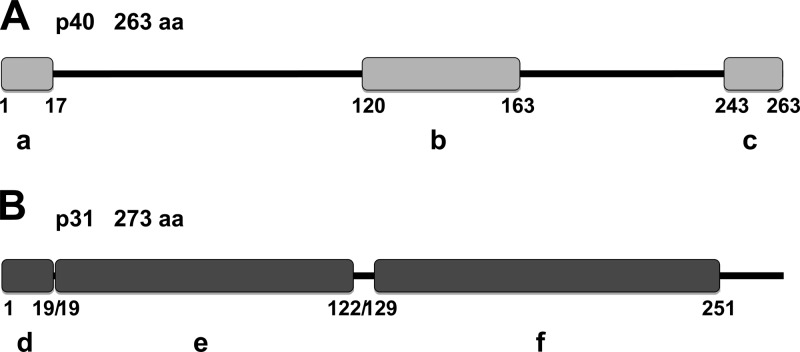
To identify potentially protective antigens to formulate an experimental vaccine against H. somni infection during BRD, we selected two OMPs and evaluated their antigenicity/immunogenicity as immunogenic proteins. Several immunodominant surface antigens of H. somni were identified previously (8), including a 40-kDa protein (p40) that was subsequently identified and cloned (50), and designated LppB for lipoprotein B (GenBank accession no. AAA72348.1). According to domain analyses performed using the Motif Scan function of MyHits (38), this protein contains a prokaryotic membrane lipoprotein lipid attachment site profile from amino acids 1 to 17, and a LysM domain from amino acids 120 to 163 that is also found in a variety of enzymes involved in bacterial cell wall degradation (25). Indeed, the structure of this domain was defined (3), and it may have a general peptidoglycan binding function. Finally, LppB contains a peptidase family M23 domain from amino acids 243 to 263 (Fig. 1A).
Fig 1.
(A) p40 protein domain structure. The prokaryotic membrane lipoprotein lipid attachment site (a), LysM domain (b), and peptidase family M23 domain (c) are indicated. (B) p31 protein domain structure. The prokaryotic membrane lipoprotein lipid attachment site (d), SmpA/OmlA family signature domain (e), and OmpA-like (outer membrane protein) domain signature (f) are indicated. The sequences were obtained from GenBank (accession numbers AAA72348.1 and AAA24941.1, respectively). Analyses were performed by Motif Scan using MyHits (35).
Another relevant immunogenic protein, p31, has also been identified (52) (GenBank accession no. AAA24941.1), which shares sequence homology with the plpD gene that encodes a 31-kDa lipoprotein (Plp4) present in Pasteurella haemolytica A1 (34), and with a 19.2-kDa antigen from Neisseria meningitidis. A domain analyses performed using Motif Scan from MyHits (38) demonstrated that p31 possesses 273 amino acids and that it contains a prokaryotic membrane lipoprotein lipid attachment site from amino acids 1 to 19 and a SmpA/OmlA family signature domain from amino acids 19 to 122. This latter domain is found in some bacterial outer membrane lipoproteins and may be involved in maintaining the structural integrity of the cell envelope (36). In addition, this protein possess an OMPA-like domain signature from amino acids 129 to 251, which is thought to be responsible for noncovalent interactions with peptidoglycan (21) (Fig. 1B).
While vaccines against H. somni have been available for several decades, traditional bacterins do not work properly and have failed to demonstrate effective protection in vaccinated animals. A multivalent vaccine for bovine bacterial respiratory disease was developed to simplify the vaccination schedule and increase the range of protection, composed of two immunogens and five bacterins (including attenuated H. somni) (5). However, similarly developed commercial vaccines produce diverse secondary effects due to the high lipooligosaccharide content of the cell walls of Gram-negative bacteria. Consequently, the generation of recombinant proteins may facilitate the formulation of recombinant vaccines against H. somni that could confer better protection against BRD.
We previously formulated an experimental vaccine using two fragments from Mannheimia haemolytica OMPs that elicited high antibody titers in rabbits and sheep and that protected mice against bacterial challenge (22). However, since BRD is a complex pathology a multivalent vaccine is more desirable, both to increase the protective range and to simplify the vaccination schedule. Hence, in the present study we selected two lipoproteins from H. somni, p31 (homologous to Plp4) and p40 (LppB), to induce antibodies with which to formulate an experimental vaccine to protect against H. somni challenge. As a vaccine adjuvant we used aluminum hydroxide [Al(OH)3], which stimulates immunity (13) by potentiating the immune response (46) and serves as a useful alternative to cholera toxin or Freund incomplete adjuvant. Our results clearly show that the combination of the p31 and p40 fusion proteins in this experimental vaccine formulation yielded high antibody titers in rabbits and sheep and protected mice from H. somni infection.
(The results presented here are included in a masters and doctoral thesis prepared by Carolina Guzmán-Brambila at the Universidad de Guadalajara, Guadalajara, Mexico.)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Escherichia coli (TOP10 or M15; Invitrogen) was used as the host to clone and propagate plasmids, which were cultured in Luria-Bertani (LB) broth supplemented with thymine (50 mg/ml) and ampicillin (100 mg/ml), chloramphenicol (25 mg/ml), or kanamycin (50 mg/ml), as necessary. H. somni, obtained from the American Type Culture Collection (strain ATCC 2336), was used to obtain bacterial DNA for PCR.
Extraction and quantification of genomic DNA and recombinant methods.
Bacterial genomic DNA was obtained using the Illustra bacteria genomic Prep Mini spin kit (GE Healthcare, United Kingdom). DNA integrity was routinely evaluated by electrophoresis in agarose gels stained with Safe DNA gel stain (Invitrogen), and its quality was evaluated by determining the absorbance ratio at A260/A280. All DNA cloning and ligation was carried out according to standard recombinant DNA techniques (2, 43).
Oligonucleotide design and PCR.
