Summary
Type I casein kinases are highly conserved among Eukaryotes. Of the two Aspergillus nidulans casein kinases I, CkiA is related to the δ/ε mammalian kinases and to Saccharomyces cerevisiæ Hrr25p. CkiA is essential. Three recessive ckiA mutations leading to single residue substitutions, and downregulation using a repressible promoter, result in partial loss-of-function, which leads to a pleiotropic defect in amino acid utilization and resistance to toxic amino acid analogues. These phenotypes correlate with miss-routing of the YAT plasma membrane transporters AgtA (glutamate) and PrnB (proline) to the vacuole under conditions that, in the wild type, result in their delivery to the plasma membrane. Miss-routing to the vacuole and subsequent transporter degradation results in a major deficiency in the uptake of the corresponding amino acids that underlies the inability of the mutant strains to catabolize them. Our findings may have important implications for understanding how CkiA, Hrr25p and other fungal orthologues regulate the directionality of transport at the ER-Golgi interface.
Introduction
In fungi, the uptake of most amino acids is mediated by transmembrane proteins belonging to the specific YAT (TC 2.A.3.10, yeast amino acid transporters) family. These transporters share a predicted topology comprising 12 transmembrane domains, which has been experimentally established for the S. cerevisiæ Gap1 (general amino acid permease, Gilstring and Ljungdahl, 2000). Members of the YAT family show, notwithstanding diverse substrate specificities, a high degree of sequence identity (André, 1995; Jack et al., 2000; Van Belle and André, 2001). In S. cerevisiæ, 18 YAT transporters have been characterized with varying degrees of detail (Jack et al., 2000). The genome of Aspergillus nidulans includes 19 predicted genes encoding transporters of this family (C. Scazzocchio, unpublished) of which two, the proline specific transporter, prnB (ANID_01732.3) and the dicarboxylic amino acid transporter, agtA (ANID_06118.3), have been functionally characterized (Sophianopoulou and Scazzocchio, 1989; Tazebay et al., 1995; Tavoularis et al., 2003; Apostolaki et al., 2009; Vangelatos et al., 2009).
The high sequence similarity extant in this family of transporters, within and across wide taxonomical boundaries, suggests that conserved structural features/mechanisms might be involved in their traffic through the endoplasmic reticulum and the Golgi, their insertion into the plasma membrane, their endocytic internalization and their delivery to the vacuole for degradation. For example, S. cerevisiæ Shr3p is a specific ER chaperone necessary for the efficient insertion of YAT transporters into the membrane (Ljungdahl et al., 1992; Kota et al., 2007), with orthologues characterized in A. nidulans, Schizosaccharomyces pombe and Candida albicans (Martínez and Ljungdahl, 2000; 2004; Erpapazoglou et al., 2006; Apostolaki et al., 2009).
The Pezizomycotina (filamentous ascomycetes) have diverged from the Saccharomycotina at least 600 million years ago (Heckman et al., 2001; Padovan et al., 2005). No systematic study of the mechanisms mediating YAT transcriptional regulation or YAT delivery to the plasma membrane has been reported for any member of this sub-phylum.
There are pointers to some differences between representatives of the Pezizomycotina and S. cerevisiæ. The endocytic internalization and degradation in the vacuole of many transporters, including YATs, is elicited by ammonium in both S. cerevisiæ (Risinger and Kaiser, 2008 and references therein) and A. nidulans, but the mechanism underlying this phenomenon does not seem to be identical (Apostolaki et al., 2009 and references therein; Gournas et al., 2010). The A. nidulans ER chaperone ShrA has a more restricted role than its yeast Shr3p counterpart (Erpapazoglou et al., 2006; Apostolaki et al., 2009). Additionally, the absence of Shr3 results in accumulation of target transporters in the ER (Ljungdahl et al., 1992), while the absence of ShrA results in their vacuolar localization (Apostolaki et al., 2009).
There are reports in early A. nidulans literature of several loci where mutations result specifically and pleiotropically in defective amino acid uptake (Kinghorn and Pateman, 1975; Sharma, 1984), suggesting that mechanisms of regulation and/or plasma membrane delivery common and specific to a number or all YAT proteins are operating and can be studied in this organism. In this study, we report that partial loss-of-function of an essential A. nidulans gene encoding a δ/ε type I casein kinase results in markedly reduced activity of most YAT proteins. To our knowledge, this is the first report implicating a casein kinase of the δ/ε isotype in the trafficking of a specific family of transporters, a finding whose significance is enhanced by recent results concerning the role of the S. cerevisiæ orthologue, Hrr25 in the ER-Golgi traffic (Lord et al., 2011).
Results
Three allelic mutations result in defective utilization of amino acids as nitrogen sources
This report focuses on three recessive mutations (designated, ckiA2, ckiA102 and ckiA1919) which result specifically in impairment of growth on several amino acids as sole nitrogen sources and in resistance to toxic amino acid analogues. The three mutants were isolated by three different screening procedures, respectively the inability to utilize δ-aminovaleric acid as a nitrogen source of puA2 strain in the presence of limiting putrescine (Herman and Clutterbuck, 1966), resistance to the toxicity of proline in a strain sensitive to semialdehyde toxicity (Arst et al., 1981) and resistance to toxic amino acid analogues. Details are provided in Supplementary Experimental procedures in the Supporting information. These mutations do not affect the utilization of nitrogen sources other than amino acids, such as purines (which A. nidulans catabolizes to ammonium), allantoin, urea, ammonium, nitrate, nitrite or acetamide. The three mutations were mapped to chromosome III, do not to complement with, and map at less than 0.2 cM, from each other (Experimental procedures). In conventional crosses ckiA102 was tentatively located at 8 cM from alX4 and 10 cM from argB2, the probable order being alX–ckiA–argB.
Interestingly, in the most recent screen (resistance to toxic amino acid analogues), only one in a total of 22 mutations defective in amino acid uptake isolated was a ckiA allele (ckiA1919). fbaA1013, one of the 21 mutations not allelic to ckiA (Apostolaki, 2003), is a down-promoter mutation in a gene encoding a fructose 1,6-biphosphate aldolase (Roumelioti et al., 2010). We arbitrarily selected five mutants (including fbaA1013), not allelic to ckiA102, and crossed each of them to the remaining twenty. Only two mutations were found to be possibly allelic to each other and none to fbaA1013. Thus, the above three ckiA mutations define only one of several loci where mutations leading to a pleiotropic defect in amino acid uptake map, demonstrating that this class is not saturated.
