Abstract
Heparin has been used clinically as an anticoagulant for over 60 years. Typically isolated from porcine intestine, heparin is a mixture of dimeric glycosidic sequences generating complex polysaccharide glycosaminoglycan chains. Recently, certain lots of heparin have been associated with an acute, rapid onset of significant side effects indicative of an allergic-type reaction. To identify potential causes for this serious rise in side effects, we examined lots of heparin that correlated with adverse events using orthogonal high resolution analytical techniques. Through comparison of these results with those obtained on reference lots, suspect lots were found to contain a highly sulfated chondroitin sulfate contaminant. Through detailed structural analysis, the contaminant was found to contain a disaccharide repeat unit of glucuronic acid linked β1→3 to a β-galactosamine. Surprisingly, the disaccharide unit contains an unusual sulfation pattern and is sulfated at the 2-O and 3-O positions of the glucuronic acid as well as at the 4-O and 6-O positions of the galactosamine. The presence of such a contaminant could elicit a biological response as highly sulfated polysaccharides, such as dextran sulfate, are known to be potent mediators of the immune system. Given the nature of the contaminant, traditional screening tests - such as those present as part of the current United States Pharmacopeia heparin monograph - cannot differentiate between affected and unaffected lots. Our analysis suggests effective screening methods that can be employed to determine whether or not heparin lots contain the contaminants reported here.
Introduction
Heparin, a complex glycosaminoglycan polysaccharide, is widely used as an anticoagulant in a number of settings, including kidney dialysis and acute coronary syndromes (1,2,3). Recently there has been marked increase in serious adverse events associated with heparin therapy, with hundreds of patients affected. These events include development of such symptoms as rash, fainting, racing heart, and other, more severe symptoms. This has resulted in a national health care crisis. Germany and other nations in the European Union are also observing a similar phenomenon, making this an international issue. The rapid onset of these symptoms suggests an anaphylactic response but the exact etiology is currently unknown. Given the clinical history of heparin, this spike in adverse events suggests the potential for contamination of heparin. However, the existent screening of heparin lots for typical biological contaminants including protein, lipids, or DNA, which, if present, may elicit such a response, indicates that there is no difference between lots which elicit adverse events from those that do not (4). Despite extensive analysis, there were no obvious differences in other potential contaminants, including lead, dioxins, and other molecular entities between lots (4). Defintive identification of how these heparin lots differ from the historical context of heparin thus becomes imperative.
Recently, several analytical tests have detected differences in suspect versus control lots (5). Screening of heparin lots by a combination of optical rotation, capillary electrophoresis, and one dimensional NMR indicates a defined pattern that can be used to distinguish suspect from control lots. In the case of capillary electrophoresis, suspect lots contain an additional, leading edge peak in addition to the broad peak associated with heparin. Proton NMR analysis indicates distinctive differences between suspect and control lots, most prominently in the acetyl region of the spectrum (2-2.2 ppm). Given the nature of these analyses, and the differences observed upon comparison of suspect versus control lots, the source of the major contaminant was surmised to be a highly sulfated “heparin-like” contaminant (5).
To understand further the structure or structures of the contaminant(s) present within specific lots of heparin, we sought to identify the major contaminant. This exercise required the use of multiple orthogonal techniques, including multidimensional NMR, to overcome the challenges inherent in the analysis of complex polysaccharides, including heparin, which in and of itself comprises a complex mixture of glycosaminoglycan chains. In doing so, we were able to determine definitively the structure of the contaminant(s), isolate it, and confirm the structural identity by comparison to a chemically synthesized standard.