Oligonucleotide sequences of the primers used for PCR were as follows. The 5′-GGTAGGCCTATGAAACTATCACGTTTTGTAT-3′ sense primer and the 5′-TAATCTCTCTTGATATAGGTAAGCTTATCA-3′ antisense primer were used to amplify an 852-bp fragment encoding p31. The 5′-TAAAGTAACGGAGAATTTACATGAA-3′ sense primer and 5′-TTAATAAAGCTTAAATTACCATATCCACG-3′ antisense primer were used to amplify an 870-bp fragment that encoded p40. Primers were designed with the StuI restriction enzyme site in both forward primers and HindIII in both reverse primers to achieve directional cloning in the expression vector.
PCR was performed using a Perkin-Elmer GeneAmp PCR System 2400 thermocycler (Perkin-Elmer, Foster City, CA). For all PCR experiments, Platinum PCR SuperMix High Fidelity (Invitrogen) was used. PCR products were visualized on 1.5% agarose gels stained with Safe DNA gel stain (Invitrogen). Optimal annealing temperatures were initially established by testing a temperature gradient. All PCRs were performed after a single denaturation step at 94°C for 5 min and involved 30 denaturation cycles at 94°C for 1 min, annealing for 1 min at 62°C for the p31 primers and at 58°C for the p40 primers, and extension at 72°C for 1 min, followed by a final extension at 72°C for 5 min.
Construction of p31-pQE 30 Xa and p40-pQE 30 Xa and molecular cloning.
To clone the PCR products, fragments were purified from agarose gels using the GFX PCR DNA and a gel band purification kit (GE Healthcare) and cloned directly using the pCR TOPO 2.1 vector (Invitrogen). TOP10 bacteria were transformed by the CaCl2 method, and positive clones were selected using the β-galactosidase reaction. Plasmid DNA was obtained from E. coli using the plasmid Prep Mini spin kit (GE Healthcare) and, for each insert, at least three clones were sequenced to verify the sequence of both genes. Only clones that precisely matched the reported sequence were used for subcloning and to express the fusion protein. Sequences were obtained in a capillary ABI Prism 310 sequencer (Applied Biosystems) and were compiled with the Chromas v2.31 and DNASTAR, Inc., software packages before they were compared to the sequences in databases using the BLAST program.
The pQE 30 Xa vector (Qiagen) was used to generate the fusion protein, which introduces a six-histidine tag and a site for factor Xa cleavage. Previously cloned plasmid DNA with the PCR inserts was digested and religated at the StuI and HindIII sites located at the ends of the p31 and p40 sequences, thereby generating two constructs expressing fusion proteins, designated p31-pQE 30 Xa and p40-pQE 30 Xa, and theoretically expressing 32.4- and 32-kDa derivatives, respectively. These constructs were transformed into E. coli M15 (Qiagen) to express the fusion proteins.
Fusion protein expression by IPTG and purification.
IPTG (isopropyl-β-d-thiogalactopyranoside) was used to express the fusion proteins. Bacterial cultures were grown overnight, of which 1 ml was used to inoculate 50 ml of LB medium and allowed to grow for 2 h. Next, 1 mM IPTG was added, and the culture was grown for a further 3 h. Finally, the crude total extracts were obtained and examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Fusion proteins were purified using the QIAexpress system (Qiagen), based on the affinity of the histidine tag included in the fusion protein for nickel-nitrilotriacetic acid (Ni-NTA) resin. Acid or basic buffers were then used to elute fusion protein from the column in 500-μl fractions and factor Xa was used to isolate the protein from the total extracts. The total protein was quantified by the Lowry method (31), and a small aliquot of the extract (5 μg of total protein) was examined by SDS-PAGE to confirm the presence of proteins corresponding to the p31 and p40 fusion proteins.
Production of anti-p31 and anti-p40 antibodies in rabbits.
To prepare the immunogen, the purified proteins in the elution buffer (8 M urea, 0.01 M Tris-HCl, 0.1 M NaH2PO4) were mixed (1/8 [vol/vol]) with the adjuvant [0.25% Al(OH)3] in sterile phosphate-buffered saline (PBS; pH 7.2). Once prepared, the immunogen was subcutaneously inoculated into New Zealand White rabbits weighing approximately 2 kg. Two rabbits were subcutaneously inoculated with each fusion protein (90 μg) on days 0, 14, and 21, and serum was collected prior to the first inoculation on day 0 (preimmune serum control) and after each inoculation (days 14 and 21). Finally, total serum was obtained by complete exsanguination of the rabbit on day 31. The crude serum was heat inactivated and stored at −20°C until it was analyzed in Western blots.
Protein electrophoresis and Western blotting.
Western blots were used to detect the production of antibodies against p31 and p40 in rabbits immunized with the purified fusion proteins. Crude extracts of total protein were obtained in lysis buffer (PBS [pH 7.2], 4% SDS), and the homogenates were then centrifuged at 14,000 × g for 15 min at 4°C. The supernatants were collected, and the protein content was determined using the Lowry method. Samples were diluted in PBS (pH 7.2) containing 4% SDS, denatured by boiling, and separated by electrophoresis on 10% acrylamide gels. The proteins were then transferred onto nitrocellulose membranes (Millipore, Bedford, MA), and the membranes were blocked overnight at 4°C with 80 g/liter of nonfat milk in 0.1% PBS–Tween–Tris-buffered saline (TBST). After the membranes were incubated with crude rabbit serum (1:1,000) for 1 h at room temperature, the blots were washed thoroughly in TBST and incubated for 1 h with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (1:5,000; Millipore). Immunoreactive proteins were detected by ECL (ECL Western blot analysis system; GE Healthcare) and analyzed using ChemiDoc (Bio-Rad Laboratories, Hercules, CA).
Vaccination and production of polyclonal antibodies in rabbits and sheep.
