SUMMARY
While diet-induced obesity has been exclusively attributed to increased caloric intake from fat, animals fed high fat diet (HFD) ad libitum (ad lib) eat frequently throughout day and night disrupting the normal feeding cycle. To test whether obesity and metabolic diseases result from HFD or disruption of metabolic cycles, we subjected mice to either ad lib or time restricted feeding (tRF) of a HFD for 8 h/day. Mice under tRF consume equivalent calories from HFD as those with ad lib access, yet are protected against obesity, hyperinsulinemia, hepatic steatosis, inflammation, and have improved motor coordination. The tRF regimen improved CREB, mTOR and AMPK pathway function and oscillations of the circadian clock and their target genes' expression. These changes in catabolic and anabolic pathways altered liver metabolome, improved nutrient utilization and energy expenditure. We demonstrate in mice that tRF regimen is a non-pharmacological strategy against obesity and associated diseases.
INTRODUCTION
In order to adapt to the daily cycles of nutrient availability, energy metabolism in animals has evolved to be cyclical. These metabolic cycles arise from cell autonomous circadian rhythms and the feeding-fasting cycle which drive genomic programs (Vollmers et al., 2009). At the molecular level, cell autonomous circadian rhythms are based on interlocked negative feedback circuits in which bHLH-PAS transcription factors BMAL1, CLOCK, NPAS2 and ROR proteins act as transcriptional activators and PER, CRY and REV-ERB function as inhibitors to produce ~24 h self-sustained rhythmic transcription of their own and target genes (reviewed in (Reddy and O'Neill, 2010)).
Feeding and fasting also drive daily rhythms in the activities of key regulators of nutrient homeostasis including AMPK, CREB and AKT (Vollmers et al., 2009). There is extensive coupling between circadian oscillator components and the feeding-fasting driven metabolic regulators. This coupling leads to coordinated oscillations at the transcript level and in the activities of a large number of neuroendocrine, signaling and metabolic pathways that temporally link discordant cellular processes.
Perturbation of circadian oscillator components leads to obesity and diabetes, illustrating the importance of this interconnection. Genetic mouse models carrying either tissue specific or whole body loss of function or hypomorphic alleles of circadian oscillator components develop impaired glucose tolerance and signs of metabolic disease. Conversely, disruption of the diurnal rhythms is commonly found in animal models of diabetes and obesity lacking specific metabolic regulators (reviewed in (Bass and Takahashi, 2010)). However, the circadian oscillator components and the metabolic regulators also control a large number of downstream effectors which do not exhibit any overt rhythms in expression (Cho et al., 2012; Feng et al., 2011; Rey et al., 2011). A number of mouse genetic models carrying whole body or tissue specific perturbation of circadian oscillators (Cho et al., 2012; Kornmann et al., 2007; Lamia et al., 2008; Marcheva et al., 2010; Preitner et al., 2002; Turek et al., 2005) or of key metabolic regulators (Andreelli et al., 2006; Herzig et al., 2003; Herzig et al., 2001; Shaw et al., 2005) exhibit no profound defect in the overt rhythms in activity or feeding under normal light:dark cycle, yet exhibit metabolic dysfunctions. Therefore, genetic models are inconclusive in addressing whether metabolic oscillations are necessary and sufficient for preventing metabolic diseases under nutritional challenge such as a high fat diet.
To test whether robust metabolic cycles can protect against nutritional challenges that predispose to obesity, we adapted a widely-used rodent model of diet-induced obesity. Mice fed high fat diet ad lib develop obesity, diabetes and metabolic syndrome. However, they also exhibit a dampened feeding- and circadian-rhythms (Kohsaka et al., 2007). Limiting access to high fat diet during day or night for up to 6 weeks shows some improvement in body weight regulation (Arble et al., 2009; Bray et al., 2010). However, since body weight and metabolic diseases are not always correlated (Ruderman et al., 1998; Wang et al., 2010), it is unclear whether time restricted feeding without changing caloric intake prevents metabolic diseases.
We subjected isogenic mice to either a diet of standard composition or one with high fat content under two food-access paradigms: ad lib or time restricted access for more than 100 days. Time restricted feeding (tRF) improved metabolic and physiologic rhythms, and protected the mice from the adverse effects of a high fat diet. The time restricted high fat fed mice showed significantly increased thermogenesis and improved rhythms in nutrient utilization, leading to reduced adiposity and liver steatosis, normal glucose tolerance, reduced serum cholesterol, increased bile acid production and improved motor function.
RESULTS
Time restricted feeding improves overt rhythms and attenuates body weight gain
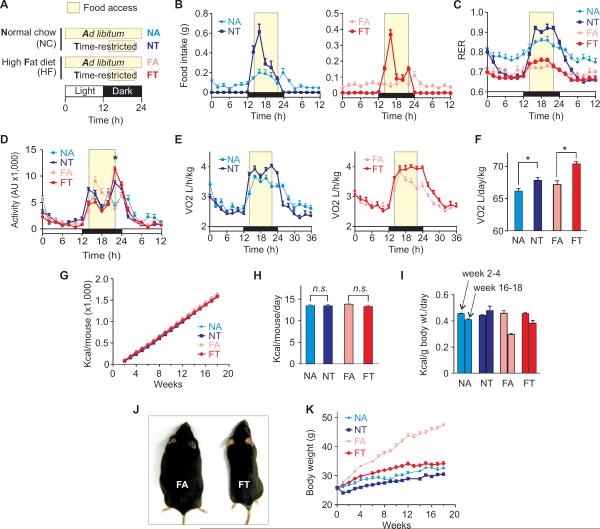
To test whether a distinct tRF regimen can prevent diet-induced obesity, we subjected 12 weeks-old male C57/BL6 mice to high fat diet (HF; 61% energy from fat) or normal chow (NC; 13% fat) under either ad lib or time restricted access to food during their natural nocturnal feeding time (Figure 1A). Mice fed normal chow under an ad lib regimen (NA) displayed diurnal rhythms in their food intake and whole body respiratory exchange ratio (RER) (Figures 1B and 1C). Food intake and RER exhibited a nocturnal increase, reflective of feeding and subsequent carbohydrate utilization, and declined during the day, consistent with lipid oxidation during fasting. Mice fed high fat diet under an ad lib regimen (FA) displayed dampened diurnal rhythms in food intake and RER. In contrast, mice fed normal chow or high fat diet under a tRF regimen (NT and FT) improved diurnal rhythms in their RER compared to their ad lib fed counterparts, with higher RER during feeding and reduced RER during fasting, indicative of increased glycolysis and fat oxidation respectively (Figure 1C). Although all four groups of mice showed overall comparable nocturnal activity, the mice on tRF regimen (FT and NT) showed increased activity and increased energy expenditure towards the end of the night (Figures 1D–1F). The average daily energy intake for individual mice in all four groups was equivalent throughout 18 weeks of the experiment (Figures 1G and 1H). When normalized for body weight, the mice on tRF paradigms, in fact, showed increased energy intake/unit body weight towards the end of the experiment (Figure 1I). Most remarkably, despite equivalent energy intake from the same nutrient source, FT mice were protected against excessive body weight gain that afflicted FA mice (Figures 1J, 1K and S1), suggesting that the temporal feeding pattern reprograms the molecular mechanisms of energy metabolism and body weight regulation.