While the cognate phenotypes are not identical, the three ckiA mutations result in strong resistance to d-serine and p-fluorophenyalanine, and in impaired growth on all canonical amino acids which can be utilized as nitrogen sources by A. nidulans, excepting glutamine and asparagine (Fig. 1), where growth is not affected. In crosses involving the ckiA mutations we confirmed the co-segregation of at least four phenotypes, resistance to d-serine, and to dl-p-fluorophenylalanine, impaired growth on glutamate and glycine, and for ckiA102, also impaired growth on proline, which rules out the possibility that the pleiotropic phenotypes scored result from more than one mutation. For all nitrogen sources tested a hierarchical order (of growth on amino acids as nitrogen sources) is observed in which ckiA102 ≤ ckiA2 < ckiA1919 < ckiA+. The phenotypes of both ckiA102 and ckiA2 do not differ between 37°C and 25°C, while that of ckiA1919 is less marked at 25°C than at 37°C (tested for utilization of glutamate and glycine and resistance to d-serine; not shown). The three alleles were also tested for their effect on the utilization of selected amino acids (glutamate, alanine, proline, phenylalanine, ornithine and threonine) as carbon sources in the presence of ammonium chloride as a nitrogen source. Ammonium chloride does not interfere with the utilization of amino acids as carbon sources. All three mutations affect amino acid utilization as carbon sources in the same hierarchical order as that described above (not shown). Finally, the mutation having the strongest phenotype, ckiA102 also results in a somewhat restricted, less dense growth on all media tested (including complete medium and minimal medium with ammonium as nitrogen source) while ckiA1919 has a subtle morphological phenotype most visible on complete medium, and ckiA2 does not have an obvious morphological phenotype.
Fig 1.

Growth phenotypes of mutant strains. The relevant genotype of each strain is shown to the left of the panel. Complete genotypes are shown in Table 1. Three wild type (wt) controls are included: CS2498, LH59 and CS1957. The mutant strains are from top to bottom: CAM45, VIE212, VIE181, CS1912, VIE180, CS1902, VIE174, VIE111, VIE114, VIE108, VIE113. CM: complete medium. Supplemented MM with glucose as carbon source was used with each of the following nitrogen sources: ammonium (NH4+), urea (Ur), asparagine (Asn), aspartate (Asp, Asp5), glutamate (Glu, Glu5), glycine (Gly). Asp5 and Glu5 contain the respective amino acids at 5 mM rather than at 10 mM. d-Ser and FPA indicate d-serine and dl-p-fluorophenylalanine on urea as nitrogen source respectively. Acet stands for acetate as a carbon source. Putative ckiA fbaA1013 double mutants were checked by outcrossing to a wild type, with recovery of both parental classes and for the strains shown in this figure, also by confirming the presence of the relevant ckiA mutation after PCR amplification of the ckiA ORF (see text for the nature of each mutation).
All three ckiA mutations impair the utilization of β-alanine and δ-aminovaleric acid, while not affecting the utilization of γ-aminobutyric acid (GABA). Most strongly affected is the utilization of aromatic and dicarboxylic amino acids and glycine. Indeed, the two more extreme ckiA mutations affect growth on 10 mM aspartate and glutamate more strongly than a deletion of the specific agtA transporter gene (Apostolaki et al., 2009), which implies that both dicarboxylic amino acids can be taken up by at least a second, lower-affinity transporter.
Null mutations in prnB, encoding the major proline permease, allow residual growth on proline as sole nitrogen source and strongly reduce proline uptake (Tazebay et al., 1995; Tavoularis et al., 2003; see Figs 1 and 2 and below), which has been interpreted as proline being taken up also by at least one other transporter. The utilization of proline is strongly affected by ckiA102 and ckiA2 and very mildly impaired by ckiA1919, the ckiA- phenotype being partially additive with that of the null prnB allele (Fig. 1; additivity not tested for ckiA2). This implies that ckiA102 and ckiA2 mutants showing markedly impaired utilization of proline are defective in the activity of both PrnB and the alternative proline uptake system.
Fig 2.
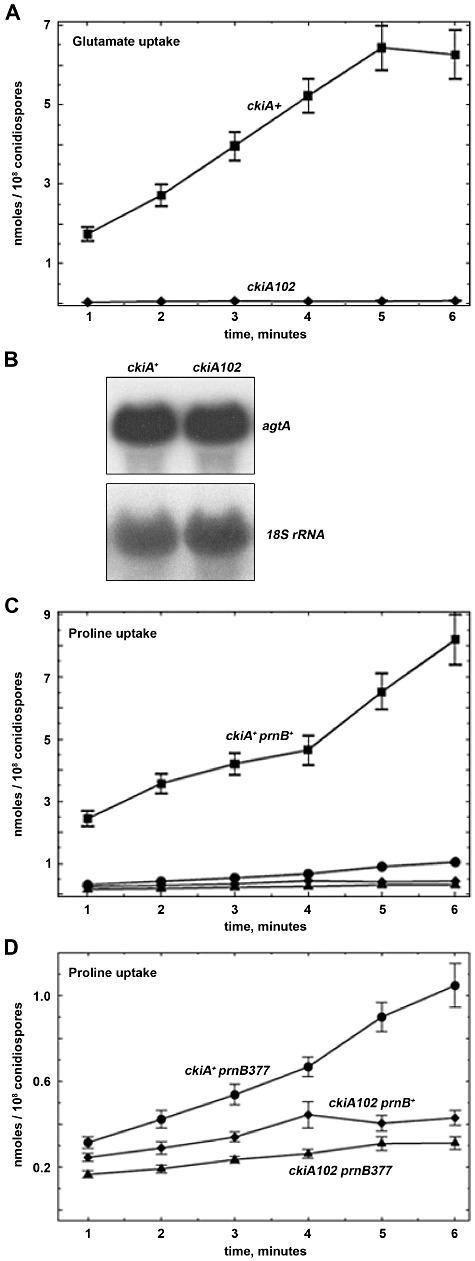
ckiA102 strains are defective in amino acid uptake. A. Glutamate uptake by ckiA+ (CS2498, squares, bars standard errors) and ckiA102 (CS1947, diamonds, error bars not shown as the uptake is almost nil) strains. B. Northern blot showing the expression of agtA in germlings pre-grown in the same conditions as for the uptake measurements (see Experimental procedures). C and D. Proline uptake by prnB+ckiA+ (CS2498, squares), prnB377 (CAM45, circles), ckiA102 (CS1947, diamonds) and ckiA102 prnB377 (CS1903, triangles, the latter two not clearly separated in C) strains. (D) Graph shows the same data as (C) for the three mutant strains, with an expanded scale in order to magnify the differences between them. Bars represent standard errors.