Twenty-five New Zealand White rabbits weighing ∼2 kg were divided into five groups (Table 1). The experimental vaccines (500 μl of each) were subcutaneously inoculated on days 0 and 14. Group A (positive control) was inoculated with a commercial bacterin (Biobac 11 vías) composed of Clostridium chauvoei, Clostridium septicum, Clostridium novyi, Clostridium sordellii, Clostridium perfringens type C, Clostridium perfringens type D, Pasteurella multocida type A, Pasteurella multocida type D, Mannheimia haemolytica serotype A1, Histophilus somni, and adjuvant [Al(OH)3]. Group B was vaccinated with the recombinant preparation containing 30 μg of recombinant p31 (rp31) and 30 μg of recombinant p40 (rp40) and complemented with the commercial vaccine Biobac 7 vías (Clostridium chauvoei, Clostridium septicum, Clostridium novyi, Clostridium sordellii, Clostridium perfringens type C, Clostridium perfringens type D, and Al(OH)3. Rabbits of group C were inoculated with a recombinant preparation composed of 60 μg of rp31 plus 60 μg of rp40 plus Biobac 7 vías. Group D (negative control) was inoculated with Biobac 7 vías, and group E (negative control) was inoculated with adjuvant alone.
Table 1.
Rabbit and sheep groups
| Group | Rabbit immunogen | Sheep immunogen | Vaccination times (days) | Sampling times (days) for blood (sera) |
|---|---|---|---|---|
| A | Biobac 11 vías | Biobac 11 vías | 0, 14 | 0, 14, 21, 28, 35, 42 |
| B | Recombinant (30 μg) | Recombinant (50 μg) | 0, 14 | 0, 14, 21, 28, 35, 42 |
| C | Recombinant (60 μg) | Recombinant (100 μg) | 0, 14 | 0, 14, 21, 28, 35, 42 |
| D | Biobac 7 vías | Biobac 7 vías | 0, 14 | 0, 14, 21, 28, 35, 42 |
| E | None [PBS + Al(OH)3] | None [PBS + Al(OH)3] | 0, 14 | 0, 14, 21, 28, 35, 42 |
For the second experiment, 10 healthy female sheep (hybrids of Pelibuey, Katahdin, and Blackbelly) weighing ∼30 kg, were housed in corrals and dewormed orally with 5% closantel. The animals were divided into five groups and vaccinated in the lateral neck region on days 0 and 14 with 2.5 ml of each formulation. The groups were as follows: group A (positive control) was immunized with the recombinant preparation alone; group B was immunized with recombinant preparation containing 50 μg of rp31, 50 μg of rp40, and Biobac 7 vías; group C was immunized with a recombinant preparation containing 100 μg of rp31, 100 μg of rp40, and Biobac 7 vías; group D (negative control) was immunized with Biobac 7 vías; and group E (negative control) with PBS and Al(OH)3.
For antibody determination, serum samples were collected on days 0, 14, 21, 28, 35, and 42 and assayed for rp31, rp40, and H. somni whole bacterial cell crude protein extract (WC) using a enzyme-linked immunosorbent assay (ELISA). Protein extracts (WC) were prepared as described previously, and 2 μg of total protein was assayed for each group.
ELISA.
Polystyrene microplates (96-well) were coated separately with H. somni WC in lysis buffer and recombinant proteins. The plates were coated with 100 ng of each protein/well diluted in carbonate buffer (0.1 M sodium carbonate, 0.1 M bicarbonate sodium [pH 9.6]), followed by incubation for 16 to 18 h at 4°C. The supernatant was removed, and the plates were washed four times in buffer (1.25 M NaCl, 250 mM Tris-HCl [pH 7.9], 1% Tween 20), blocked with 2% nonfat milk in 0.1% TBST, and incubated at 37°C for 1 h. Subsequently, the plates were washed, and the WC, rp31, and rp40 samples were applied at a 1:800 dilution. After incubation for 1 h at 37°C, the microplates were washed four times, followed by incubation with the secondary antibody (HRP-conjugated anti-rabbit IgG [1:1,000]; Millipore) for 1 h at 37°C. After o-phenylenediamine (OPD; Sigma) was added as a peroxidase substrate, the plates were incubated for 5 min at room temperature, and the absorbance was then read at 415 nm. The results were reported as the median values from independent determinations each performed in triplicate and corrected versus the background control. To determine the levels of anti-rp31 and anti-rp40 for the sheep IgG antibodies, the previous steps were repeated diluting the serum 1:800 and using HRP-conjugated rabbit anti-sheep IgG (H+L, 1:1,000; Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD) as the secondary antibody.
Bacterial challenge in BALB/c mice.
As previously demonstrated (17, 18, 45), H. somni causes septicemia when inoculated intraperitoneally in mice. Therefore, we selected this model to test for the efficacy of our experimental vaccine.
Seventy-five male BALB/c mice weighing 26 to 32 g were housed under controlled conditions of temperature, humidity, and lighting. The mice were divided into five groups and injected intraperitoneally on days 0 and 14 with 250 μl of each formulation. Group A (positive control) was immunized with Biobac 11 vías. Group B was immunized with a recombinant preparation containing 10 μg of rp31, 10 μg of rp40, and Biobac 7 vías. Group C was immunized with a recombinant preparation containing 20 μg of rp31 plus 20 μg of rp40 plus Biobac 7 vías, and the negative control groups D and E were immunized with Biobac 7 vías and PBS-Al(OH)3, respectively. Since the final objective was to formulate and test a combined experimental vaccine, we did not include groups to test each recombinant protein individually.
All mice were challenged 28 days after the initial immunization with approximately 3.3 × 108 CFU of virulent H. somni (ATCC 2336), injected intraperitoneally, and their survival was monitored for 10 days. Necropsies were performed to verify the effect of the bacterial challenge.
Animal care and use.
The present study complied with all Institutional Guidelines and the Official Mexican Regulations (35) for the production, care, and use of laboratory animals. The protocol was approved by the local Animal Ethics Committee. Every effort was made to minimize animal suffering and the number of animals used.
Statistical analyses.