Results
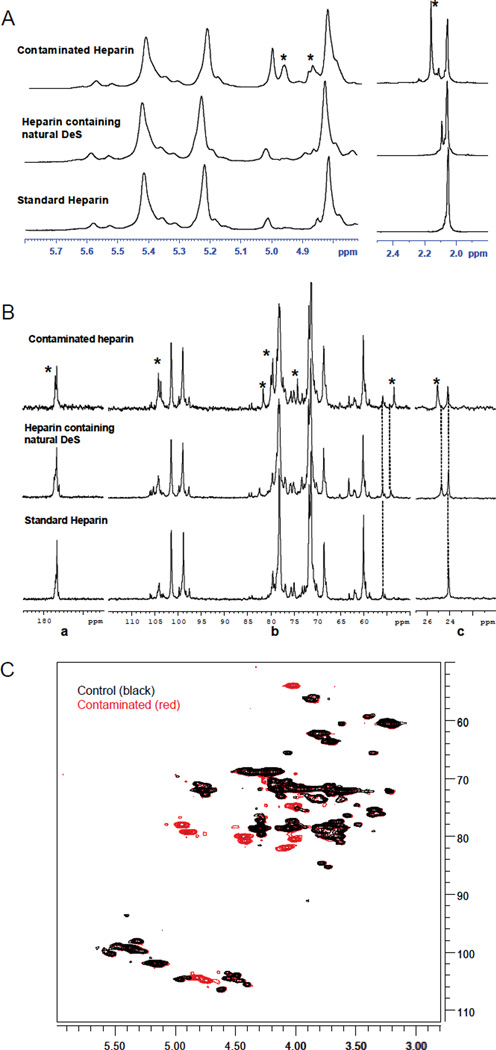
For this study, lots that were associated with adverse clinical reactions (designated S1–S6) were examined as well as two control lots of heparin (designated C1 and C2) not associated with adverse events. Initial analysis of S1–S6 by one-dimensional NMR indicated that all of these samples exhibited an unusual series of N-acetyl signals (Figure 1A, Supplemental Information Figure 1). For example, particularly evident in the proton spectrum of S1 is the signal at 2.16 ppm corresponding to an N-acetyl group different from that of heparin (2.04 ppm). This N-acetyl signal is also distinct from that of dermatan sulfate, a known impurity in heparin samples (2.08 ppm). To complement and extend the proton analysis, carbon (100MHz) NMR spectroscopy was performed. Comparison of the carbon spectra of S1 and a control sample (C1) indicates the presence of several additional signals not normally associated with heparin structural signatures (Figure 1B). The acetyl signal at 25.6 ppm together with the signal at 53.5 ppm is indicative of the presence of an N-acetyl galactosamine of unknown structure, but again distinct from the N-acetyl galactosamine contained within dermatan sulfate, with corresponding signals at 24.8 ppm and 54.1 ppm, respectively. Other signals are visible in the ring and anomeric regions of the carbohydrate moiety. The latter signals (103–105 ppm) agree with a beta configuration of glycosidic linkages for the contaminant.
Figure 1.
(A) Comparison of anomeric and acetyl regions of the proton spectra of standard heparin, heparin containing natural dermatan sulfate (DeS) and contaminated heparin. (B) Comparison of carbonyl (a), sugar (b) and N-acetyl regions (c) of the carbon spectra of standard heparin, heparin containing natural dermatan sulfate, and contaminated heparin. Signals due to the contaminant are highlighted by asterisks. (C) HSQC spectrum of the contaminated sample S1 overlaid on control sample C1.
Multidimensional Heteronuclear Single Quantum Coherence (HSQC) spectra were also collected on the S1 and C1, to separate the observed signals in two dimensions – the carbon and proton – to further identify the number and type of any major contaminants. Ten major signals were observed in sample S1 which were not observed in C1. These same signals were observed in samples S2–S6 (Supplemental Information Figure 2). In addition, these results and those from other two dimensional experiments, including TOCSY, COSY and ROESY, were also consistent with the basic findings outlined above, namely that the major contaminant consists of a polymeric repeat of N-acetyl galactosamine linked to glucuronic acid through exclusively beta linkages.
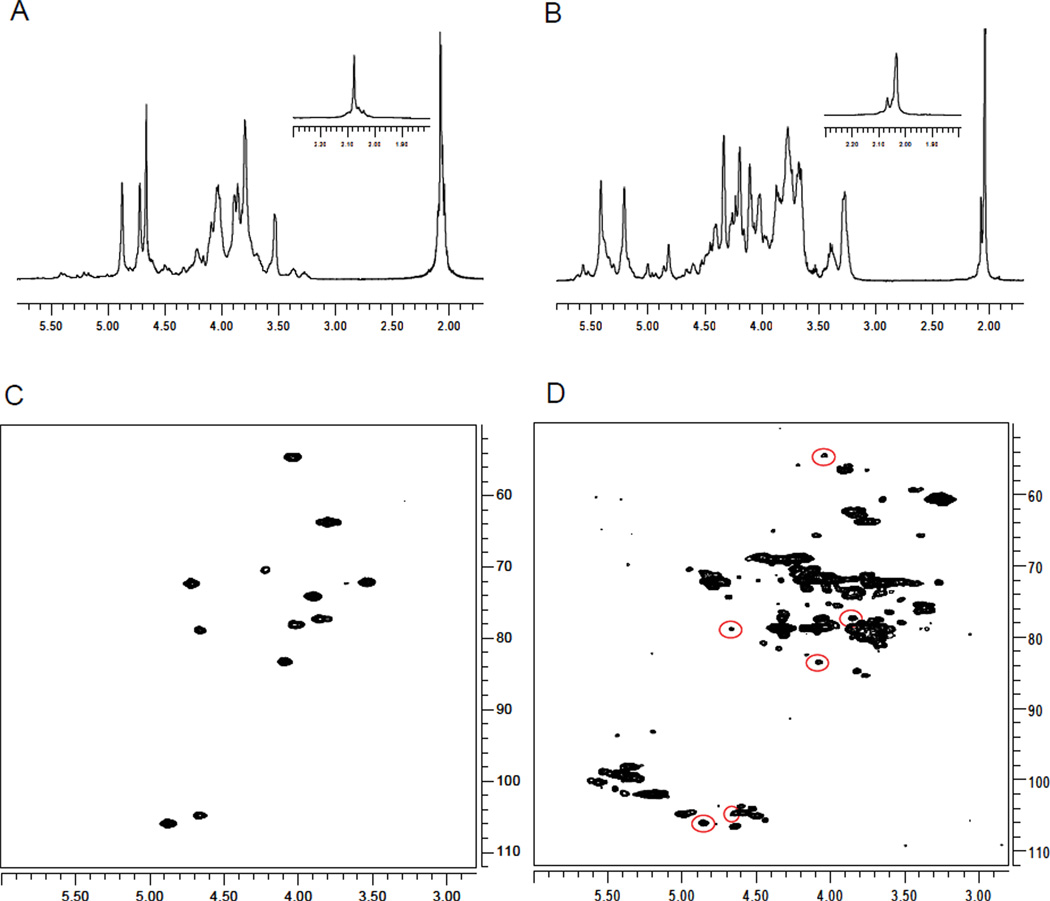
Much of the analysis outlined above focused on sample S1, as it possessed signals in the N-acetyl region at 2.04 ppm (arising from heparin) and at 2.16 (arising from the unknown contaminant). In addition to this unusual signal at ~2.16 ppm, some samples (S2–S6 and C2) possessed an additional N-acetyl peak (Figure 2B) which had a chemical shift of 2.08 ppm which indicate the presence of another contaminant that is distinct from the one described above. Given the observed chemical shifts, it is likely that this species is dermatan