Figure 1. Time of feeding shapes diurnal pattern of whole-body metabolism and influences body weight gain.
(A) Schematic outline of four feeding regimens used in this study. Time restricted fed mice were allowed access to food from ZT13 through ZT21. Food availability is indicated by light beige boxes. (B) Food ingested, (C) respiratory exchange ratio (CO2 exhaled/O2 inhaled), (D) average activity (+ SEM, n = 4 mice) and (E) whole body energy expenditure as measured by volume of O2 consumed in 2 h bins plotted against time. Since the high fat (HF) diet is energy rich (5.51 Kcal/g), the mice on HF diet consume equivalent amount of energy as the mice on normal chow (NC) (3.36 Kcal/g). (F) Area under the curve analyses of energy expenditure (from Figure 1E) (+ SEM, n = 4, *p < 0.05). Given the differences in body composition, metabolic activities in different organs and heterogeneity of substrate uses in different groups of mice, both food intake and energy expenditures were expressed relative to individual animal or unit body weight. (G) Cumulative average energy intake or (H) average daily energy intake (+ SEM, n = 24 over 17 weeks) by individual mice on NC or HF diet is not significantly different (n.s., p > 0.05) under ad lib or tRF paradigm. The near equivalent average energy intake from NC and HF diet is not different from several published studies. Voluntary energy intake is often independent of dietary fat content both in rodents and human twins (examples include but are not limited to (Bray et al., 2010; Fujisaka et al., 2011; Hosooka et al., 2008; Kennedy et al., 2007; Lin et al., 2000; Saltzman et al., 1997; Samuel et al., 2004)), and these studies also show irrespective of caloric intake, voluntary ingestion of high fat diet predisposes to obesity, diabetes and related diseases. The higher proportion of energy intake from fat is usually considered the cause for diet-induced obesity in these studies. (I) Average energy intake (+ SEM) normalized to unit body weight shows no difference at the beginning of the experiment. In the subsequent weeks with gradual increase in body weight, this value progressively declines. By the end of 16–18 weeks, mice on tRF consume more energy/unit body weight than the ad lib counterparts. (J) Representative FT mice were remarkably leaner than the FA mice. (K) Average body weight (+ SEM, n = 20 – 32 mice). Also see Figure S1.
Temporal feeding pattern shapes rhythms in CREB, mTOR and AMPK activities and in circadian oscillator
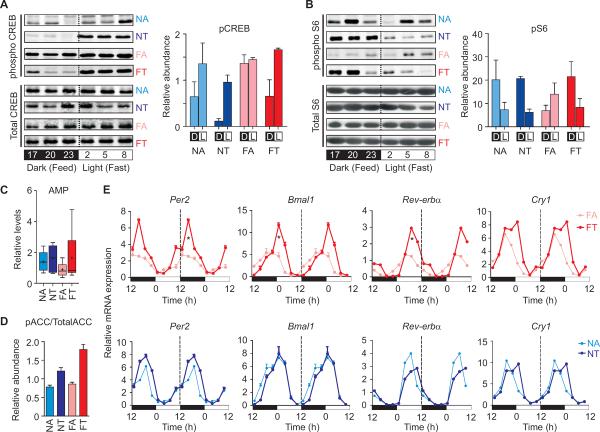
A high fat diet fed under an ad lib regimen perturbs metabolic regulators, including CREB, mTOR and AMPK, and contributes to metabolic diseases (Altarejos and Montminy, 2011; Inoki et al., 2011). Hepatic CREB phosphorylation is elevated during fasting, consistent with the role of active pCREB in gluconeogenesis (Herzig et al., 2001), while insulin and mTOR-stimulated pS6 levels (Um et al., 2006) are elevated during feeding. In NA mice, the diurnal rhythm in food intake (Figure 1B) induced hepatic CREB phosphorylation during daytime fasting and increased pS6 levels during nighttime feeding (Figures 2A and 2B). In FA mice, the perturbed circadian feeding pattern blunted pCREB and pS6 oscillations and led to constitutively elevated pCREB and reduced pS6 levels. In contrast, in the FT mice, the tRF imposed a diurnal rhythm in food intake, thereby restoring the daytime peak in pCREB and nighttime peak in pS6. The diurnal rhythm in food intake and associated daily fasting period under the FT paradigm increased the hepatic AMP levels relative to that seen in the FA paradigm (Figure 2C). Upon allosteric activation by AMP, AMPK, a master regulator of energy metabolism, phosphorylates and deactivates one of the rate limiting enzymes of fatty acid oxidation, acetyl CoA carboxylase (ACC) (Davies et al., 1990). Thus, increased AMP and phospho-ACC (pACC, relative to total ACC) (Figures 2C, 2D, S2A–S2C and Table S1) in the livers of FT mice reflected increased AMPK activity relative to the livers of FA mice.
Figure 2. Time restricted feeding improves diurnal rhythms in metabolic regulators and the circadian oscillator.
Representative immuno blots and densitometry quantification of the immuno blots (average (+ SEM, n = 3) during feeding and fasting in the tRF mice which coincide with night and day, respectively) for (A) transcriptionally active phospho-Ser133-CREB and (B) phospho-S6 in the mouse liver. (C) Whisker plot showing the AMP level in the liver of FT mice is significantly higher than that in FA mice. (D) Proportion of phospho-ACC (pACC) relative to total ACC from mouse liver (Also see Figure S2). (E) Double-plotted average (+ SEM, n = 4) mRNA levels of circadian oscillator components Per2, Bmal1,Rev-erbα, Cry1 and additional clock components (Figure S2D) in liver at different times of the day. Transcript levels were measured by qRT-PCR and normalized to Gapdh RNA levels. Broken line separates double plotted data. Also see Figure S2.
During fasting, AMPK phosphorylates a circadian repressor CRY (Lamia et al., 2009) and targets it for subsequent degradation, thus preventing it from repressing the CLOCK:BMAL1-target genes, including Rev-erbα, Per and Cry. During feeding, mTOR activity indirectly modulates Per expression(Giebultowicz and Kapahi, 2010). Under the FA regimen, perturbed total and/or diurnal change in active CREB, mTOR and AMPK reduced the mRNA levels and/or dampened the oscillations of circadian clock components (Per1, Per2, Cry1, Bmal1, Clock, Rorα Rev-erbα and an immediate output target Dbp) in the liver (Figures 2E and S2D). Under the FT regimen, the imposed feeding rhythms resulted in improved oscillations of circadian clock components in the liver, with increased peak to trough ratio of mRNA levels. NA mice show a robust feeding rhythm and clear oscillations in clock gene expression, which is moderately improved by feeding consolidation in the NT mice.