The Fisher exact test was used to evaluate the protection against H. somni, and the results are presented as a Kaplan-Meier curve. ELISA results were compared by analysis of variance.
RESULTS
Induction of rp31-pQE 30 Xa and rp40-pQE 30 Xa expression.
PCR fragments from of each gene were initially cloned into pCR 2.1 TOPO (Invitrogen), digested and the gel-purified StuI-HindIII restriction fragments were subcloned into the pQE-30 Xa expression vector (Qiagen). The fusion proteins were expressed in M-15 E. coli (Invitrogen) transformed by heat shock.
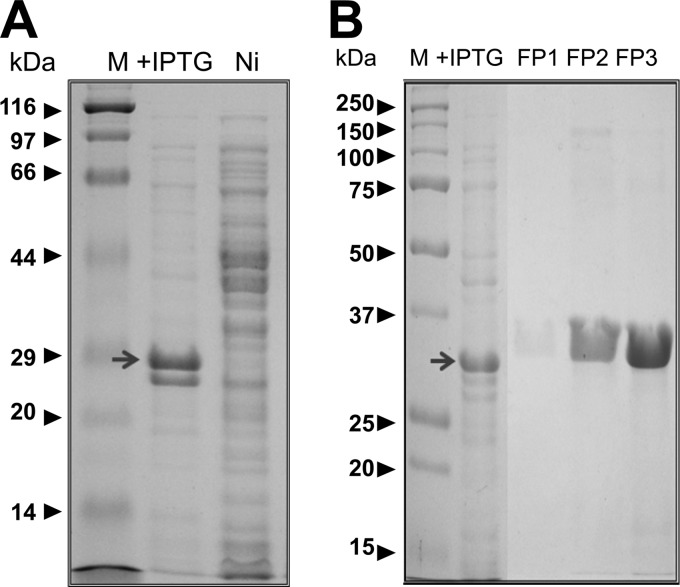
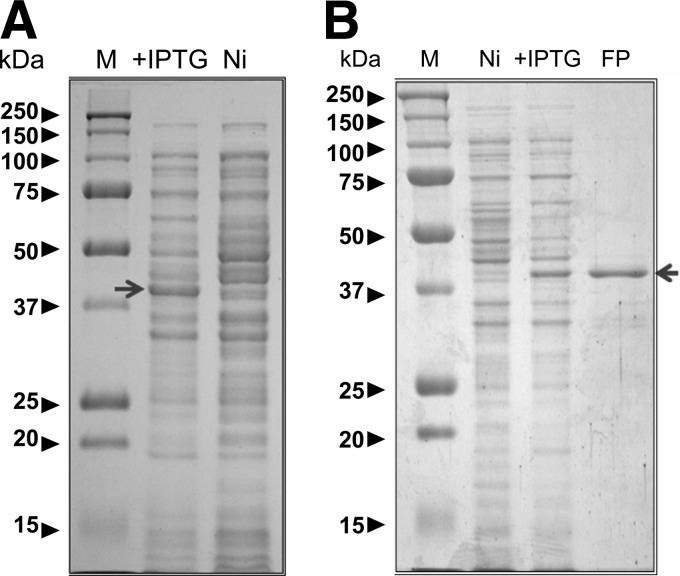
In the case of p31, the whole protein was cloned (Fig. 1A) as a 283-amino-acid polypeptide to generate a fusion protein with a theoretical molecular mass of 32 kDa that included the histidine tag and the Xa factor recognition site. The fusion protein was clearly induced by IPTG, and its electrophoretic migration corresponded well with the theoretical molecular mass (Fig. 2A). The construct used for p40 (LppB) contained 289 amino acids and included the tag and protease recognition site. The resulting fusion protein had a theoretical molecular mass of 32 kDa. Although this protein was also clearly induced by IPTG, it had a slightly higher electrophoretic mobility than expected (Fig. 3A).
Fig 2.
Expression and purification of p31-pQE 30 Xa. (A) Induction of fusion protein expression; (B) protein purification after elution from the column.
Fig 3.
Expression and purification of p40-pQE 30 Xa. (A) Induction of fusion protein expression; (B) protein purification after elution from the column.
Fusion protein purification and antibody production.
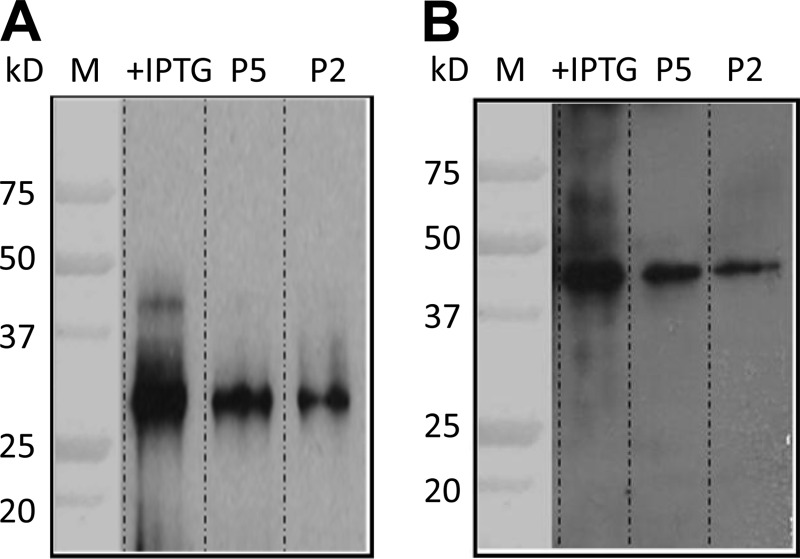
Fusion proteins were purified from the crude extracts using the QIAexpress assay system (Qiagen), and they were analyzed by SDS-PAGE (Fig. 2B and 3B). Three New Zealand White rabbits were immunized with 30 μg of each protein (a total of 90 μg) on days 0, 14, and 21 after collecting preimmune serum prior to the first immunization as an internal control. Ten days after the last immunization the rabbits were sacrificed and exsanguinated to obtain the final serum sample. All samples were analyzed in Western blots of total protein extracts, with or without the induced proteins (Fig. 4A [p31] and B [p40]).