sulfate, a known natural impurity of heparin (9,10). To confirm the identity of this species a 2D 1H-13C HSQC experiment was performed on sample C2 (Figure 2D) and the results compared to those obtained on sample C1 that did not show this split. A comparison of the chemical shifts observed in sample C2 but not in sample C1 indicates that these are similar to those reported in the literature (9) and suggest that the additional peaks in the HSQC of C2 can be assigned to dermatan sulfate. This assignment was confirmed by comparison to 1H and HSQC data obtained on a standard of dermatan sulfate (Figure 2A,C). Similar to the analysis completed on sample C2, the presence of dermatan sulfate was confirmed in samples S2, S3, S4, S5, and S6. Therefore, these samples, but not sample S1, contain both dermatan sulfate as well as the unusual contaminant.
Figure 2.
Comparison between 1H-NMR spectra of (A) dermatan sulfate and (B) sample C2, which contains dermatan sulfate. Inset for panels A and B specifically shows the N-acetyl region of the spectrum. (C) HSQC spectra of dermatan sulfate and (D) sample C2. Peaks representative of dermatan sulfate are highlighted.
Finally, to confirm the findings from NMR analysis of the samples, we conducted enzymatic digestion of S1–S6 and C1–C2 with either a combination of heparinases or heparinases plus Δ4,5 glycuronidase and 2-O sulfatase followed by separation and analysis by HPLC. Digestion with the heparinases reduces heparin to its component di, tri- and tetrasaccharides and imparts a Δ4,5 bond monitorable at 232 nm; completed in conjunction with treatment with glycuronidase and sulfatase this digest permits the identification of minor species, including those disaccharides containing a modified galacturonic acid (6,7). Thus, concomitant use of a matrix of enzymes, especially in conjunction with LC-MS/MS analysis allows for the complete separation, identification, and quantification of heparin components in the mixture (8). We find that the total area under the curve of digested S1–S6 are significantly less than those of the control samples, indicating that the major contaminant is not significantly digested by the heparinases (Table 1). Furthermore, the relative quantities of the individual heparin components are similar between the controls and suspect samples, with only minor differences.
Table 1.
Total Area Under the Curve for Heparinase Digests of S1–S6, C1–C2
| Sample | Total Area |
|---|---|
| C1 | 2.52E+07 |
| C2 | 2.70E+07 |
| S1 | 1.74E+07 |
| S2 | 9.50E+06 |
| S3 | 1.94E+07 |
| S4 | 1.59E+07 |
| S5 | 1.91E+07 |
| S6 | 1.96E+07 |
To assign the unknown compound present in the contaminated heparin samples, we attempted to isolate the contaminant using a variety of methods. First, given the overall properties of the contaminant, elucidated through NMR, CE and HPLC analysis, we reasoned that this material could be differentially precipitated upon addition of an organic solvent. Partial purification of the contaminant was indeed achieved through the addition of increasing amounts of ethanol to an aqueous solution of S1. Similarly, because the contaminant contains N-acetylhexosamine (and not N-sulfohexosamine), it was also purified by degradation of heparin by nitrous acid and isolation of the remaining components.