Hepatic glucose metabolism is improved under time restricted feeding
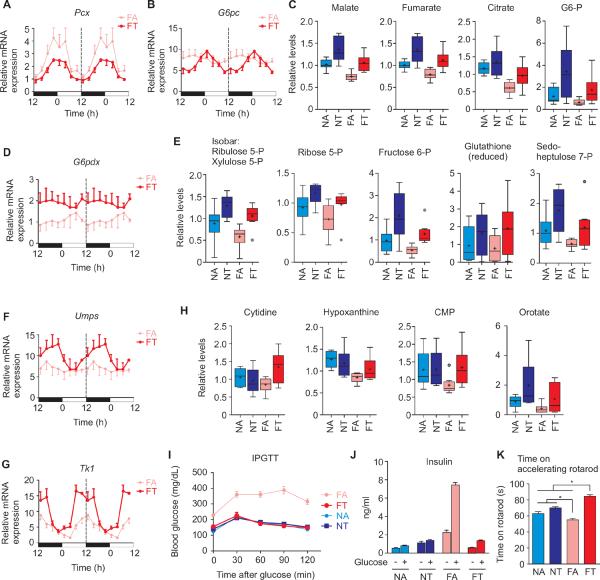
Coordination among circadian oscillator components and metabolic regulators helps maintain glucose homeostasis and anabolic metabolism in the liver. Rhythmic Cry expression and pCREB oscillation synergistically suppress gluconeogenic gene expression (Vollmers et al., 2009; Zhang et al., 2010). Accordingly, in the livers of FT mice, improved CRY expression and suppressed nocturnal pCREB(Figures 2A and 2E) reduced the mRNA levels of gluconeogenic CREB targetgenes Pyruvate carboxylase (Pcx) and Glucose-6-phosphatase (G6pc) (Figures 3A and 3B) which encode enzymes that mediate the committing step in gluconeogenesis. With reduced Pcx and G6pc gene expression, pyruvate is likely used in the TCA cycle, since the levels of several TCA cycle intermediates (e.g. malate, fumarate and citrate) are increased in the livers of FT mice (Figure 3C and Table S1). InIn parallel, reduced dephosphorylation of glucose-6-phosphate (G6-P) contributed to lowering blood glucose by decreasing the release of free glucose. The increased G6-P in the FT liver (Figure 3C) is likely diverted to the pentose phosphate cycle (PPC).
Figure 3. Time restricted feeding alters glucose metabolism and restores normal glucose tolerance.
qRT-PCR estimates of mRNA levels of gluconeogenic CREB targets (A) Pcx and (B) G6pc in the liver show reduced expression during feeding in the liver of FT mice. This is accompanied by (C) increased levels of G6-P and TCA cycle metabolites, malate, fumarate and citrate. (D) Elevated mRNA level of G6pdx in the livers of FT mice correlated with (E) a rise in pentose phosphate cycle (PPC) metabolites ribulose-5-P, ribose 5-P, fructose 6-P and sedoheptulose 7-P and higher levels of reduced glutathione. tRF regimen also improved oscillation of (F) Umps and (G) Tk1 mRNA in liver and resultant improvement in (H) nucleotide metabolites (also see Figures S3 and S4A) which are produced from PPC intermediates. (I) IPGTT shows normal glucose tolerance in FT mice. Average blood glucose levels (+ SEM, n = 6 mice) in overnight fasted and after glucose infusion are shown. (J) Levels of blood insulin after overnight fasting or 1 h after glucose infusion. (K) Time spent (average + SEM, n = 6, *p < 0.05) on an accelerating rotarod.
mTOR induces the expression of glucose-6-phosphate dehydrogenase (G6pdx) (Duvel et al., 2010), whose protein product is the rate limiting enzyme of the PPC and is activated by accumulation of its substrate G6-P. In turn, the PPC is a major source of NADPH which reduces glutathione. In the livers of mice under tRF, induced expression of G6pdx along with elevated G6-Pled to increased activity of the PPC as measured by higher levels of PPC intermediates and of reduced glutathione (Figures 3D, 3E and S3).
The pentose sugars of PPC and intermediates of TCA cycle are substrates for both de novo and salvage pathways of nucleotide biosynthesis. Several genes encoding enzymes of purine and pyrimidine biosynthesis and nucleotide salvage pathways are the direct targets of the circadian activator BMAL1 (Uridine monophosphate synthase (Umps) and Thymidine kinase1 (Tk1)) and/or exhibit rhythmic expression patterns (phosphoribosylpyrophosphate synthetase (Prps-1,-2), phosphoribosyl pyrophosphate amidotransferasePpat) and Cad) (Hughes et al., 2009; Nakahata et al., 2009; Ramsey et al., 2009; Rey et al., 2011; Vollmers et al., 2009). In the FT livers, increased Bmal1 mRNA led to a parallel increase in the mRNA levels of Umps and Tk1 as well as elevated levels of both purine and pyrimidine metabolites (Figures 3F–3H, S3 and S4A). Hence, these coordinated changes in gene expression and metabolites show that the tRF regimen temporally reprograms glucose metabolism away from gluconeogenesis towards glycolysis, reduced glutathione and anabolic pathways. Accordingly, FT mice did not display the hallmarks associated with glucose intolerance found in diet-induced obesity, instead showing glucose tolerance and insulin levels comparable to the control NA mice (Figures 3I and 3J). The overall improvement in metabolic state also paralleled improved motor coordination in the mice under tRF paradigms (Figure 3K).