Fig 4.
(A and B) Western blot analysis of rabbit serum. Serum containing antibodies against p31 (A) and p40 (B). M, molecular mass marker; +IPTG, total bacterial extract in which expression was induced; P5, 5 μg of the purified fusion protein; P2, 2 μg of purified fusion protein.
ELISA analysis of the antibody response.
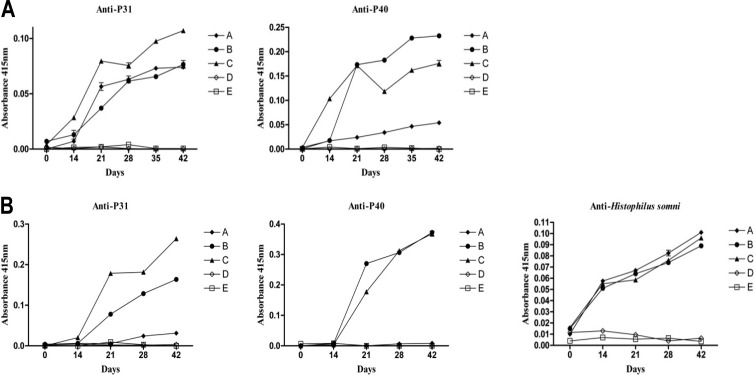
Antibodies against H. somni, rp31 and rp40, were evaluated by ELISA in two experimental models (rabbits and sheep). Vaccination of rabbits with commercial bacterin (Biobac 11 vías) and recombinant vaccine formulations stimulated detectable levels of anti-p31 and anti-p40 antibodies (Fig. 5A), with significant increases in antibody production observed from day 21. For rp31, the antibody response was significantly greater in group C (rp31, 60 μg) than in groups A and B, whereas no antibody production was observed in groups D or E (negative controls). For rp40, the antibody response was significantly greater in group B (rp40, 30 μg) than in groups A and C, also was higher in group C (rp40, 60 μg) than in group A (commercial vaccine).
Fig 5.
Results from ELISA analysis. (A) Rabbits. Group A (positive control) was inoculated with a commercial bacterin (Biobac 11 vías [see Materials and Methods]). Group B was vaccinated with a recombinant preparation containing 30 μg of recombinant p31 (rp31) and 30 μg of recombinant p40 (rp40) and complemented with the commercial vaccine Biobac 7 vías. Group C was inoculated with a recombinant preparation containing 60 μg of rp31, 60 μg of rp40, and Biobac 7 vías. Negative control groups D and E were inoculated with Biobac 7 vías and PBS-adjuvant [Al(OH)3], respectively. (B) Sheep. Group A (positive control) was immunized with the recombinant preparation alone. Group B was immunized with a recombinant preparation containing 50 μg of rp31, 50 μg of rp40, and Biobac 7 vías. Group C was immunized with a recombinant preparation containing 100 μg of rp31, 100 μg of rp40, and Biobac 7 vías. Negative control groups D and E were inoculated with Biobac 7 vías and PBS-adjuvant [Al(OH)3], respectively.
In the second experiment (Fig. 5B), serum samples from sheep were analyzed for rp31 and rp40 and for WC H. somni. Increases in anti-p31 and anti-rp40 were clearly observed from day 21 in groups B (rp31 + rp40, 50 μg) and C (rp31 + rp40, 100 μg) (recombinant vaccine formulations), with greater responses in the latter group for p31, when comparing with controls but also with commercial vaccine (group A). Increased levels of anti-WC H. somni were detected from day 14 in all groups treated with the commercial and recombinant vaccine formulations, at a similar levels until day 42.
Immunization of rabbits and sheep with experimental vaccines complemented with recombinant proteins strongly stimulated the dose-dependent production of antibodies against p31 and p40 (Fig. 5). Hence, the recombinant proteins used in these experiments are clearly immunogenic and can be used to formulate vaccines against BRD.
Bacterial challenge.
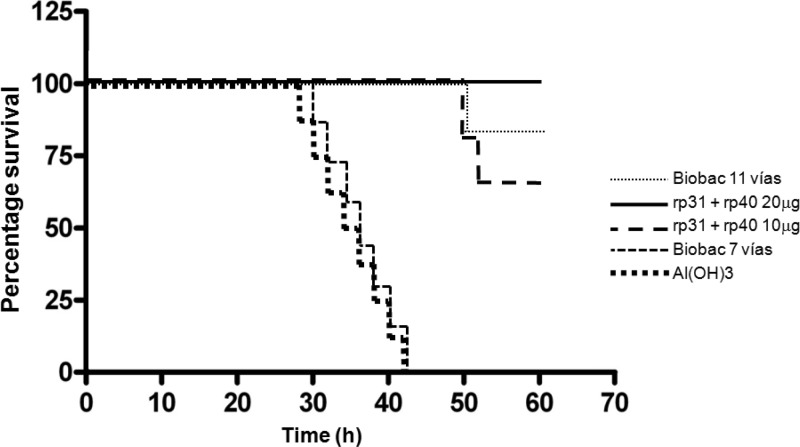
Protection against bacterial challenge with pathogenic H. somni was analyzed by the Fisher exact test, and the results are summarized on a Kaplan-Meier curve (Fig. 6). Mice were monitored for 10 days after challenge with 3.3 × 108 CFU of virulent H. somni, and protection was observed in groups A, B, and C as follows: group A (Biobac 11 vías), 93% survival; group B (immunized with a low dose of recombinant vaccine (rp31 + rp40, 10 μg of each), 87% survival; and group C (immunized with a high dose of recombinant vaccine (20 μg of rp31 + rp40), 100% survival. No protective effects were observed in the negative control groups (D and E), in which no survival was observed following the challenge. The protective effect of the commercial (Biobac 11 vías) and recombinant vaccines (containing rp31 and rp40) was significantly greater (P < 0.0001) than that of the negative controls (Biobac 7 vías and adjuvant).