Double mutants carrying each of the three ckiA alleles and fbaA1013 were constructed (Fig. 1). The results of these tests are consistent with ckiA and fbaA not acting sequentially in the same pathway. ckiA alleles do not affect growth on acetate, a characteristic of the fbaA1013 mutation (Roumelioti et al., 2010). Double mutants show restricted growth on complete medium, a phenotype somewhat more restricted than that of fbaA1013, more clearly shown for the ckiA2 fbaA1013 double mutants. On all nitrogen sources tested, the double mutant shows the phenotype of the more extreme parent. This is most noticeable on proline, where fbaA1013 strains are hardly affected but ckiA2 fbaA1013 and ckiA102 fbaA1013 are as affected as the ckiA parent. Weak additivity can be seen for growth on glutamate and aspartate at 10 mM for the ckiA2 and ckiA102 fbaA1013 double mutants. The ckiA fbaA1013 double mutants show more restricted growth on d-serine and p-fluorophenylalanine than either parent strain, this possibly due to the restricted colony morphology seen on all media for strains carrying fbaA1013.
ckiA102 results in defective glutamate and proline uptake
The pleiotropic phenotypes of ckiA mutants are most readily explained by impaired amino acid uptake. Consistent with this interpretation is the fact that we have been unable to isolate ckiA102 argB2 and ckiA102 leuA2 recombinants (argB2 and leuA2 result in arginine and leucine auxotrophies respectively) in crosses where these markers were segregating. The three ckiA mutations result in a drastic impairment of dicarboxylic amino acid utilization and ckiA2 and ckiA102 also in significant impairment of proline utilization. Thus, we tested the activities of the well characterized AgtA (Apostolaki et al., 2009) and PrnB transporters (Tazebay et al., 1995; Tavoularis et al., 2003 and references therein) to investigate the basis of these defects. Figure 2A shows that a strain carrying ckiA102 is unable to take up glutamate at the concentration tested. Figure 2C and D shows the same strain being more drastically affected for the uptake of proline than a prnB deletion mutant, which implies that ckiA102 affects both the specific proline transporter, PrnB, and alternative proline transporter(s). Partial additivity is seen for the ckiA102 prnB377 double mutant (Fig. 2D).
Identification of the ckiA gene
The ckiA gene was identified by complementation of the ckiA102 mutation with a genomic library constructed in a self-replicating vector (Osherov and May, 2000) and with pools from the minimal ordered compressed cosmid library of the third chromosome of A. nidulans (see Experimental procedures). From the first library, the smallest plasmid insert that complemented the ckiA102 mutation contained the whole open reading frame of ANID_04563.1 (http://www.broadinstitute.org/annotation/genome/aspergillus_group/MultiHome.html) encoding a casein kinase I, together with 322 bp of its upstream region and a partial sequence of the adjacent ANID_04562.1 gene. ckiA102-complementing cosmid L12G07 cross-hybridized with the above insert and one subclone containing ANID_04563.1 plus 1053 bp upstream and 595 bp downstream of the ORF also complemented ckiA102 (see Experimental procedures). Moreover, ANID_04563.1 is located in contig 78, chromosome III, between ANID_04409.1 (argB in contig 76) and ANID_04603.1 (alX, in contig 79; Hamari et al., 2009) in agreement with the position determined for ckiA in the genetic map. These findings and the identification within ANID_04563.1 of the three ckiA mutations (see below) established unambiguously that ckiA is ANID_04563.1.
The predicted ANID_04563.1 ORF encodes 367-amino-acid residues and is interrupted by two introns near the 5′ end of the coding region. An almost complete cDNA sequence was obtained (M. Billini and V. Sophianopoulou, unpublished; accession number HQ 661153) that experimentally confirmed the presence and position of these introns and established the position of the initiation codon. Northern blots showed an mRNA of about 2.5 kb in both ckiA+ and ckiA102 strains (not shown). The mRNA steady state levels are nearly identical in mycelia grown on either ammonium or glutamate as nitrogen sources, and this throughout the morphological transition from isotropic growth to young hyphae. The ckiA mRNA is also present in ungerminated conidiospores (Fig. S1).
ckiA encodes a δ/ε casein kinase: phylogeny of fungal casein kinases I
CkiA shows a striking amino acid sequence identity with mammalian δ/ε and related fungal casein kinases I as illustrated in Fig. 3. Two structures of close CkiA relatives have been solved, those of S. pombe Cki1 (Xu et al., 1995) and of Rattus norvegicus Casein kinase 1-δ (Longenecker et al., 1996). The structures comprise two lobes, which together form the catalytic domain. The amino-terminal lobe contains five antiparallel β-sheets interrupted by an α-helix (Fig. 3A), while the carboxy-terminal lobe consists in a succession of α-helixes. CkiA can be readily modelled on these structures (Fig. 3B), the predicted structure showing an identical succession of β-sheets and α-helices to that seen in the solved structures (Fig. 3A; see below and Discussion). The three signature motifs of casein kinases I (Longenecker et al., 1996) are conserved in CkiA. The first, LLGPSLEDL, is the linker peptide between the β and α lobes of the protein. The second, SINTHLGIEQSRRDDLE, spans the α−D and α−E helixes. It contains in CkiA two conservative substitutions (in the residues underlined above) shared with other fungal orthologues (Fig. 3). This motif comprises the SIN (Ser-Ile-Asn) sequence that in all type I casein kinases replaces the Ala-Pro-Glu sequence present in protein kinases of other families. The third motif LPWQGLKA, fully conserved in CkiA, is comprised in the loop between helices α−E and α−F. A putative nuclear entry signal, typical of casein kinases I, and a kinesin–tubulin interaction domain complete the inventory of conserved signatures (Fig. 3A).
Fig 3.
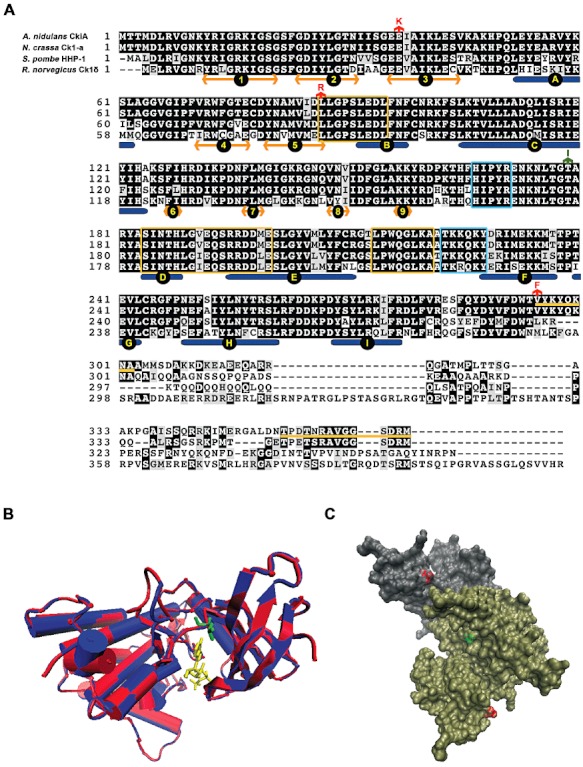
A. Alignment of A. nidulans CkiA with selected orthologues (see text). Alignment carried out with MUSCLE and visualized with box-shade. Under the alignment the succession of β-sheets and α-helices as deduced from known structures is shown (Longenecker et al., 1996). Double headed golden arrows, β-sheets (numbered from 1–9), rounded-ended blue rectangles, α-helixes (A to I). The sequences boxed in yellow represent the signature motifs of casein kinase I proteins (see text), the sequences boxed in cyan, the kinesin homology domain and downstream from it the SV40 T-antigen nuclear localization sequence (Gross and Anderson, 1998). The yellow lines between the sequences of the A. nidulans and N. crassa orthologues show sequences that are conserved in the Pezizomycotina but not among other ascomycetes (see text). The three amino acid substitutions described in this article are indicated by red arrows above the A. nidulans CkiA sequence. In green we indicate the position and mutant change of the hrr25-2 (rst2-1) suppressor of S. cerevisiæ (Murakami et al., 1999; see Discussion). B. Model of the CkiA protein. In red, CkiA, Val8-Thr294, in blue S. pombe Cki1 Asn6-Leu298 (PDB 1cnsA). In yellow ATP molecule. The substrate lies in a groove between the β-sheet lobe and α-helix lobe of the molecule. Leucine 87 is shown in green. We show the comparison with the S. pombeγ-like homologue, rather than the closer rat δ-homologue (Longenecker et al., 1996; see Fig. 4) as only the former has been crystallized with the bound ATP (Xu et al., 1995). C. Surface representation of the rat casein kinase I δ dimer (Longenecker et al., 1996) showing the surface residues discussed in this article. Subunit A in grey, subunit B in tan. In red Glu 34, corresponding to Glu 37 of CkiA, in green Thr 176, corresponding to Thr 176 of Hrr25. Modelling with I-Tasser, images made with VMD/NAMD/BioCoRE/JMV/other software support. VMD/NAMD/BioCoRE/JMV/ is developed with NIH support by the Theoretical and Computational Biophysics group at the Beckman Institute, University of Illinois at Urbana-Champaign.