Temporal feeding pattern determines lipid homeostasis
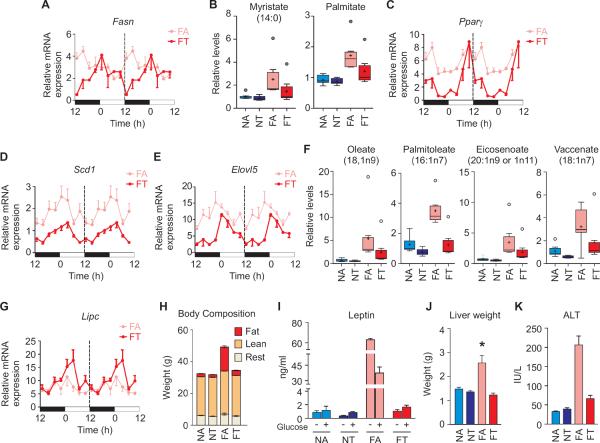
The circadian oscillator components interact with metabolic regulators to maintain lipid homeostasis. In the livers of FT mice, increased levels of the transcriptional repressor Rev-erbα (Figure 2E), led to reduced expression of its direct target and a key lipogenic gene, fatty acid synthase (Fasn) (Cho et al., 2012) (Figure 4A). Reduced Fasn mRNA levels (Figure 4A) and increased relative phospho-ACC levels (Figure 2D) are known to contribute to a decline in the level of several of long chain free fatty acids (Harwood, 2004) including myristate and palmitate (Figure 4B and Table S1). In parallel, increased Per2 expression in FT liver (Figure 2E) acts as an inhibitor of already-reduced levels of Pparγ (Figure 4C) (Grimaldi et al., 2010), further attenuating PPARγ-driven lipogenic gene expression. Repression of the PPARγ target gene Stearoyl coA desaturase1 (Scd1) (Figure 4D), which codes an enzyme mediating fatty acid desaturation, along with the reduced mRNA levels of the unsaturated fatty acid elongase Elovl5 (Figures 4E and S3) paralleled with >50% decline in several unsaturated fatty acids, including oleate (18:1n9), palmitoleate (16:1n7), vaccenate (18:1n7) and eicosenoate (20:1n9 or 11) (Figures 4F and S4B). Altogether, we observed reduced fatty acid synthesis, elongation and desaturation in the livers of FT mice compared to those of the FA mice.
Figure 4. Time restricted feeding alters fatty acid metabolism and prevents obesity and liver pathology.
(A) Reduced hepatic mRNA levels of REV-ERBα target gene Fasn in the livers of FT mice contributes to (B) the reduction in free fatty acids myristate and palmitate. Reduced mRNA levels of (C) Pparγ, (D) Scd1 and (E) Elovl5 in the liver of FT mice (also see Figure S3) accompanied reduced levels of several unsaturated fatty acids including (F) oleate, palmitoleate, eicosenoate and vaccinate (also see Figure S4B). Reduced malonyl-carnitine, increased BHBA (see also Figure S4B) and (G) increased Lipc mRNA are indicative of increased β-oxidation. (H) Body composition analyses by MRI illustrates tRF prevents excessive whole-body fat accumulation in mice fed HF diet. Average weights of fat, lean and remaining body mass are shown. (I) Increased levels of leptin after overnight fasting (−) and after glucose administration (+) indicative of increased adipose tissue in FA mice are absent in the FT mice. (J) tRF also prevents enlarged liver and (K) liver damage indicated by increased serum ALT.
Fatty acid synthesis inhibits mitochondrial β-oxidation. Malonyl-CoA, a product of ACC activity in the first step of fatty acid synthesis, allosterically inhibits mitochondrial carnitine palmitoyltransferase (CPT). CPT is essential for the transit of long chain fatty acids and acylcarnitine esters into the mitochondria for β-oxidation. Increased hepatic malonylcarnitine levels in FA mice, but not in FT mice (Figure S4B), are indicative of specific disruption of fatty acid oxidation caused by impaired entry of fatty acids into the mitochondria. Conversely, increased hepatic lipase (Lipc) expression (Figure 4G) along with 3-hydroxybutyrate (BHBA) (Figure S4B), one of the end products of β-oxidation, in FT mice relative to FA mice indicated that tRF enhanced lipolysis and β-oxidation, further contributing to reduction in liver free fatty acids.
Time restricted feeding prevents excessive body weight gain, hepatosteatosis and liver damage
Among mice fed normal chow, the tRF regimen resulted in moderately lower weight (30.5 ± 0.4 g. ± SEM, n = 24) than the ad lib regimen (32.6 ± 0.4 g) (Figure 1K). In contrast, tRF remarkably reduced obesity in mice fed a high fat diet. Mice with ad lib access to high fat diet (FA) attained an average body weight of 47.4 g ± 0.7 g by 18 weeks in the feeding regimen (Figure 1K) while FT mice weighed 28% less (34.2 ± 0.6 g) and were comparable to the NA group. Most of the extra body weight in the FA mice was due to increased adiposity (Figure 4H). FA mice showed 70% more fat deposits than those in the FT mice (FA 18.0 ± 1.03 g and FT 4.3 ± 1.23 g, p < 0.05). Consequently, hyperleptinemia associated with diet-induced obesity in FA mice was absent in FT mice (Figure 4I).
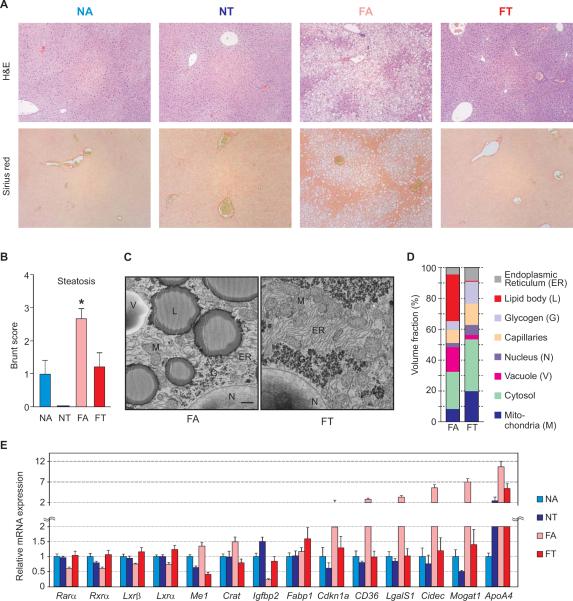
FT mice were also protected from the hepatomegaly and elevated serum alanine aminotransferase (ALT) levels that are associated with obesity-induced hepatic steatosis (Figures 4J and 4K). To further characterize the pathologic state and steatosis of the liver in these four groups, we used a Brunting scoring system under blinded conditions. Tissue samples from the FT mice had significantly less hepatic steatosis compared to those from the FA mice (Figures 5A and 5B). In addition, volume analyses of serial block-face scanning electron microscope images of the liver samples revealed that livers from the FT mice did not have the profound increase in intracellular fat deposits, reduced mitochondrial density and reduced endoplasmic reticulum that were characteristic of the liver samples from the FA mice (Figures 5C, 5D and Table S2).
Figure 5. Time restricted feeding prevents liver diseases.
(A) Representative histopathology (upper panel, H&E, Scale bars 200 μm) and Sirius red (lower panel, Scale bars 200 μm) of the liver. Steatohepatitis was scored by a histopathologist who was blinded to the source condition of each sample using a semi-quantitative method derived from Brunt et al. (Brunt et al., 2004) measuring the degree of steatosis (0–3), ballooning degeneration (0–2), lobular (0–3) and portal (0–2) inflammation and fibrosis (0–4). (B) Average Steatosis score (+ SEM, n = 4 mice) of liver sections. (C) Representative scanning electron microscope image of a liver from FA mice shows large lipid droplets and vacuoles that are reduced in FT liver. Scale bar = 1 μm. (D) Measurement of several volume fractions show FT regimen prevents the decline in density of mitochondria and ER under ad lib HF diet (FA). Measurements are from volume rendering obtained from serial block-face images of the liver. See also Table S2. (E) Gene expression signature of diet-induced obesity in the mouse liver is attenuated by tRF. Average expression (+ SEM) from 8 different time points (Figure S2A) are shown in bar-graphs. Significant differences between FA and FT (*p < 0.05) were found. Temporal expression profiles of these genes are shown in Figure S5.