Fig 6.
Result of challenge test in BALB/c mice. A Kaplan-Meier curve shows the percent survival over time.
DISCUSSION
BRD is generally detected in cattle raised on farms with poor or nonexistent cattle health management plans (20), but many feedlots, calf raising facilities, and other facilities with excellent management also still have a considerable BRD problem (14, 15, 20, 33, 47). Therefore, the use of vaccines as a preventative measure is important to prevent the spread of this disease (40). Although several commercial vaccines were developed prior to the 1990s (reviewed in reference 41), most caused secondary adverse reactions (12), emphasizing the need to develop recombinant vaccines in which the presence of more than one antigen may increase efficacy. The results presented here demonstrate that a recombinant experimental vaccine containing protein fragments of two OMP H. somni antigens, p31 (Plp4) and p40 (LppB), induced high antibody titers in mice and sheep. Furthermore, this vaccine formulation protected mice from septicemia after bacterial challenge. In addition, the use of Al(OH)3 as an adjuvant potentiated the immune response, thereby maximizing the potency and efficacy of the antigens, which are generated in limited amounts (13), and enhancing the immune response (46).
Protective effects of antibodies against the H. somni OMPs p31 (Plp4) and p40 (LppB).
The immunogenic potential of some H. somni proteins has been previously assessed, demonstrating that they can produce partial protection against bacterial infection. Among antigens most probably involved in stimulating host defense as well as immunopathology, OMPs are relevant as virulence factors (6, 9).
Proteins on the cell surface undoubtedly play an important role in H. somni virulence and host immunity (reviewed in reference 44). Because Gram-negative bacteria exhibit a high degree of genomic variability in some of its proteins, conserved OMPs became relevant immunogenic targets that are able to elicit cellular mechanisms of host defense involving the antigen-induced release of cytokines from lymphocytes and the resulting activation of macrophages with the ability to kill the pathogen.
For example, a 40-kDa OMP from H. somni was proposed as a candidate protective protein against pneumonia in calves following active immunization (19). However, according to a recent analysis of genes and gene products putatively involved in H. somni strain 2336 virulence, this protein corresponds to a different OMP than the p40 (LppB) used here (see Table 2 in reference 44). A 78-kDa OMP antigen that was also shown to be consistently and intensely immunoreactive in Western blots of H. somnus WC reacted with convalescent-phase serum from cattle with experimental H. somnus pneumonia (26). However, this antigen failed to elicit protective effects (19).
An immunoglobulin binding protein A (IbpA) containing a Fic motif involved in the virulence of several pathogens (42, 53) was recently described (30, 55) as a viable vaccine candidate in the bovine host. Immunization with the IbpA DR2 subunit from H. somni was demonstrated to partially protect against bacterial infection in a natural host (16, 30). However, further studies will be necessary to determine the immunogenic properties of the different virulence factors involved in H. somni infection, since the evidence for protection against pneumonia by current vaccines remains controversial (29, 48).
Although further immunogenic studies in beef cattle are required, the results presented here demonstrate for the first time that the antibody response in rabbits and in sheep against two H. somni OMPs, p31 (Plp4) and p40 (LppB), is relevant. In addition, when combined with a commercial vaccine for other bacterial diseases (Biovac 7 vías), recombinant fragments of p31 (Plp4) and p40 (LppB), which appear to be conserved structural proteins of the outer bacterial membrane, exert a protective effect against H. somni bacterial challenge in mice.
p31 and p40 structure and possible functional implications.
The electrophoretic mobility observed for the p40 fusion protein corresponded to that originally reported for this protein (8, 50), although it was slightly higher than theoretically expected. This protein is rich in proline (8%), asparagine (8.0%), and isoleucine (9.7%) compared to the average amino acid composition of vertebrate proteins (54), a profile that may influence its electrophoretic mobility. The fusion protein also contains some putative peptidoglycan binding sites (32), which may modify its relative molecular mobility. In addition, the increased electrophoretic mobility described here may be partially due to the addition of a histidine tag and the factor Xa recognition site. Nonetheless, when the clones were sequenced they precisely matched the reported sequence, and thus the induced fusion protein corresponds to p40.
It is of interest to determine whether the organization of the distinct domains in these two proteins are implicated in the effects of H. somni on endothelial cells and in the aggregation of platelets to form thrombi in blood vessels (19). These phenomena induce endothelial cell proinflammatory responses and platelet internalization (27), and they trigger cytoskeletal alterations that increase the permeability of the endothelium (4), resulting in the redistribution of PECAM 1 on the surfaces of bronchial endothelial cells (51).
Further studies are thus required with antibodies that identify p31 (Plp4) and p40 (LppB), such as those described here, that can be used to study their effects at the cellular level (e.g., by analyzing the antibody neutralization of these two proteins in vitro).
Finally, the basis of viral/bacterial synergism and the manner in which cattle respond to the virulence strategies of bacterial pathogens remain poorly understood (10). The ability of H. somni to resist leukocytes while creating a proinflammatory and procoagulation environment at the endothelial cell surface and the ability of M. haemolytica to circumvent leukocyte antibacterial activity via its leukotoxin LKTA probably contribute to the intense inflammation that characterizes BRD (10). As such, it seems feasible to propose that a vaccine that combines antigenic surface proteins from M. haemolytica (e.g., different fragments of LktA and Plp) (7, 22) and H. somni (e.g., p31 and p40, as demonstrated here) may be useful in preventing infection and reducing the incidence of BRD. Further experiments would be needed to test this hypothesis.
ACKNOWLEDGMENTS
This study was partially supported by CONACYT grants 2007-C01-71496 to B.F.-S., 2009-111203 and 2010-129167 to D.O.-S., and 237076 to C.G.-B.