A phylogeny of the casein kinases I proteins is shown in Fig. 4. The class I casein kinases of the ascomycetes fall into two clades. In all available Pezizomycotina proteomes one homologue (exceptionally two; see Fig. 4) of each clade is present. One clade includes the Yck proteins of S. cerevisiæ and comprises proteins that all have a palmitoylation motif in their carboxy-terminus. This motif is also present in membrane-anchored γ-mammalian Casein kinase I isoform (Davidson et al., 2005). The S. cerevisiæ enzymes are anchored to the plasma (Yck1 and Yck2) or vacuolar (Yck3) membranes via a covalently bound fatty acid (Vancura et al., 1994; Roth et al., 2002; Sun et al., 2004). The homologous A. nidulans ANID_05757.1, designated ckiB (see Fig. 4) is also membrane-bound (S. Amillis, T. Schinko, J. Strauss and C. Scazzocchio, unpublished).
Fig 4.
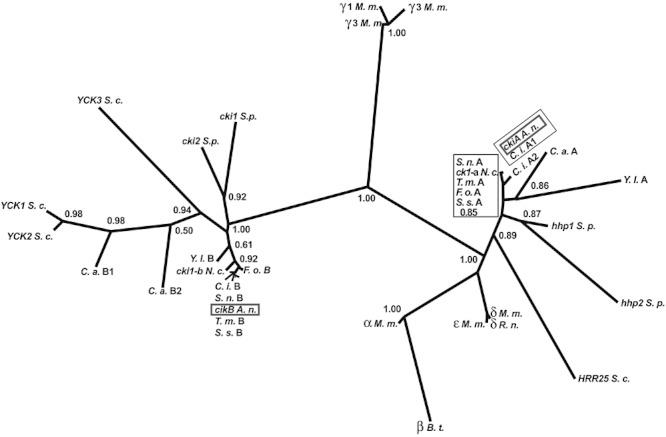
Phylogeny of the ascomycete casein kinase I proteins. This tree includes casein kinases I from representative species of the ascomycetes. All the isoforms of Mus musculus (M. m.) are also included, together with the unique isoform β of Bos taurus (B. t.) and the isoform δ of Rattus norvegicus (R. n.) mentioned in various contexts in the text. Characterized genes of A. nidulans, N. crassa, S. cerevisiæ and S. pombe are indicated by their standard genetic symbols followed by the abbreviation of the species. Other ORFs are indicated by the following convention: A two letter abbreviation of the species followed by ‘A’ for the homologues of ckiA, HRR25, hhp1 and hhp2, which never have a palmitoylation signal in the their carboxy-terminus, and ‘B’ for the homologues of YCK which always carry such a signal. When more than one ‘A’ or ‘B’ homologue is present for a given species, this indicated by a number following the letters A or B. The ascomycete species included are: Taphrinomycotina: Schizosaccharomyces pombe (S. p.); Saccharomycotina, Saccharomycetales: Saccharomyces cerevisiæ (S. c.), Candida albicans (C. a.), Yarrowia lypolitica (Y. l.). Pezizomycotina: Eurotiales, Aspergillus nidulans (A. n.), Onygenales, Coccidioides immitis (C. i.), Sordariales, Neurospora crassa (N. c.), Hypocreales, Fusarium oxysporum (F. o.), Helotiales, Sclerotina sclerotium (S. s.), Pleosporales, Stagonospora nodorum (S. n.), Pezizales, Tuber melansporum (T. m.). ORF sequences were obtained from the appropriate databases (see Supplementary Experimental procedures for accession numbers) and aligned with T-Coffee. The phylogenetic tree was redrawn from an original PhyMyl (Guindon et al., 2010) tree obtained after curation of the alignment with G-blocks and visualized with Drawtree (see Experimental procedures). The decimals in the nodes indicate the values of the Approximate Probability Ratio Test (aLTR; Anisimova and Gascuel, 2006). For clarity's sake some of the values for the minor nodes are not included. The boxed homologues are not separated in this tree, the 0.85 value refers to the aLTR separating the two boxed branches containing the Pezizomycotina homologues in clade ‘A’. The non-boxed homologues in a column in the ‘B’ clade are separated by extremely short branches, they correspond to up to bottom to the five short branches, left to right. Note the very high branch support for all the major nodes. The two A. nidulans genes are highlighted by a grey box. Note the striking conservation of sequences within the Pezizomycotina, including Tuber melanosporum, a basal species (Spatafora et al., 2006; see Fig. 4), which has diverged from the crown orders of the Pezizomycotina (including all the other Pezizomycotina represented in the figure) about 300 million years ago (Berbee and Taylor, 2007; Lucking et al., 2009).
Members of the second clade are proteins related to the metazoan δ and ε casein kinases, which do not carry a palmitoylation signal. This group includes the Hhp1 and Hhp2 paralogues of S. pombe, Hrr25p of S. cerevisiæ, CK1a of N. crassa and CkiA of A. nidulans. Two conserved carboxy-terminal amino acid sequence motifs are characteristic of proteins of the Pezizomycotina, being absent in the homologues of other fungi (see Fig. 3 for a comparison with the S. pombe orthologue). The carboxy-terminal motif VYKYQKNA is completely conserved in all available protein sequences of the Pezizomycotina (the Thr preceding this sequence is conserved also in other ascomycetes, but not in the mammalian isotype δ sequences). The VYKQKNA motif follows a universally conserved sequence, which in the crystal structure of the rat enzyme is located downstream of helix α−I and is structurally disordered (see Fig. 3A). This motif is linked by a less conserved K/N rich sequence to the most carboxy-terminal conserved motif, TP(D/E)T(N/S)RAVGGSDRM specific of δ/ε-like casein kinases of the Pezizomycotina (see the comparison of CkiA with the N. crassa orthologue in Fig. 3A). In addition to these conserved motifs, the S. cerevisiæ CkiA orthologue Hrr25p carries a long sequence in its carboxy-terminus which is absent from the S. pombe homologues and from all the Pezizomycotina proteins (not shown). Breitkreutz et al., (2010, Supporting information) identified within this non-conserved Hrr25 sequence two phosphorylated peptides, located between residues 310 and 388. Finally, it is worth noting that there is a striking amino acid sequence conservation both within the ckiA-like (δ/ε-related) and ckiB-like (possibly γ-related) clades of all the Pezizomycotina which contrasts with the sequence divergence seen in other ascomycetes (Saccharomycotina, Taphrynomycotina), even between paralogues of the same species (see Fig. 4).