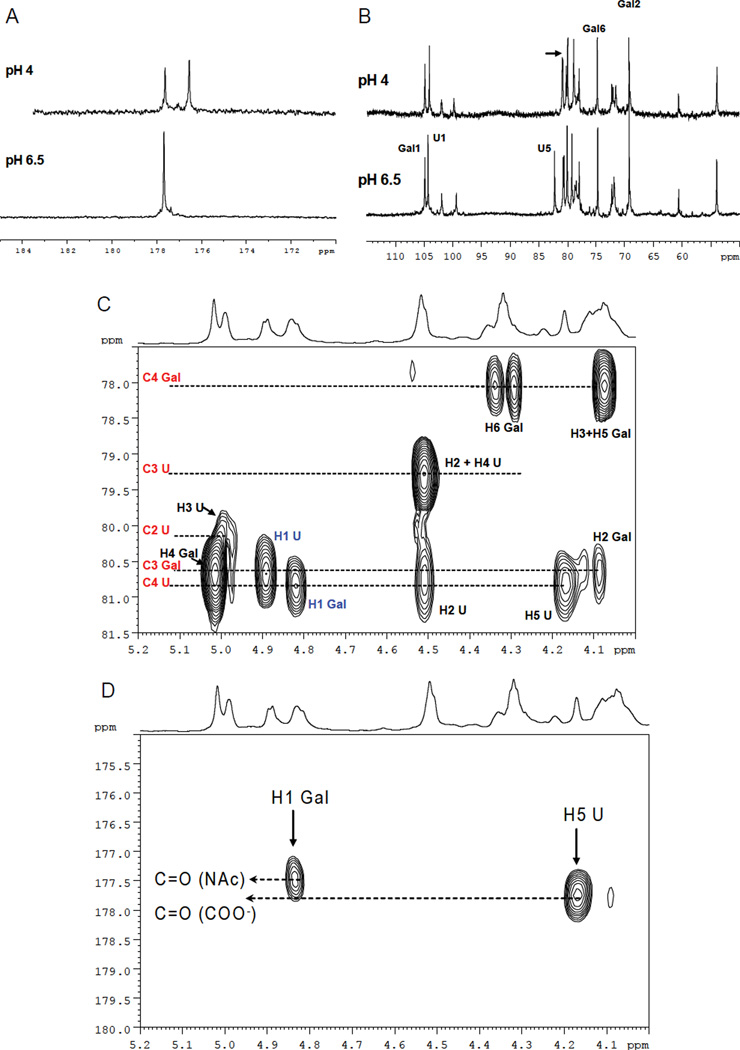
After isolation, the proton spectrum reveals a residual heparin content of about ≥ 30% (depending on the isolation method employed) as determined by 1D NMR (data not shown). At neutral pH values, one apparent signal arises at 177.6 ppm characteristic of carbonyl groups. Acidification of a solution of the product (Figure 3A) reveals two distinct carbonyl signals consistent with the carbonyl group of an N-acetyl function and the protonated form of carboxylic acid respectively. Similar shifts were not observed for any other signals except C-5 of uronic acid, which has known sensitivity to the ionization state of the carboxylic acid (Figure 3B). To further characterize the isolated sample, homonuclear (COSY and TOCSY, data not shown) and heteronuclear (HSQC, HSQC-TOCSY and HMBC) 2D-NMR spectroscopy were employed (Figure 3C,D). These analyses indicate the presence of two types of residues where chemical shift patterns agree with one type of monosaccharide being 4,6-O-sulfo-N-acetyl-galactosamine (Gal) and the other being a 2,3-O-sulfo-glucuronic acid (U). In addition to confirming the assignments of the sugar moieties, the HMBC spectrum demonstrates the correlation across the glycosidic linkages which indicate the presence of a β 1–4 linkage between Gal and U and a β1–3 linkage between U and Gal. The long-range correlations between H1 of Gal and H5 of U with two different carbonyl groups (177.5 and 177.8 ppm) due to the N-acetyl of galactosamine and the carboxylic group of glucuronic acid confirm the structure.
Figure 3.
(A) Carbonyl region of carbon spectrum of the contaminant measured at pH 6.5 and 4. (B) Sugar region of the carbon spectrum of the contaminant measured at pH 6.5 and 4. (C) A portion of 600 MHz HMBC spectrum of contaminant. Intramolecular two and three bonds proton-carbon correlations are shown in black; interglycosidic proton-carbon correlations are indicated in blue. (D) Portion of 600MHz HMBC spectrum of contaminant. Long range correlation between the H1 of Gal with C=O of the acetyl group and H5 of U with the carboxylic group are shown.