The liver disease in FA mice was associated with markers of inflammation. The increased pool of free fatty acids in the livers of FA mice included proinflammatory long chain n-6 fatty acids dihomolinoleate (20:2n6) and arachidonate (20:4n6) (Figure S4C). Oxidation of arachidonate and linoleic acid in an oxidative environment marked by decreased glutathione in the livers of the FA mice further increased the levels of pro-inflammatory eicosanoids: 15-Hydroxyeicosatetraenoic acid (HETE), 5-HETE and 13-HODE. In contrast, the suppressed lipogenic program along with a glutathione-enriched cellular environment in the livers of the FT mice attenuated the levels of proinflammatory lipids (Figure S4C).
Additional gene expression signatures often associated with hepatic inflammation and fatty liver disease (Figures 5E and S5) were either reversed or attenuated under a tRF regimen. Changes in hepatic expression of nuclear hormone receptors Rarα, Rxrα, Lxrα and Lxrβ, a characteristic of diet-induced obesity (Kohsaka et al., 2007), are prevented upon tRF. Increased expression of several genes encoding key enzymes for lipid metabolism, including Crat (cytoplasmic carnitine acyltransferase), Me1 (Malic enzyme producing reducing NADPH for fat synthesis) and Mogat1 (Monoacylglycerol O-acyltransferase 1) in the FA mice return to normal chow fed levels in the FT group. Similarly, higher expression of lipid droplet-associated and lipolysis inhibitor gene Cidec (Puri et al., 2007), triglyceride storage associated protein CD36 (Koonen et al., 2007) and plasma triglyceride marker ApoA4 (Talmud et al., 2002) in the liver of FA mice are reduced in the FT mice. The expression of the cell cycle regulator Cdkn1a (p21) and Lgals1 (or Galectin-1), a marker of hepatocellular carcinoma and metastasis (Camby et al., 2006), are elevated in FA group, but are reduced under tRF. High fat ad lib feeding (FA) reduced expression of antidiabetic gene Igfbp2 (Hedbacker et al., 2010), while tRF elevated expression irrespective of the nutrient source. Additionally, Fabp1 (Fatty acid binding protein 1) which binds to and clears potentially toxic unesterified long chain fatty acids from the cytoplasm (Atshaves et al., 2010) exhibits a moderate increase in expression in the liver of FT mice.
Time restricted feeding improves bile acid production, adipose tissue homeostasis and alleviates inflammation
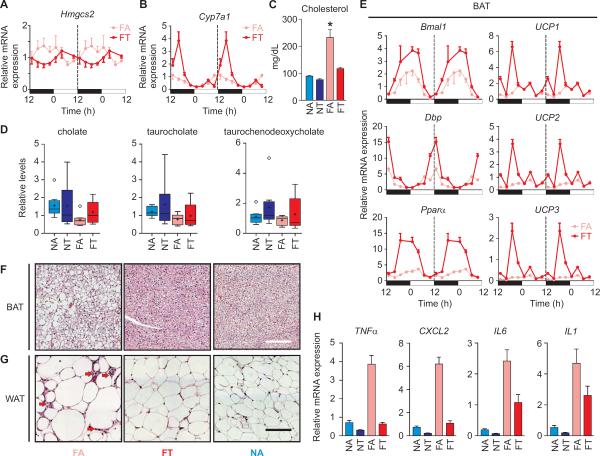
Hepatic fatty acid metabolism contributes to cholesterol and bile acid homeostasis. Both the diurnal rhythms in food intake and the clock component Rev-erbα are known to participate in the diurnal production of cholesterol and bile acids through transcriptional regulation of lipid homeostasis regulator Srebp1c and several rate-limiting enzymes including Hmgcs2 and Cyp7a1 (Cho et al., 2012; Le Martelot et al., 2009). Distinct feeding rhythms along with improved Rev-erbα rhythms in the livers of the FT regimen mice altered expression of Srebp1c, Hmgcs2 and Cyp7a1 (Figures 6A, 6B, S3 and S5). Increased peak levels of Cyp7a1 mRNA, which encodes the rate limiting step in bile acid production from cholesterol, elevated hepatic bile acids and contributed to a decrease in serum cholesterol levels in the FT mice (Figures 6C, 6D and S6 and Table S1). Although the liver is efficient in reabsorbing bile acids, increased postprandial hepatic bile acids spill over into circulation and raise energy expenditure in brown adipose tissue (BAT) by inducing expression of the uncoupling proteins (Watanabe et al., 2006). Indeed, in BAT derived from FT mice, we observed increased and rhythmic UCP expression (Figure 6E) which paralleled nighttime energy consumption as measured by whole body oxygen use (Figures 1E and 1F) as well as improved circadian oscillator function and increased PPARα expression (Figure 6E).
Figure 6. Time restricted feeding modulates bile acid metabolism, energy expenditure and inflammation.
tRF regimen (A) suppressed expression of Hmgcs2 and (B) increased expression of Cyp7a1 in the liver, which accompanied (C) a reduction in serum cholesterol and (D) increase in hepatic bile acids. Relative levels of representative bile acids are shown (also see Figure S6). (E) qRT-PCR measurements of relative mRNA levels of Bmal1, Dbp and Pparα show robust circadian oscillation in the BAT of FT mice (also see Figure S3 for NA and NT gene expression). Increased expression of UCP1-3 during the late night correlates with increased energy expenditure in FT mice (Figure 1). H&E stained sections of (F) BAT or (G) WAT show adipocyte hypertrophy in FA mice is prevented in the FT mice. Scale bar = 50 μm. Arrows indicate infiltrating cells that are most likely macrophages. Reduction in infiltrating macrophages also correlates with a reduction in (H) the mRNA levels of several pro-inflammatory cytokines in the WAT of FT mice. Average (+ SEM, n = 8, *p < 0.05) mRNA levels of pro-inflammatory cytokines TNFα, CXCL2, IL6 and IL1 in the WAT are reduced under tRF paradigm.
Elevated β-oxidation and reduced fatty acid synthesis in the liver coupled with increased BAT energy expenditure observed in the FT mice prevented the adipocyte hypertrophy common to BAT and white adipose tissue (WAT) derived from the FA mice (Figures 6F and 6G). Furthermore, inflammation marked by extensive infiltration of macrophages and expression of proinflammatory genes, including TNFα, IL6 and CXCL2 that are generally found in the WAT of the FA mice, were attenuated in the FT mice (Figure 6H). Even in mice fed normal diet, tRF reduced the expression of inflammatory cytokines in the WAT. In summary, the tRF paradigm affected multiple tissues and improved whole body energy homeostasis, and reduced inflammation.