Identification of the ckiA mutations
The ckiA gene of strains carrying each of the three mutant alleles was sequenced. ckiA2 is a G to A transition in nucleotide 109 (numbering from the A of the ATG initiation codon), resulting in a Glu37 to Lys substitution; ckiA1919 is a T to G transversion in nucleotide 260 resulting in a Leu87 to Arg substitution; ckiA102 is a G to T transversion in nucleotide 838 resulting in a Val295 to Phe substitution. The mutated residues are indicated in Fig. 3. Glu37 is conserved across all species in the δ/ε kinases (but not in the Yck/ANID_05757.3 fungal clade, nor in mammalian class I casein kinases outside the δ/ε clade). Leu87 is universally conserved. In the S. pombe CkI, it is one of the residues that form the hydrophobic pocket where the adenine ring of the ATP is lodged into the solved structure (Xu et al., 1995; see Discussion; Fig. 3B). Val295 is the N-terminal residue of the most carboxy-terminal conserved motif specific for the δ-like casein kinases of the Pezizomycotina.
ckiA is an essential gene
We attempted to delete ckiA by replacement of its coding region by a ‘split’pantoB marker in a pantoB100 strain and, when nkuΔ strains became available (Nayak et al., 2006), by the A. fumigatus riboB marker to complement riboB2. Only heterokaryotic transformants (and in the nkuA+ strain also heterologous insertions) were obtained, indicating that ckiA is essential (O'Donnell et al., 1991; Osmani et al., 2006). A recent systematic initiative to delete all A. nidulans kinases by insertion of the A. fumigatus pyrG marker confirmed that ckiAΔ can be rescued in heterokaryons but not in homokaryons (Osmani and de Souza, http://www.fgsc.net/Aspergillus/KO_Cassettes.htm).
We also demonstrated that the lethal ckiAΔ phenotype is recessive in a heterozygous diploid and that ckiAΔ strains cannot be recovered by haploidization: diploid strain LH61 (relevant markers, yA+ pantoB100 argB+/yA2 pantoB100 argB2), homozygous for pantoB100 and heterozygous for makers on chromosomes I (yA) and III (argB) (ckiA is located on chromosome III) was transformed with the ckiAΔ::pantoB+ deletion cassette. Five pantothenic acid prototrophs analysed by Southern blots were all shown to be heterozygous for the ckiA+ and the ckiAΔ alleles (Fig. S2). Two of these, T12 and T15, were haploidized. No pantoB+ (i.e. ckiAΔ::pantoB+) strain was obtained among a total of 150 haploid sectors recovered. The 100 haploid sectors arising from T12 were all pantoB100 and argB+ whereas the 50 haploid strains isolated from T15 were all pantoB100 and argB2, indicating that the ckiAΔ deletion event in T12 had occurred in coupling with argB2 while the one in T15 had occurred in coupling with argB+. Thus these results also establish that the failure to recover ckiAΔ strains is due to its lethality rather than to a possible co-lethality of ckiAΔ and argB2. The chromosome I yA marker segregated randomly in the haploid sectors. In summary, several independent lines of evidence establish beyond question that ckiA is an essential gene. This is of some importance as earlier reports claimed that S. cerevisiæ HRR25 is not essential (Hoekstra et al., 1991; Wang et al., 1996; Murakami et al., 1999), in stark contradiction with a recent article (Lord et al., 2011). The strain deleted for both the two S. pombe paralogues is viable (Dhillon and Hoekstra, 1994; Bimbóet al., 2005) but the N. crassa orthologue seems to be essential (Görl et al., 2001, systematic deletion data in http://www.dartmouth.edu/~neurosporagenome/knockouts_completed.html).
CkiA localizes both to the cytoplasm and nucleus
A ckiA–gfp fusion was constructed by gene replacement, resulting in a strain where the fusion protein, expressed at physiological levels, is the only source of ckiA. This strain is wild type for growth on all amino acids tested, is as sensitive as the wild type to d-serine and p-fluorophenylalanine and does not show the somewhat restricted growth shown by the ckiA102 strains on utilizable nitrogen sources, leading us to conclude that the fusion protein is fully functional. Both epifluorescence (Fig. 5A) and confocal microscopy (not shown) showed that CkiA–GFP is present throughout the cytoplasm and in the nuclei. We never observed any association of CkiA–GFP to the membrane, the septum or to the tip of the growing hyphæ. This intracellular distribution was identical on all nitrogen sources tested (ammonium, urea, glutamate, aspartate, proline, glycine, not shown) and did not change either with the morphological switch from conidia to hyphae (results not shown). However, it is worth noting that the protein was detectable in quiescent conidia, where we had also detected ckiA mRNA (see above; Fig. S1). The localization of CkiA–GFP in conidia and conidiophores is shown in Fig. S3.
Fig 5.
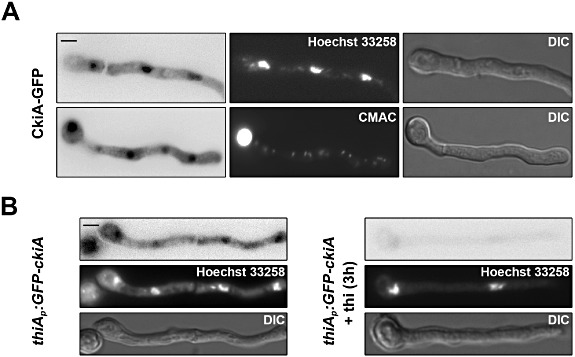
Intracellular localization of CkiA–GFP. A. Localization of CkiA–GFP in germlings. Two germlings are shown, the top one is counterstained with Hoechst 33258, the bottom one with CMAC. Germlings of strain VIE172 grown in MM for 16 h on urea as sole nitrogen source. B. Epifluorescence detection of GFP-CkiA driven by the thiA promoter in germlings (strain VIE047) grown in MM for 14 h on proline as sole nitrogen source and then shifted to proline in the absence and presence of thiamine (thi) for the additional time indicated. Counterstaining with Hoechst 33258 is also shown. Scale Bars: 5 µm.