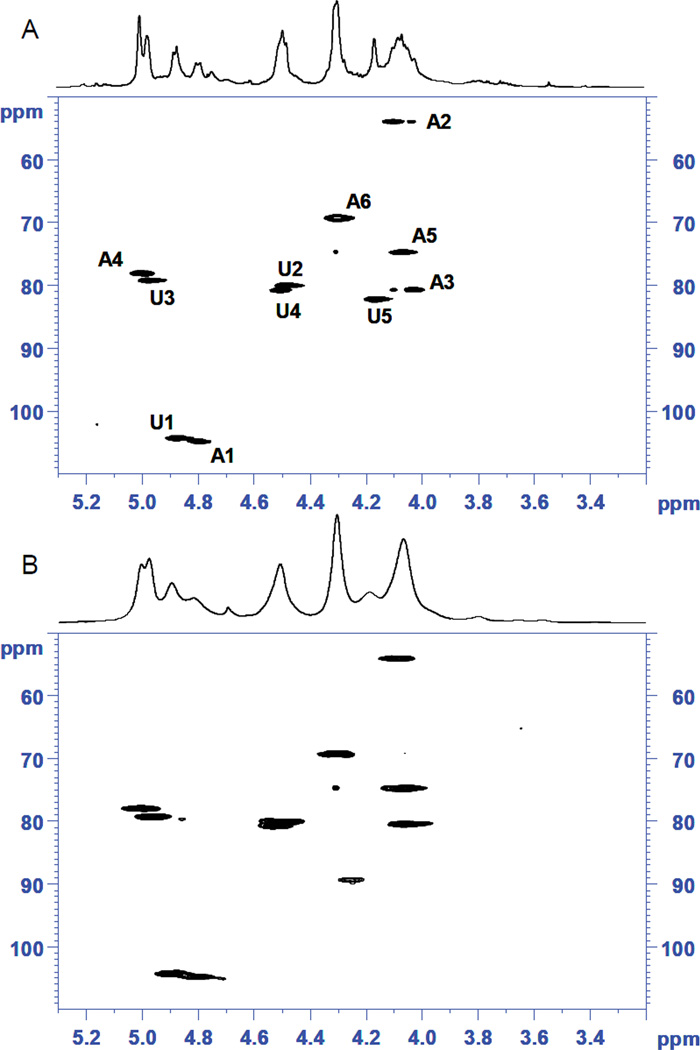
The assignment of the contaminant as an oversulfated chondroitin sulfate containing a tetrasulfated glucuronic acid linked to N-acetylglucosamine was surprising, as this is an unusual structure. To ensure the accuracy of this assignment, a standard was prepared by oversulfation of chondroitin sulfate using well-established chemistry, analysis of this sample by 2D NMR and careful comparison of the HSQC both to the literature (11) as well as to the HSQC spectrum obtained for the isolated contaminant. The comparison of the HSQC spectra for the synthesized standard compared to the isolated contaminant (Figure 4) confirmed that the major contaminant consists of per-O-sulfated chondroitin sulfate, where all of the hydroxyl groups of both the uronic acid and galactosamine residues bear sulfate substituents. The proton chemical shifts of the contaminant (Table 1) are furthermore in agreement with those assigned to fully sulfated chondroitin (degree of sulfation of 4) published by Maruyama and co-workers in 1998 (11). These results were independently confirmed by distinct labs participating in this study. The final derived structure of the major contaminant present in heparin is shown in Figure 5.
Figure 4.
(A) 1H-NMR and HSQC spectra of the isolated contaminant. Signals for 4,6-O-sulfo-N-acetyl-galactosamine (A) and of 2,3-O-sulfo-glucuronic acid (U) are labelled. (B) 1H-NMR and HSQC spectra of the chemically synthesized oversulfated chondroitin.
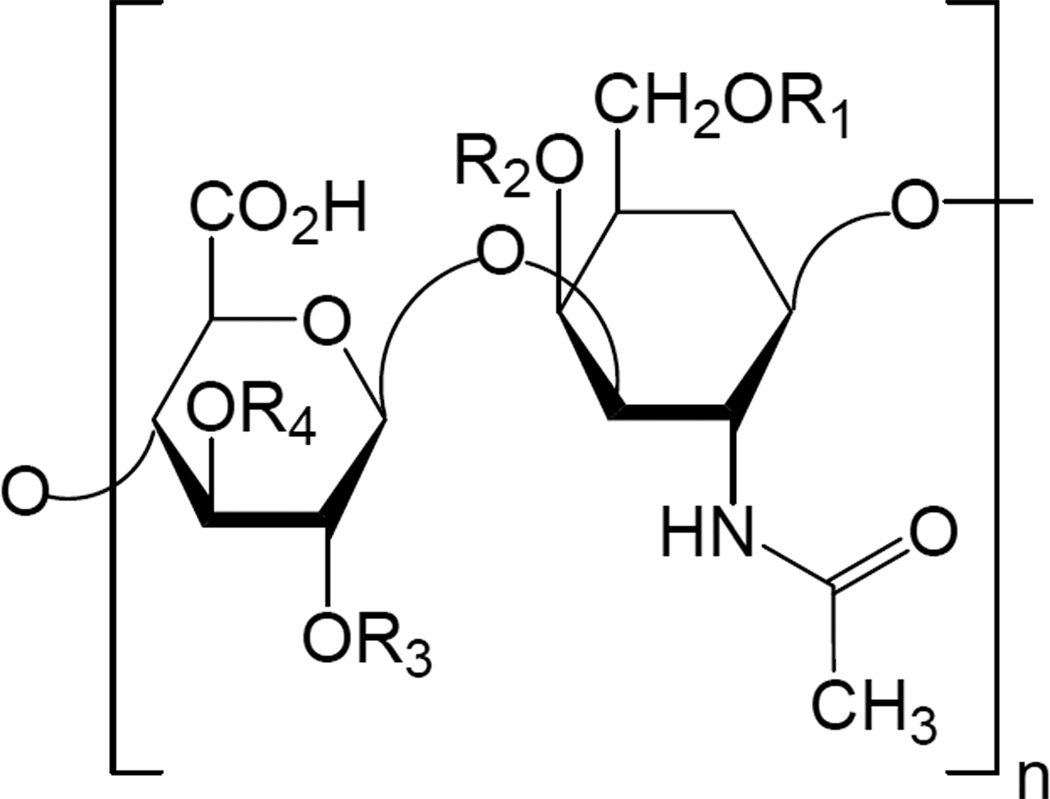
Figure 5.