We thank Bio-Zoo laboratories for the use of their field-testing facilities.
Footnotes
Published ahead of print 12 September 2012
REFERENCES
- 1. Angen Ø, Ahrens P, Kuhnert P, Christensen H, Mutters R. 2003. Proposal of Histophilus somni gen. nov., sp. nov. for the three species incertae sedis “Haemophilus somnus,” “Haemophilus agni,” and “Histophilus ovis.” Int. J. Syst. Evol. Microbiol. 53:1449–1456 [DOI] [PubMed] [Google Scholar]
- 2. Ausubel F, et al. 2001. Current protocols in molecular biology. John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 3. Bateman A, Bycroft M. 2000. The structure of a LysM domain from Escherichia coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299:1113–1119 [DOI] [PubMed] [Google Scholar]
- 4. Behling-Kelly E, McClenahan D, Kim KS, Czuprynski CJ. 2007. Viable “Haemophilus somnus” induces myosin light-chain kinase-dependent decrease in brain endothelial cell monolayer resistance. Infect. Immun. 75:4572–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cho YS, et al. 2008. Safety and efficacy testing of a novel multivalent bovine bacterial respiratory vaccine composed of five bacterins and two immunogens. J. Vet. Med. Sci. 70:959–964 [DOI] [PubMed] [Google Scholar]
- 6. Confer AW. 2009. Update on bacterial pathogenesis in BRD. Anim. Health Res. Rev. 10:145–148 [DOI] [PubMed] [Google Scholar]
- 7. Confer AW, et al. 2009. Immunity of cattle following vaccination with a Mannheimia haemolytica chimeric PlpE-LKT (SAC89) protein. Vaccine 27:1771–1776 [DOI] [PubMed] [Google Scholar]
- 8. Corbeil LB, Kania SA, Gogolewski RP. 1991. Characterization of immunodominant surface antigens of Haemophilus somnus. Infect. Immun. 59:4295–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Corbeil LB. 2007. Histophilus somni host-parasite relationships. Anim. Health Res. Rev. 8:151–160 [DOI] [PubMed] [Google Scholar]
- 10. Czuprynski CJ. 2009. Host response to bovine respiratory pathogens. Anim. Health Res. Rev. 10:141–143 [DOI] [PubMed] [Google Scholar]
- 11. Ellis JA. 2001. The immunology of the bovine respiratory disease complex. Vet. Clin. N. Am. Food Anim. Pract. 17:535–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ellis JA, Yong C. 1997. Systemic adverse reactions in young Simmental calves following administration of a combination vaccine. Can. Vet. J. 38:45–47 [PMC free article] [PubMed] [Google Scholar]
- 13. Exley C, Siesjo P, Eriksson H. 2010. The immunobiology of aluminum adjuvants: how do they really work? Trends Immunol. 31:103–109 [DOI] [PubMed] [Google Scholar]
- 14. Fulton RW. 2009. Bovine respiratory disease research (1983–2009). Anim. Health Res. Rev. 10:131–139 [DOI] [PubMed] [Google Scholar]
- 15. Gagea MI, et al. 2006. Diseases and pathogens associated with mortality in Ontario beef feedlots. J. Vet. Diagn. Invest. 18:18–28 [DOI] [PubMed] [Google Scholar]
- 16. Geertsema RS, et al. 2011. IbpA DR2 subunit immunization protects calves against Histophilus somni pneumonia. Vaccine 29:4805–4912 [DOI] [PubMed] [Google Scholar]
- 17. Geertsema RS, Kimball RA, Corbeil LB. 2007. Bovine plasma proteins increase virulence of Haemophilus somnus in mice. Microb. Pathog. 42:22–28 [DOI] [PubMed] [Google Scholar]
- 18. Geertsema RS, et al. 2008. Protection of mice against H. somni septicemia by vaccination with recombinant immunoglobulin binding protein subunits. Vaccine 26:4506–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gogolewski RP, Leathers CW, Liggitt HD, Corbeil LB. 1987. Experimental Haemophilus somnus pneumonia in calves and immunoperoxidase localization of bacteria. Vet. Pathol. 24:250–256 [DOI] [PubMed] [Google Scholar]
- 20. Griffin D. 2010. Bovine pasteurellosis and other bacterial infections of the respiratory tract. Vet. Clin. N. Am. Food Anim. Pract. 26:57–71 [DOI] [PubMed] [Google Scholar]
- 21. Grizot S, Buchanan SK. 2004. Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Mol. Microbiol. 51:1027–1037 [DOI] [PubMed] [Google Scholar]
- 22. Guzmán-Brambila C, et al. 2012. LKTA and PlpE small fragments fusion protein protect against Mannheimia haemolytica challenge. Res. Vet. Sci. 93:1293–1300 [DOI] [PubMed] [Google Scholar]
- 23. Harris FW, Janzen ED. 1989. The Haemophilus somnus disease complex (hemophilosis): a review. Can. Vet. J. 30:816–822 [PMC free article] [PubMed] [Google Scholar]
- 24. Howard MD, et al. 2011. Genetics and molecular specificity of sialylation of Histophilus somni lipooligosaccharide (LOS) and the effect of LOS sialylation on Toll-like receptor-4 signaling. Vet. Microbiol. 153:163–172 [DOI] [PubMed] [Google Scholar]
- 25. Joris B, et al. 1992. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol. Lett. 70:257–264 [DOI] [PubMed] [Google Scholar]
- 26. Kania SA, Gogolewski RP, Corbeil LB. 1990. Characterization of a 78-kilodalton outer membrane protein of Haemophilus somnus. Infect. Immun. 58:237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuckleburg CJ, McClenahan DJ, Czuprynski CJ. 2008. Platelet activation by Histophilus somni and its lipooligosaccharide induces endothelial cell proinflammatory responses and platelet internalization. Shock 29:189–196 [PMC free article] [PubMed] [Google Scholar]