Phenotype of a ckiA conditional mutation
The recessivity of the classical ckiA mutations strongly indicates that they result from a partial loss-of-function. To buttress this proposal and to investigate the effects of CkiA depletion we constructed a conditional ckiA allele by placing the ckiA gene under the control of the thiA promoter, which is repressible by thiamine (M. Mathieu, A. Rincón, and C. Scazzocchio, unpublished; Calcagno-Pizarelli et al., 2011). We constructed strains carrying the gfp-tagged agtA gene (Apostolaki et al., 2009), together with the thiAp–ckiA transgene, with or without a FLAG epitope tag in the amino-terminus of CkiA. Additionally, we constructed a thiAp–gfp–ckiA strain carrying the gfp sequence in the amino-terminus of ckiA, which shows a repression-sensitive localization of the protein to cytoplasm and nucleus (Fig. 5B). Figure 6A shows the phenotype of the thiAp::ckiA agtA–gfp strain (identical phenotypic responses were seen for the other strains, not shown). In the presence of as little of 5 nM thiamine, a diminution of growth on glutamate, aspartate and glycine as nitrogen sources is noticeable together with a moderate resistance to d-serine and p-fluorophenylalanine (Fig. 6A; not shown for aspartate). At 25 nM thiamine, the phenotype of this strain is very similar to that of ckiA2 and ckiA102, but in addition, a restricted growth on other nitrogen sources (such ammonium and nitrate, only shown for ammonium) is seen, as expected from the lethal phenotype of the deletion. The effect of thiamine on ammonium (another utilizable nitrogen source) is steadily more pronounced with increasing thiamine concentrations, but no complete inhibition of growth is seen even at 100 µM thiamine (shown for 10 µM in Fig. 6A), indicating that the repression of the thiA promoter is not complete. We used the thiAp–flag–ckiA strain to determine experimentally the degree of CkiA downregulation by Western blots. These experiments showed that while thiamine repression was efficient and CkiA levels rapidly (within 1–2 h) decreased after shifting cells to thiamine, even after 3 h of repression by 10 µM thiamine, a faint band CkiA::FLAG could be seen in Western blots (Fig. 6B), in agreement with growth tests indicating that repression is incomplete.
Fig 6.
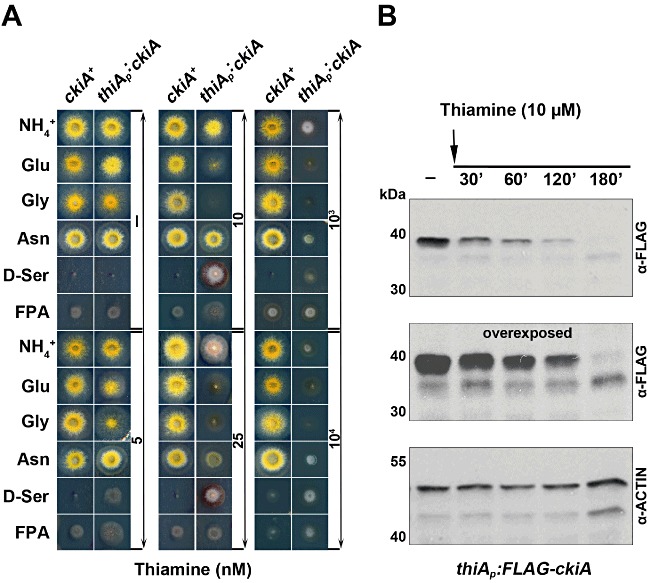
Phenotype of a ckiA conditional mutation. A. Growth tests of strains LH59 (wt) and VIE050 (thiAp–ckiA) carried out on supplemented MM with nitrogen sources or inhibitors indicated as in Fig. 1. Only relevant genotypes are shown. For complete genotypes see Table 1. Adjacent to each panel, thiamine concentrations in nM. Growth tests were carried out at 37°C and pH 6.8 for 48 h. B. Depletion of the FLAG-CkiA protein after the addition of 10 µM thiamine. Mycelia were grown for 14 h on ammonium as sole nitrogen source before addition of thiamine.
ckiA mutant alleles and CkiA depletion result in miss-routing of amino acid transporters into the vacuole
Northern blots demonstrated that steady state mRNA of agtA (encoding the specific dicarboxylic amino acid transporter of A. nidulans; Apostolaki et al., 2009) were not affected by the phenotypically strongest mutation, ckiA102 (Fig. 2), despite the fact that the mutation markedly impairs growth on dicarboxylic amino acids (Fig. 1). Thus, we further investigated the effect of the ckiA mutations on levels and subcellular localization of GFP-tagged AgtA. Plasma membrane levels of AgtA are exquisitely regulated through transcriptional and post-transcriptional mechanisms (Apostolaki et al., 2009). In cells cultured on GABA, AgtA is efficiently synthesized and delivered to the plasma membrane. However, if these cells are shifted to ammonium (the preferred nitrogen source for A. nidulans), agtA transcription is efficiently shut-off and the pool of plasma membrane-resident permease is internalized by endocytosis, sorted into the multivesicular body (MVB) pathway and delivered to the vacuole for degradation (Apostolaki et al., 2009; Abenza et al., 2010; Calcagno-Pizarelli et al., 2011). As a result of the balance between these two mechanisms, in ckiA+ strains cultured on GABA, AgtA–GFP localizes mainly to the cell membrane and secondarily to the vacuole (Apostolaki et al., 2009; Fig. 7, top panels). In contrast, in strains carrying ckiA102, AgtA–GFP is present exclusively in the vacuole, resembling the wild-type situation after shifting cells to ammonium. ckiA2 and ckiA1919 had a milder effect: some residual fluorescence of AgtA–GFP can be seen in the membrane of ckiA2 and even more clearly of ckiA1919 strains (Fig. 7). Thus, the severity of the phenotypes seen in growth tests inversely correlates with the relative AgtA levels at the plasma membrane (Figs 1 and 7).
Fig 7.
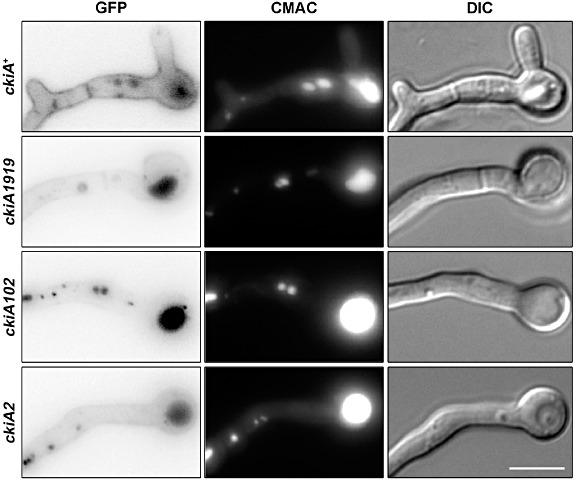
Localization of AgtA–GFP in ckiA mutants. Epifluorescence images of AgtA–GFP in strains carrying each of the three ckiA alleles (left panel). CMAC and differential interference contrast microscopy (DIC) images are shown on the middle and right panels respectively. Strains AMC132, CAM51, AMC314 and AMC129 were grown in MM containing GABA as nitrogen source for 16 h at 25°C. Scale Bar (5 µm) shown to the right of the DIC panel.