Structure of the deduced disaccharide sequence of the major contaminant where R1,R2,R3, and R4 are all sulfated.
Discussion
Several factors required the use of multiple approaches to ensure an accurate structural determination of the oversulfated chondroitin contaminant. First, heparin as a polydisperse mixture of saccharides is chemically complex which makes careful interpretation of results a necessity to avoid misinterpretation. This is especially important when addressing the multiple isomeric possibilities within the chains of a complex mixture, and necessitates the use of orthogonal techniques, including using an enzyme matrix and multidimensional NMR. Second, oversulfated materials, such as the chondroitins, co-purify with heparin renderng isolation challenging and they are also resistant to enzymatic digestion techniques (11, 12).
These complexities are exhibited in a number of ways, including the fact that structurally distinct species may nonetheless have overlapping signals and properties and thus may be “masked” within a single analysis. A non-integrated approach can therefore potentially lead to a false conclusion, especially when differentiating between heparin, dermatan sulfate, and oversulfated chondroitin sulfate in any given sample. In this scientific study we took care to devise experimental techniques which could distinguish among the multiple possibilities, including between dermatan (a known contaminant of heparin) and oversulfated chondroitin sulfate.
The structure of the contaminant, which contains a tetrasulfated disaccharide repeat, is highly unusual. First, the presence of a 3-O sulfated glucuronic acid is rare, only observed in specific contexts within certain organisms (13). In addition, a tetrasulfated disaccharide repeat unit has not been isolated to date from animal tissues. It is highly unlikely that the contaminant reported here is found biologically. This finding raises the question of where in the heparin production process the contaminant was introduced. Several possibilities exist, including after purification of the heparin drug substance or at the crude stage prior to purification. Finally, chemically synthesized tetrasulfated disaccharide repeat units of chondroitin sulfate are known to exhibit a high degree of anti-IIa activity (11), which could also explain how, contaminated heparin very easily, passed activity screens, including whole blood coagulation tests. Further investigation is warranted to understand how this material was introduced.
With respect to the biological ramifications of this finding, the presence of an oversulfated chondroitin sulphate within a heparin preparation could provoke increased side effects as highly sulfated polysaccharides, such as dextran sulfate, have been shown to be potent mediators of the immune response, including hypersensitivity (16, 17). The structural determination of this contaminant provides a way to identify a plausible biological role, either direct or indirect, in the onset of such adverse clinical events. Furthermore, it is unclear whether the significant amounts of dermatan sulfate present in these samples may have contributed in any fashion to some of the observed events.
Taken together, the orthogonal nature of the experiments provides strong support to our conclusion. Additionally, it provides a scientific underpinning for screening methods that could be employed to monitor the heparin supply and ensure the absence of oversulfated chondroitin sulfate contamination. For example, using the structural information presented here, it is possible now to design reference standards that ensure accuracy of analysis and quantification for a given analytical method, as well as assess its specificity, and devise an experimental protocol to clearly define the nature and extent, if any, of contamination in a given heparin lot. Finally, the ramifications of these findings are broad and extend beyond scientific considerations to include clinical and health policy issues. Given the importance of these findings – at the intersection of science, technology, and drug regulation – this study offers a potential model to approach, effectively and swiftly, similar challenging issues that may arise in the future. This is especially critical in our global age where a variety of drug substances and products are produced in different parts of the world.
Methods
Materials
Chondroitin sulfate type A from whale cartilage, pyridine-sulphur trioxide complex, tributylamine, dry N,N dimethylformamide, pyridine, methanol, dimethylsulfoxide, NaNO2, NaBH4, 2,2-dimethyl-2-silapentane-5-sulfonate sodium salt, and sodium trimethyl-sylil-propionate were purchased from Sigma-Aldrich (St. Louis, MO, USA). Tetrabutylammonium chloride was purchased from Fluka (St. Louis, MO, USA). 99.9% D2O and 99.96% D2O were obtained from Cambridge Isotope laboratories (Andover, MA). The following eight lots of unfractionated heparin were tested: S1, S2, S3, S4, S5, S6, C1, and C2.