DISCUSSION
Obesity is a major health challenge in many developed countries, reaching global pandemic proportions (Finucane et al., 2011). The prevalence of obesity has risen unabated for the last 4 decades currently affecting 35.5% of the population of United States and is projected to affect as much as 50% of the population in another 4 decades (King, 2011). The morbidity and mortality of its associated metabolic diseases, as well as its economic impact (Hammond and Levine, 2010), has made finding novel treatments for obesity an imperative of multiple national health agencies. Lifestyle modification is the first line intervention in the treatment of obesity due to ease of access, cost and superiority over pharmacotherapy or surgery (McTigue et al., 2003). The focus of currently recommended modification has been altering nutrition. In a murine model, here we introduce a lifestyle intervention thatcan prevent obesity as well as its associated metabolic disorders by preserving natural feeding rhythms without altering nutrition intake.
Although it has long been assumed that the cause of adiposity associated with mouse models of diet-induced obesity is nutritional, there is an emerging suggestion that the temporal spreading of calorie intake could be contributing as well. Under ad lib access to food, a high fat diet blunts the diurnal feeding rhythms more severely than a standard diet (Figure 1B). Therefore, mice fed high fat diet ad lib have a short fasting period and a long feeding window. This feeding pattern perturbs metabolic pathways entrained by both circadian- and feeding- rhythms. The temporal disruption in cellular metabolic processes, in combination with the nutrient quality, predisposes the organism to obesity and metabolic diseases.
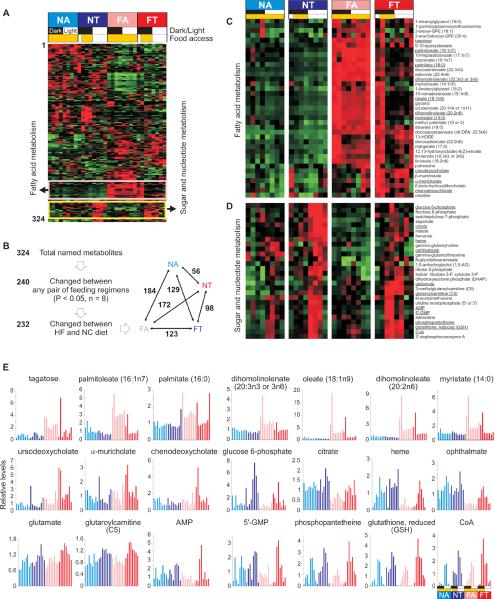
The tRF regimen entrained the circadian clock and metabolic regulators to fixed feeding times and prevented high fat diet-induced disruption of the normal cellular metabolic program. Compared to their normal chow counterparts, the beneficial effect of tRF on the gene expression (Figures 2–6, S3 and S5) and metabolic signatures is more pronounced in mice fed a high fat diet. Liver metabolome analyses detected 324 named metabolites common to all four groups of mice, of which 240 (74%) changed between at least one pair of feeding regimen (Figures 7A, 7B, Table S1 and http://metabolites.salk.edu), thus highlighting both nutrient quality and daily feeding pattern are important determinants of the liver metabolic homeostasis. Among mice fed a standard diet, time of food access changed the overall levels of 56 metabolites, while 123 metabolites changed between mice under FA and FT regimen. Both the average levels and diurnal oscillations of these metabolites are defined by the time of feeding (Figures 7C–7E and S7).
Figure 7. Time of feeding exerts a larger effect on liver metabolites in mice on a high fat diet.
(A) Heat-map rendering of normalized levels of 324 liver metabolites at eight different time points in the liver of NA, NT, FA and FT groups of mice. The metabolites were clustered by hierarchical clustering. (B) Summary of metabolite changes in the liver highlights the larger effect of temporal feeding pattern when animals were fed high fat diet. For statistical analyses, all eight time-points for each feeding regimen were treated as replicates. Number of metabolites that changed between any two of the six different contrasts is shown in the bottom panel. (C) Heatmap rendering of a subset of metabolites of a cluster enriched for fatty acids and (D) another cluster enriched in intermediates of energy- and anabolic- metabolism. Tissues were harvested at ZT14, 17, 20, 23, 2, 5, 8 and 11. The time of food access is indicated in yellow boxes. Red = high, Green = low. Steady state levels of several of the metabolites at 8 different time points representing one full day from (C) and (D) marked with underlined text are shown in (E). Normalized values presented in Table S1 are plotted against time.
Implicit in our findings is that the control of energy metabolism is a finely-tuned process that involves an intricate network of signaling pathways and transcriptional effectors, including nutrient sensing mechanisms and the circadian system. tRF acted on these interwoven networks and moved their state towards that of a normal feeding rhythm. In addition to nutrient metabolism, pathways regulating steady state levels of signaling molecules and co-factors in liver such as bile acids, sterols, riboflavin, heme and coenzyme-A (Figures 7E, S7 and Table S1) are also affected by time feeding. molecules affect the of These signaling and cofactors likely functions of multiple other organs, partly explaining how feeding rhythms can indirectly have systemic consequences. The beneficial effects of tRF was also evident in mice fed normal chow, thus implying tRF might be improve metabolism under diverse nutritional challenges.
Although a number of clinical studies have shown perturbation of light:dark or sleep:wake cycle (e.g. shiftwork) has adverse metabolic consequences in humans, there is very little information on the perturbation of eating rhythms in the participants of these studies. Hence, the contribution of frequent feeding and reduced daily fasting period to obesity, type 2 diabetes and other adult-onset metabolic diseases is unclear. Current public health surveys on human nutrition emphasize on the quality and quantity of nutrition with no evidence-based method in place to monitor temporal pattern of food intake. More studies are necessary to define the relationship between temporal eating and obesity in humans. The results presented in our study with mice suggests that time restricted feeding could be a novel, non-pharmacological intervention in humans that could prevent obesity and its associated metabolic disorders.
EXPERIMENTAL PROCEDURES
Animals
All animal experiments were carried out in accordance with the guidelines of Institutional Animal Care and Use Committee of the Salk Institute. The feeding regimen experiments were repeated on four independent batches of mice and representative datasets are presented here. Whole body indirect calorimetry was carried out on a subset of mice at 4, 8 or 12 weeks and all other measurements were carried out on a subset of mice at least 12 weeks after the initiation of the feeding regimens.