We also checked the localization of the proline transporter PrnB (see Figs 1 and 2), which is also delivered from the plasma membrane to the vacuole when cells cultured on proline are shifted to ammonium (Apostolaki et al., 2009). In strains carrying ckiA102 cultured on proline PrnB–GFP is localized exclusively to the vacuole (Fig. 8). All the above data strongly indicate that miss-routing of plasma membrane transporters to the vacuole is the common defect underlying the pleiotropic effects of ckiA- mutations on the utilization of amino acids.
Fig 8.
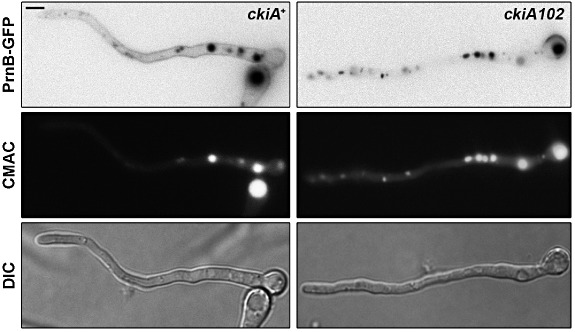
Localization of PrnB–GFP in a ckiA102 mutant. Germlings of strain TpA4 and VIE178 were grown on ammonium as nitrogen source for 12 h and then shifted to MM containing 10 mM proline for additional 4 h. Both counterstaining with CMAC and DIC images are shown. Scale Bars: 5 µm.
We then investigated whether depletion of CkiA results in vacuolar miss-routing of the AgtA transporter. In thiAp–ckiA cells shifted to GABA (which leads to agtA expression), the absence of thiamine (i.e. CkiA-sufficient conditions) leads to plasma membrane (and secondarily in the vacuolar lumen) localization of AgtA–GFP, as in the wild type (Fig. 9A transfer to GABA, compare with Fig. 7, top panel). In contrast, if cells were shifted to GABA in the presence of thiamine (which results in CkiA downregulation), virtually all GFP fluorescence appeared into the vacuolar lumen after 6 h (Fig. 9A, transfer to GABA + thiamine). The vacuolar luminal localization of GFP (attached to the cytosolic C-terminus of AgtA) demonstrates that the transporter is sorted into the MVB pathway, but does not address whether the vacuolar GFP accumulation reflects endocytic or biosynthetic traffic (or both) of AgtA to the vacuole. Thus we carried out a second set of experiments in which we investigated the fate of AgtA in thiAp–ckiA cells pre-cultured on GABA that were transferred to medium containing urea, with or without thiamine. Urea represses strongly (albeit not completely, see below) agtA transcription but, at variance with ammonium, does not promote the endocytic internalization of the transporter (Apostolaki et al., 2009). Under these conditions (i.e. AgtA–GFP synthesis shut-off with or without CkiA downregulation), AgtA–GFP can still be seen in the membrane even after 6 h in the presence of thiamine (CkiA-downregulated), strongly suggesting the AgtA molecules that are already in the membrane are not internalized towards the vacuoles as a consequence of CkiA depletion. Repression of agtA transcription under identical conditions of those used for the experiment of Fig. 9 is shown in Fig. S4. Therefore these experiments suggest that the strong vacuolar GFP fluorescence seen in cells cultured on GABA following downregulation of CkiA largely results from direct (i.e. without passing through the plasma membrane) biosynthetic traffic of AgtA to the vacuoles.
Fig 9.
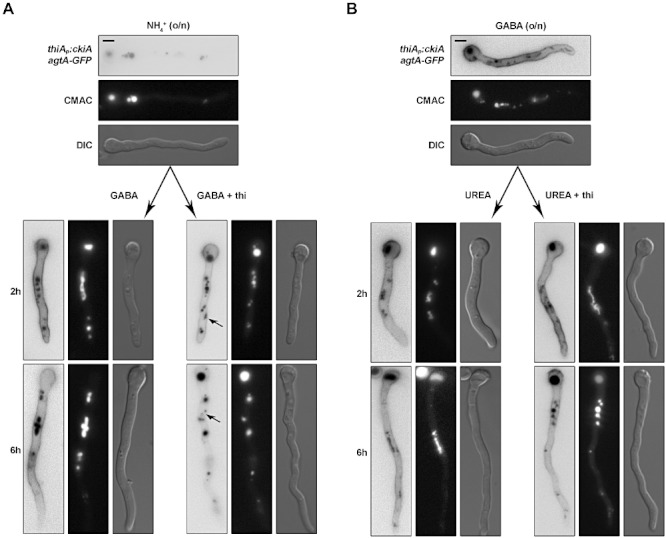
Localization of AgtA–GFP in a ckiA conditional mutant. Strain VIE050 was grown for 14–16 h in the presence of ammonium (NH4+ o/n) or GABA (GABA o/n) as nitrogen sources and then shifted to supplemented MM containing GABA or Urea as nitrogen sources, with or without 10 µM thiamine (+ thi) for the additional time indicated. For details on growth conditions, see Experimental procedures. Arrows indicate structures that are not stained by CMAC. Scale Bars: 5 µm.
The ckiA alleles and CkiA depletion both result in proteolysis of the AgtA transporter
Delivery of AgtA to the vacuole should result in its proteolytic degradation. We thus investigated, using Western blots, whether ckiA mutations also result in degradation of an AgtA–(HA)3 fusion protein expressed from a gene replaced allele (Apostolaki et al., 2009). In strains carrying ckiA2 or ckiA102 the AgtA–(HA)3 steady state levels were markedly reduced, implying that the protein is degraded in the vacuole and that the fluorescence seen in the vacuole is mostly free GFP which is notoriously resistant to degradation (see below). In strains carrying the ckiA1919 allele the signal is intermediate between that seen in the ckiA+ strain and that seen in strains carrying ckiA2 and ckiA102, in line with the less extreme phenotype of this allele (Fig. 10; not shown for ckiA2).
Fig 10.
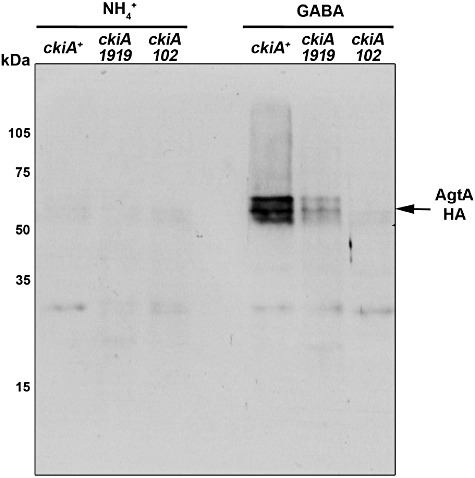
AgtA-3HA protein levels in ckiA mutants. Mycelia of LH121 (ckiA+), LH127 (ckiA1919), CAM13 (ckiA102) were pre-grown for 14 h at 30°C on ammonium as sole nitrogen source and transferred to either ammonium (NH4+) or γ-aminobutyric acid (GABA) for an additional 3 h incubation at 37°C. The ckiA allele used is indicated above each track. Loading was monitored by Coomassie blue staining and membranes were reacted with anti-HA primary antibodies and stained as described in Experimental procedures. Northern blots (not shown) carried out in parallel showed an identical pattern of de-repression of agtA after transfer to GABA (Apostolaki et al., 2009) for wild type and mutant strains. In a separate analogous experiment LH121 (ckiA+) was compared with AMC264 (ckiA2). The latter behaved exactly as ckiA102 (not shown).