NMR analysis
Samples for 1H-NMR or bidimensional NMR analysis were dissolved in 0.7 mL of D2O (99.9%) and freeze-dried repeatedly to remove exchangeable protons or directly measured without any treatment to preserve potential volatile components. The thoroughly dried samples were dissolved in 0.7 mL of D2O (99.96 %). Before spectra acquisition samples were sonicated for 60s to remove air bubbles. Spectra were obtained at 303K or 308K employing a 600 MHz Varian VNMRS spectrometer or a 600 MHz Bruker Avance 600 spectrometer, both equipped with a 5 mm triple resonance inverse cryoprobe. Monodimensional 1H spectra were obtained with presaturation of residual HOD, for 32–128 scans. Bidimensional DQF-COSY and 2D-TOCSY spectra were acquired using 32 scans per series of 2048 × 512 data points. HSQC and HSQC-TOCSY spectra were recorded with carbon decoupling during acquisition with 320 increments for 12–64 scans. The polarization transfer delay was set with a 1JC-H coupling value of 155 Hz. Heteronuclear multiple bond correlation (HMBC) spectrum was recorded with 320 increments of 64 scans for each, without carbon decoupling and with two-fold low-pass J-filter to suppress one-bond correlations (15). The delay for evolution of long range couplings was set with a Jlr of 8 Hz. Samples for 13C-NMR analysis were dissolved in D2O (99.9%) at 40 mg/mL and analysis was performed at 303K with a 400 MHz Bruker spectrometer equipped with a multinuclear probe.
Composition Analysis by Ion-pair RPHPLC
Samples were constituted in water and digested under two different conditions. The first digest employed an enzyme cocktail of Heparinases I (500 mIU), II (400 mIU), and III (500 mIU) at 30°C, for 16 hr. A portion of this digest was further treated with 2-O-sulfatase (1000 mIU) and Δ4,5-glycuronidase (2000 mIU) for 6 hr, at 30 °C to obtain the second digest. Each digested material was passed through a Ni++ spin column (Qiagen, Germantown, MD, USA) and analyzed by ion-pair RPHPLC similar to as previously described (8). Elution was monitored by UV detection at 232 nm.
Isolation of major contaminant
300 mg of sample S1, dissolved in 1.5 ml of water, were added with absolute ethanol up to the appearance of a white precipitate (ethanol 23%, v/v). Precipitated material was separated by centrifugation for 5 min at 5000 rpm on a Labofuge 200 (Heraeus). In addition to the above, sample S1 was treated with nitrous acid (18). A solution of the sample (500 mg) was dissolved in 20 ml of H2O and cooled at 4 °C. After addition of 140 mg of NaNO2 dissolved in 1 ml of water, the pH was adjusted to 1.7 with 0.1 M HCl. The solution was stirred at 4 °C for 20 min, and additional 100 mg NaNO2 were added stirring the solution for other 20 min. The solution pH was then brought to 7. Solid NaBH4 (200 mg) was added in several portions under stirring. After 2 h, the pH was adjusted to 4 with 0.1 M HCl and the solution was neutralized with 0.1 M NaOH. The product, obtained by precipitation with 4 volumes of methanol, was recovered by centrifugation, dissolved in water and freeze-dried.
Chemical sulfonation of chondroitin sulfate
Fully sulfated chondroitin sulfate was prepared from chondroitin sulfate according to Maruyama et al (11). Chondroitin sulfate (108 mg) was converted into its tributylamine salt and dissolved in dry N,N dimethylformamide (1 mL). After addition of 159 mg of pyridine-sulfur trioxide complex the solution was heated for 1 h at 40 °C . The reaction was interrupted by addition of 2 ml of water and the product was precipitated at 4°C by addition of 35 mL of an ethanol solution saturated with sodium acetate. The product recovered by centrifugation was dissolved in water, dialysed and recovered by freeze-drying.
Supplementary Material
Table 2.