- 28. Kumar R, et al. 2012. RNA-seq-based transcriptional map of bovine respiratory disease pathogen “Histophilus somni 2336.” PLoS One 7:e29435 doi:10.1371/journal.pone.0029435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Larson RL, Step DL. 2012. Evidence-based effectiveness of vaccination against Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni in feedlot cattle for mitigating the incidence and effect of bovine respiratory disease complex. Vet. Clin. N. Am. Food Anim. Pract. 28:97–106 [DOI] [PubMed] [Google Scholar]
- 30. Lo KL, et al. 2012. Antibody responses of calves to Histophilus somni recombinant IbpA subunits. Comp. Immunol. Microbiol. Infect. Dis. 35:453–459 [DOI] [PubMed] [Google Scholar]
- 31. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 32. Marchler-Bauer A, et al. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39:225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McVey DS. 2009. BRD research needs in the next 10-20 years. Anim. Health Res. Rev. 10:165–167 [DOI] [PubMed] [Google Scholar]
- 34. Nardini PM, Mellors A, Lo RY. 1998. Characterization of a fourth lipoprotein from Pasteurella haemolytica A1 and its homology to the OmpA family of outer membrane proteins. FEMS Microbiol. Lett. 165:71–77 [DOI] [PubMed] [Google Scholar]
- 35. Norma Oficial Mexicana 2008. Especificaciones técnicas para la producción, cuidado y uso de los animales de laboratorio. Document NOM-062-ZOO-1999. Modified 26 November 2008 Senasica, Mexico City, Mexico: www.senasica.gob.mx/default.asp?doc=743 [Google Scholar]
- 36. Ochsner UA, Vasil AI, Johnson Z, Vasil ML. 1999. Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA. J. Bacteriol. 181:1099–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. O'Toole D, Allen T, Hunter R, Corbeil LB. 2009. Diagnostic exercise: myocarditis due to Histophilus somni in feedlot and backgrounded cattle. Vet. Pathol. 46:1015–1017 [DOI] [PubMed] [Google Scholar]
- 38. Pagni M, et al. 2007. MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 35:W433–W437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pérez-Romero N, Aguilar-Romero F, Arellano-Reynoso B, Díaz-Aparicio E, Hernández-Castro R. 2011. Isolation of Histophilus somni from the nasal exudates of a clinically healthy adult goat. Trop. Anim. Health Prod. 43:901–903 [DOI] [PubMed] [Google Scholar]
- 40. Potter A, Gerdts V, Littel-van den Hurk SD. 2008. Veterinary vaccines: alternatives to antibiotics? Anim. Health Res. Rev. 9:187–199 [DOI] [PubMed] [Google Scholar]
- 41. Rice JA, Carrasco-Medina L, Hodgins DC, Shewen PE. 2007. Mannheimia haemolytica and bovine respiratory disease. Anim. Health Res. Rev. 8:117–128 [DOI] [PubMed] [Google Scholar]
- 42. Roy CR, Mukherjee S. 2009. Bacterial FIC proteins AMP up infection. Sci. Signal. 2:pe14. [DOI] [PubMed] [Google Scholar]
- 43. Sambrook J, Fritsch EF, Maniatis T. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, New York, NY [Google Scholar]
- 44. Sandal I, Inzana TJ. 2010. A genomic window into the virulence of Histophilus somni. Trends Microbiol. 18:90–99 [DOI] [PubMed] [Google Scholar]
- 45. Sanders JD, Bastida-Corcuera FD, Arnold KF, Wunderlich AC, Corbeil LB. 2003. Genetic manipulation of immunoglobulin binding proteins of Haemophilus somnus. Microb. Pathog. 34:131–139 [DOI] [PubMed] [Google Scholar]
- 46. Seubert A, Monaci E, Pizza M, O'Hagan DT, Wack A. 2008. The adjuvants aluminum hydroxide and MF59 induce monocyte and granulocyte chemoattractants and enhance monocyte differentiation toward dendritic cells. J. Immunol. 180:5402–5412 [DOI] [PubMed] [Google Scholar]
- 47. Snowder GD, Van Vleck LD, Cundiff LV, Bennett GL. 2006. Bovine respiratory disease in feedlot cattle: environmental, genetic, and economic factors. J. Anim. Sci. 84:1999–2008 [DOI] [PubMed] [Google Scholar]
- 48. Srikumaran S, Kelling CL, Ambagala A. 2007. Immune evasion by pathogens of bovine respiratory disease complex. Anim. Health Res. Rev. 8:215–229 [DOI] [PubMed] [Google Scholar]
- 49. Taylor JD, et al. 2010. The epidemiology of bovine respiratory disease: what is the evidence for predisposing factors? Can. Vet. J. 51:1095–1102 [PMC free article] [PubMed] [Google Scholar]
- 50. Theisen M, Rioux CR, Potter AA. 1993. Molecular cloning, nucleotide sequence, and characterization of lppB, encoding an antigenic 40-kilodalton lipoprotein of Haemophilus somnus. Infect. Immun. 61:1793–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tiwari R, Sullivan J, Czuprynski CJ. 2009. PECAM-1 is involved in neutrophil transmigration across Histophilus somni-treated bovine brain endothelial cells. Microb. Pathog. 47:164–170 [DOI] [PubMed] [Google Scholar]
- 52. Won J, Griffith RW. 1993. Cloning and sequencing of the gene encoding a 31-kilodalton antigen of Haemophilus somnus. Infect. Immun. 61:2813–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Worby CA, et al. 2009. The fic domain: regulation of cell signaling by adenylylation. Mol. Cell 34:93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wilkins MR, et al. 1996. From proteins to proteomes: large-scale protein identification by two-dimensional electrophoresis and amino acid analysis. Biotechnology (NY) 14:61–65 [DOI] [PubMed] [Google Scholar]
- 55. Zekarias B, et al. 2010. Histophilus somni IbpA DR2/Fic in virulence and immunoprotection at the natural host alveolar epithelial barrier. Infect. Immun. 78:1850–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]