Feeding schedule and diets
Eight weeks-old male C57BL/6J mice from Jackson Laboratory were group housed (3–5 mice/cage) under a 12h light:12h dark schedule for 4 weeks to adapt to the housing condition. They were fed normal chow (LabDiet 5001; 29% protein, 13% fat, 58% carbohydrates) or high fat diet (LabDiet 58Y1; 18% protein, 61% fat, 21% carbohydrates) either with unrestricted (ad lib) or temporally restricted access to food (tRF) (See Figure 1A). Under tRF, mice were allowed access to food between ZT13 (1 h after lights off) and ZT21 (3 h before lights on). Food access was regulated by transferring mice daily between cages with food and water and cages with water only. To control for mouse handling, ad lib fed mice were also transferred between feeding cages at the same time. Weekly food intake was measured by monitoring the weight of the remaining food.
Metabolic cages
Whole body metabolic states were tested by indirect calorimetry in a CLAMS system (Columbus Instruments) for 2 days after 4 days of habituation following manufacturer's instructions. Light and feeding conditions were kept the same as in the home cages.
Glucose tolerance
Mice were fasted for 16 h and fasted glucose was measured using a Glucometer (One Touch Ultra™) by tail-bleeds. Subsequently, mice were intraperitoneally injected with 1 g glucose/kg of body weight, and blood glucose was measured in intervals of 30 min for 2 h.
Insulin and Leptin ELISAs
In a second group, overnight fasted mice were intraperitoneally injected with 1 g of glucose/kg of bodyweight and retro-orbital blood was collected after 1 h. Insulin (Crystal Chem #90080) and Leptin (Millipore) ELISAs were measured in the blood of fasted and glucose-injected mice following manufacturer's instructions.
ALT and Cholesterol
Mice were fasted for 16 h starting from ZT21, and blood was collected retro-orbitally. ALT and Cholesterol were assayed by IDEXX laboratories.
Rotarod
Mice were placed on accelerating (10 – 70 rpm) rotarods for up to 180 sec. Mice were tested on subsequent days. On the first day, mice were given 5 trial runs for habituation. Data was collected from 3 runs on the second day after 2 trials runs.
Histology and Electron Microscopy
Sections (6 μm) of formaldehyde fixed liver, WAT and BAT were stained following standard H&E. Liver sections were also stained using Sirius Red method. See Supplemental Information for the details.
Body composition
Body fat and lean mass of live mice was assessed using a mouse MRI (Echo Medical Systems) following manufacturer's protocol.
Transcript protein and metabolite analyses
Three to four mice in each feeding group were sacrificed every 3 h over 24 h period, and individual liver, WAT and BAT were flash frozen. Aliquots of frozen tissues were used for immuno blot and qRT-PCR analyses carried out as described earlier (Vollmers et al., 2009). Frozen liver aliquots were analyzed for detection and relative quantification of metabolites by Metabolon following published methods (Evans et al., 2009).
Statistical tests
To account for diurnal variations in liver metabolites, samples collected at 8 different time points throughout the day were analyzed and treated as replicates in ANOVA tests. For a given metabolite, the measured levels across all samples were median normalized to 1 and missing data point (if any) were imputed with the minimum values. Results were plotted in whisker plots where the box denotes the middle 80 percentile with mean (+) and median (−). Error bars denote maximum and minimum of distribution with extreme data points marked (o). The relative levels of all detected metabolites, changes in different feeding groups and the associated statistical test results are presented in Table S1. Metabolic cage data, body weight, food consumed and qRT-PCR results were analyzed by Student's t test (one tailed or two tailed based on sample types). Only in cases where the average values appear close, significant differences at p < 0.05 are denoted with “*”. Average (+ SEM.) values are shown in figures.
Supplementary Material
RESEARCH HIGHLIGHTS
-
-
Time restricted feeding (tRF. 8h/day) improves clock and nutrient sensor functions
-
-
tRF prevents obesity, diabetes and liver diseases in mice on a high fat diet
-
-
Nutrient-type and time of feeding determine liver metabolome and nutrient homeostasis
-
-
tRF raises bile acid production, energy expenditure and reduces inflammation
ACKNOWLEDGMENTS
We thank Drs. Marc Montminy and Ron Evans for helpful comments and advice on the work and for sharing their research equipment. We also thank Hiep Le, Sheena Keding, Chrissta Maracle and Ishika Arora for technical support. The IMOD stereology plug-in was developed by Andrew Noske and we thank him for providing instruction and guidance. This work was partially supported by the Pew Scholars Program in Biomedical Sciences, NIH grant DK091618, Sanofi Discovery Innovation Grant and Anderson Foundation support to SP, JSPS fellowship to MH, Blasker Science & Technology Grant Award to CV, NIH T32 DK07202 training grant to AZ and NCRR grant 5P41RR004050 to ME.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors state no commercial conflict of interest.
REFERENCES
- Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreelli F, Foretz M, Knauf C, Cani PD, Perrin C, Iglesias MA, Pillot B, Bado A, Tronche F, Mithieux G, Vaulont S, Burcelin R, Viollet B. Liver adenosine monophosphate-activated kinase-alpha2 catalytic subunit is a key target for the control of hepatic glucose production by adiponectin and leptin but not insulin. Endocrinology. 2006;147:2432–2441. doi: 10.1210/en.2005-0898. [DOI] [PubMed] [Google Scholar]
- Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 2009;17:2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atshaves BP, Martin GG, Hostetler HA, McIntosh AL, Kier AB, Schroeder F. Liver fatty acid-binding protein and obesity. The Journal of nutritional biochemistry. 2010;21:1015–1032. doi: 10.1016/j.jnutbio.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray MS, Tsai JY, Villegas-Montoya C, Boland BB, Blasier Z, Egbejimi O, Kueht M, Young ME. Time-of-day-dependent dietary fat consumption influences multiple syndrome parameters in mice. Int J Obes (Lond) 2010;34:1589–1598. doi: 10.1038/ijo.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunt EM, Neuschwander-Tetri BA, Oliver D, Wehmeier KR, Bacon BR. Nonalcoholic steatohepatitis: histologic features and clinical correlations with 30 blinded biopsy specimens. Human pathology. 2004;35:1070–1082. doi: 10.1016/j.humpath.2004.04.017. [DOI] [PubMed] [Google Scholar]
- Camby I, Le Mercier M, Lefranc F, Kiss R. Galectin-1: a small protein with major functions. Glycobiology. 2006;16:137R–157R. doi: 10.1093/glycob/cwl025. [DOI] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, Ditacchio L, Atkins AR, Glass CK, Liddle C, Auwerx J, Downes M, Panda S, Evans RM. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012 doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SP, Sim AT, Hardie DG. Location and function of three sites phosphorylated on rat acetyl-CoA carboxylase by the AMP-activated protein kinase. Eur J Biochem. 1990;187:183–190. doi: 10.1111/j.1432-1033.1990.tb15293.x. [DOI] [PubMed] [Google Scholar]
- Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Analytical chemistry. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane MM, Stevens GA, Cowan MJ, Danaei G, Lin JK, Paciorek CJ, Singh GM, Gutierrez HR, Lu Y, Bahalim AN, Farzadfar F, Riley LM, Ezzati M. National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet. 2011;377:557–567. doi: 10.1016/S0140-6736(10)62037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaka S, Usui I, Kanatani Y, Ikutani M, Takasaki I, Tsuneyama K, Tabuchi Y, Bukhari A, Yamazaki Y, Suzuki H, Senda S, Aminuddin A, Nagai Y, Takatsu K, Kobayashi M, Tobe K. Telmisartan improves insulin resistance and modulates adipose tissue macrophage polarization in high-fat-fed mice. Endocrinology. 2011;152:1789–1799. doi: 10.1210/en.2010-1312. [DOI] [PubMed] [Google Scholar]
- Giebultowicz J, Kapahi P. Circadian clocks and metabolism: the nutrient-sensing AKT and TOR pathways make the link. Curr Biol. 2010;20:R608–609. doi: 10.1016/j.cub.2010.05.052. [DOI] [PubMed] [Google Scholar]
- Grimaldi B, Bellet MM, Katada S, Astarita G, Hirayama J, Amin RH, Granneman JG, Piomelli D, Leff T, Sassone-Corsi P. PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell metabolism. 2010;12:509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond RA, Levine R. The economic impact of obesity in the United States. Diabetes, metabolic syndrome and obesity : targets and therapy. 2010;3:285–295. doi: 10.2147/DMSOTT.S7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood HJ., Jr. Acetyl-CoA carboxylase inhibition for the treatment of metabolic syndrome. Curr Opin Investig Drugs. 2004;5:283–289. [PubMed] [Google Scholar]
- Hedbacker K, Birsoy K, Wysocki RW, Asilmaz E, Ahima RS, Farooqi IS, Friedman JM. Antidiabetic effects of IGFBP2, a leptin-regulated gene. Cell metabolism. 2010;11:11–22. doi: 10.1016/j.cmet.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Herzig S, Hedrick S, Morantte I, Koo SH, Galimi F, Montminy M. CREB controls hepatic lipid metabolism through nuclear hormone receptor PPAR-gamma. Nature. 2003;426:190–193. doi: 10.1038/nature02110. [DOI] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Hosooka T, Noguchi T, Kotani K, Nakamura T, Sakaue H, Inoue H, Ogawa W, Tobimatsu K, Takazawa K, Sakai M, Matsuki Y, Hiramatsu R, Yasuda T, Lazar MA, Yamanashi Y, Kasuga M. Dok1 mediates high-fat diet-induced adipocyte hypertrophy and obesity through modulation of PPAR-gamma phosphorylation. Nature medicine. 2008;14:188–193. doi: 10.1038/nm1706. [DOI] [PubMed] [Google Scholar]
- Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Kim J, Guan KL. AMPK and mTOR in Cellular Energy Homeostasis and Drug Targets. Annu Rev Pharmacol Toxicol. 2011 doi: 10.1146/annurev-pharmtox-010611-134537. [DOI] [PubMed] [Google Scholar]
- Kennedy AR, Pissios P, Otu H, Xue B, Asakura K, Furukawa N, Marino FE, Liu FF, Kahn BB, Libermann TA, Maratos-Flier E. A high-fat, ketogenic diet induces a unique metabolic state in mice. Am J Physiol Endocrinol Metab. 2007;292:E1724–1739. doi: 10.1152/ajpendo.00717.2006. [DOI] [PubMed] [Google Scholar]
- King D. The future challenge of obesity. Lancet. 2011;378:743–744. doi: 10.1016/S0140-6736(11)61261-0. [DOI] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell metabolism. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Koonen DP, Jacobs RL, Febbraio M, Young ME, Soltys CL, Ong H, Vance DE, Dyck JR. Increased hepatic CD36 expression contributes to dyslipidemia associated with diet-induced obesity. Diabetes. 2007;56:2863–2871. doi: 10.2337/db07-0907. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, Thompson CB, Evans RM. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Sasso GL, Moschetta A, Schibler U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Thomas TC, Storlien LH, Huang XF. Development of high fat diet-induced obesity and leptin resistance in C57Bl/6J mice. Int J Obes Relat Metab Disord. 2000;24:639–646. doi: 10.1038/sj.ijo.0801209. [DOI] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, Lopez JP, Philipson LH, Bradfield CA, Crosby SD, JeBailey L, Wang X, Takahashi JS, Bass J. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTigue KM, Harris R, Hemphill B, Lux L, Sutton S, Bunton AJ, Lohr KN. Screening and interventions for obesity in adults: summary of the evidence for the U.S. Preventive Services Task Force. Annals of internal medicine. 2003;139:933–949. doi: 10.7326/0003-4819-139-11-200312020-00013. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Puri V, Konda S, Ranjit S, Aouadi M, Chawla A, Chouinard M, Chakladar A, Czech MP. Fat-specific protein 27, a novel lipid droplet protein that enhances triglyceride storage. J Biol Chem. 2007;282:34213–34218. doi: 10.1074/jbc.M707404200. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, O'Neill JS. Healthy clocks, healthy body, healthy mind. Trends Cell Biol. 2010;20:36–44. doi: 10.1016/j.tcb.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderman N, Chisholm D, Pi-Sunyer X, Schneider S. The metabolically obese, normal-weight individual revisited. Diabetes. 1998;47:699–713. doi: 10.2337/diabetes.47.5.699. [DOI] [PubMed] [Google Scholar]
- Saltzman E, Dallal GE, Roberts SB. Effect of high-fat and low-fat diets on voluntary energy intake and substrate oxidation: studies in identical twins consuming diets matched for energy density, fiber, and palatability. The American journal of clinical nutrition. 1997;66:1332–1339. doi: 10.1093/ajcn/66.6.1332. [DOI] [PubMed] [Google Scholar]
- Samuel VT, Liu ZX, Qu X, Elder BD, Bilz S, Befroy D, Romanelli AJ, Shulman GI. Mechanism of hepatic insulin resistance in non-alcoholic fatty liver disease. J Biol Chem. 2004;279:32345–32353. doi: 10.1074/jbc.M313478200. [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Lamia KA, Vasquez D, Koo SH, Bardeesy N, Depinho RA, Montminy M, Cantley LC. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmud PJ, Hawe E, Martin S, Olivier M, Miller GJ, Rubin EM, Pennacchio LA, Humphries SE. Relative contribution of variation within the APOC3/A4/A5 gene cluster in determining plasma triglycerides. Hum Mol Genet. 2002;11:3039–3046. doi: 10.1093/hmg/11.24.3039. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um SH, D'Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell metabolism. 2006;3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Liu H, Blanton WP, Belkina A, Lebrasseur NK, Denis GV. Brd2 disruption in mice causes severe obesity without Type 2 diabetes. Biochem J. 2010;425:71–83. doi: 10.1042/BJ20090928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, Schoonjans K, Bianco AC, Auwerx J. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, Brenner DA, Montminy M, Kay SA. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nature medicine. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.