Chemical shifts observed for contaminant by two independent laboratories and comparison to literature data on oversulfated chondroitin sulfate
| Monosaccharide | Lab 1a |
Lab 2b |
1H (literature)c Fully O- sulfonated CS |
||
|---|---|---|---|---|---|
|
1H (observed) |
13C (observed) |
1H (observed) |
13C (observed) |
||
| GalNAc4,6S | |||||
| H1/C1 | 4.77 | 105.0 | 4.79 | 104.8 | 4.86 |
| H2/C2 | 4.06 | 54.0 | 4.10 | 54.0 | 4.10 |
| H3/C3 | 4.05 | 80.7 | 4.03 | 80.7 | 4.10 |
| H4/C4 | 4.98 | 78.0 | 5.00 | 78.0 | 5.02 |
| H5/C5 | 4.05 | 74.8 | 4.07 | 74.7 | 4.06 |
| H6,6’/C6 | 4.28 | 69.3 | 4.30 | 69.2 | 4.29 |
| N-Acetyl | 2.12d | 25.6 | 2.16e | 25.6 | 2.16 |
| G2,3S | |||||
| H1/C1 | 4.87 | 104.5 | 4.87 | 104.3 | 4.97 |
| H2/C2 | 4.47 | 80.0 | 4.49 | 80.0 | 4.53 |
| H3/C3 | 4.95 | 79.3 | 4.98 | 79.2 | 4.94 |
| H4/C4 | 4.46 | 80.9 | 4.51 | 80.8 | 4.55 |
| H5/C5 | 4.12 | 82.0 | 4.17 | 82.2 | 4.20 |
Chemical shifts are measured at 303K and referenced to external 2,2-dimethyl-2-silapentane-5-sulfonate sodium salt (DSS)
Chemical shifts are measured at 308K and referenced to external sodium trimethyl-sylil-propionate (TSP) (which resonates 0.12 ppm upfield of DSS (14))
Measured at 303 K and referenced to TSP (11).
Additional minor signals observed at 2.20 and 2.07 ppm
Additional minor signals observed at 2.23 and 2.11 ppm
Acknowledgements
The authors would like to thank Sucharita Roy for work on the chemical synthesis of oversulfated chondroitin sulfate standards and Scott Bailey for assistance in the analysis of selected samples by one- and two-dimensional NMR. We thank This work was supported in part by the National Institute of General Medical Sciences grant GM57073 (R.S).
References
- 1.Capila I, Linhardt RJ. Angew. Chem. Int. Ed. Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 2.Lepor NE. Rev. Cardiovasc. Med. 2007;8(Suppl. 3):S9–S17. [PubMed] [Google Scholar]
- 3.Fischer KG. Hemodial. Int. 2007;11:178–189. doi: 10.1111/j.1542-4758.2007.00166.x. [DOI] [PubMed] [Google Scholar]
- 4.Heparin investigation Update by Baxter. http://www.medicalnewstoday.com/articles/100760.php.
- 5.FDA Communication: Information on Heparin Sodium Injection. http://www.fda.gov/cder/drug/infopage/heparin/default.htm.
- 6.Myette JR, Shriver Z, Kiziltepe T, McLean MW, Venkataraman G, Sasisekharan R. Biochemistry. 2002;41:7424–7434. doi: 10.1021/bi012147o. [DOI] [PubMed] [Google Scholar]
- 7.Myette JR, Shriver Z, Claycamp C, McLean MW, Venkataraman G, Sasisekharan R. J. Biol. Chem. 2003;278:12157–12166. doi: 10.1074/jbc.M211420200. [DOI] [PubMed] [Google Scholar]
- 8.Kuberan B, Lech M, Zhang L, Wu LZ, Beeler DL, Rosenberg RD. J. Am. Chem. Soc. 2002;124:8707–8718. doi: 10.1021/ja0178867. [DOI] [PubMed] [Google Scholar]
- 9.Holme KR, Perlin AS. Carbohydr. Res. 1989;186:301–312. doi: 10.1016/0008-6215(89)84044-3. [DOI] [PubMed] [Google Scholar]
- 10.Guerrini M, Bisio A, Torri G. Semin. Thromb. Hemost. 2001;27:473–482. doi: 10.1055/s-2001-17958. [DOI] [PubMed] [Google Scholar]
- 11.Maruyama T, Toida T, Imanari T, Yu G, Linhardt RJ. Carbohydr. Res. 1998;306:35–43. doi: 10.1016/s0008-6215(97)10060-x. [DOI] [PubMed] [Google Scholar]
- 12.Linhardt RJ. In: Current Protocols in Molecular Biology. Varki A, editor. vol. 2. Boston: Wiley Interscience; 1992. pp. 17.13.17–17.13.32. [Google Scholar]
- 13.Kinoshita A, Yamada S, Haslam SM, Morris HR, Dell A, Sugahara K. J. Biol. Chem. 1997;272:19656–19665. doi: 10.1074/jbc.272.32.19656. [DOI] [PubMed] [Google Scholar]
- 14.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 15.Cicero O, Barbato G, Bazzo R. J. Magn. Reson. 2001;148:209–213. doi: 10.1006/jmre.2000.2234. [DOI] [PubMed] [Google Scholar]
- 16.Hadding U, Dierich M, König W, Limbert M, Schorlemmer HU, Bitter-Suermann D. Eur. J. Immunol. 1973;3:527–529. doi: 10.1002/eji.1830030817. [DOI] [PubMed] [Google Scholar]
- 17.Kang OH, Kim DK, Choi YA, Park HJ, Tae J, Kang CS, Choi SC, Nah YH, Lee HK, Lee YM. Int. J. Mol. Med. 2006;18:893–899. [PubMed] [Google Scholar]
- 18.Cifonelli JC. Carbohydr. Res. 1968;8:233–242. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.