We next used thiAp–ckiA strains expressing N-terminally FLAG-tagged CkiA kinase and AgtA–GFP, to monitor simultaneously the depletion of CkiA and the eventual degradation of the transporter. When AgtA–GFP synthesis was derepressed by shifting cells to GABA, if the medium lacked thiamine, both full-length AgtA–GFP fusion protein and GFP-containing degradation products were detected, in agreement with microscopy data (Fig. 11, lanes 2–4). However, in the presence of thiamine (CkiA-downregulated), only degradation products, of which the most conspicuous is free GFP, were seen (Fig. 11, lanes 5–7). On the other hand, when agtA–gfp was pre-derepressed for a period of time before adding both thiamine (to downregulate CkiA) and urea (to stop agtA transcription), full-length AgtA–GFP was clearly visible (Fig. 11, lanes 10 and 11), in complete agreement with the localization experiments in which plasma membrane-resident AgtA was unaffected by CkiA depletion. Thus these data strongly suggest that CkiA plays a role in a sorting step that precedes the delivery of AgtA to the plasma membrane.
Fig 11.
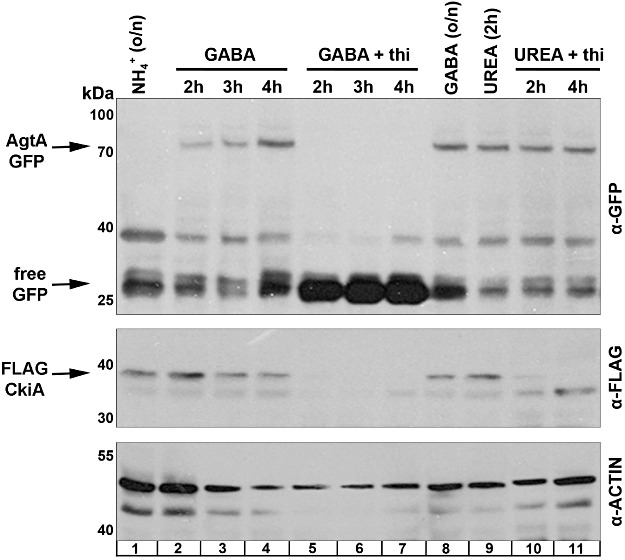
Degradation of AgtA–GFP after CkiA depletion. Strain VIE051, which carries agtA–gfp driven by its own promoter and ckiA-flag, driven by the thiA promoter was grown for 14 h at 30°C before transfer and/or addition of thiamine. For the eight tracks to the left of the panel, mycelia were grown for 14 h on ammonium (NH4+ o/n) and transferred to either GABA or GABA + 10 µM thiamine and grown for the time indicated. For the four tracks to the right of the panel mycelia was pre-grown for 14 h on GABA (GABA o/n) and then shifted to urea or urea + 10 µM thiamine for the time indicated. AgtA and its degradation products were monitored with an anti-GFP antibody, CkiA with an anti-FLAG antibody and total loading by an anti-actin antibody. For details see Experimental procedures.
Discussion
YAT localization and/or function are affected by mutations in numerous A. nidulans genes
Both recessive ckiA mutations and depletion (as a result of thiamine-mediated repression) of the CkiA protein result in defective utilization of a number of amino acids. This work strongly indicates that the partial CkiA deficiency resulting from any of the above mutant conditions affects a common mechanism specifically regulating intracellular trafficking of YAT transporters, leading to their miss-routing to the vacuole and subsequent degradation. ckiA mutants are able to grow on GABA, Gln and Asn, an ability shared by all the 22 non-ckiA mutants. The apparent YAT specificity of CkiA might explain why the utilization of GABA as a nitrogen source is unaffected by the mutations, as the GABA permease does not belong to the YAT family (Hutchings et al., 1999). Less straightforward is the interpretation of the finding that Gln and Asn utilization is also unaffected, as in S. cerevisiæ Asn and Gln are taken up by YAT transporters (Schreve et al., 1998; Regenberg et al., 1999). As the transporters responsible for their uptake in A. nidulans have not been identified, it is plausible that these belong to a different family.
Kinghorn and Pateman (1975) identified four loci where mutations result in inability to utilize glutamate. In three of these, aauB, aauC and aauD, mutations appear to affect a number of different amino acid transporters pleiotropically. As they map to chromosomes VII, II and VIII respectively, none of these loci can be ckiA (III) or fbaA (VI; Roumelioti et al., 2010). Sharma (1984) also identified phenotypically similar mutations mapping to chromosome VIII, whereas we have identified by transposon mutagenesis (Li Destri Nicosia et al., 2001) two insertional mutations mapping to chromosomes V and VII respectively, phenotypically very similar to ckiA mutations (Apostolaki, 2003). Thus, mutations in at least six (almost certainly more, see Results) loci affect amino acid uptake pleiotropically.
Two mutations identified in this study have a highly specific effect on YAT localization
The growth phenotype of ckiA1919 (Leu87Arg) is quite similar to that obtained when very low concentrations of thiamine (5–10 nM) are used to repress thiAp–ckiA partially, indicating that the efficient delivery of YAT transporters to the plasma membrane is extremely sensitive to ckiA deficiency. Leu87 is in the hydrophobic pocket that accommodates the purine ring of ATP (Fig. 3B). Arguably introduction of a positively charged, bulky arginine residue in this pocket might increase the Kd for ATP and thus impair kinase activity. Such impairment is definitely mild because ckiA1919 affects YAT trafficking without affecting viability or general growth rate.
A null ckiA mutation is lethal. Thus the two more extreme classical mutations can be more readily interpreted as loss-of-function mutations specifically affecting YAT trafficking. These mutations do not (ckiA2) or only slightly (ckiA102) affect colony morphology or growth on nitrogen or carbon sources other than amino acids. The inability of the cognate mutants to utilize amino acids resembles that seen in thiAp–ckiA strains at thiamine concentrations that, in contrast with ckiA2 and ckiA102, also result in severely restricted growth on ammonium or on other utilizable nitrogen sources. Further evidence that ckiA2 and ckiA102 specifically affect YAT trafficking is their resistance phenotype to d-serine and p-flourophenylalanine. Again, full resistance to these toxic analogues is not accompanied by a strong growth defect, in contrast with the markedly restricted growth seen when CkiA is depleted. ckiA2 (Glu37Lys) involves an N-terminal residue, universally conserved in casein kinases I of the δ/ε isotype, which lies in the surface of the modelled protein (Fig. 3C). Different surface residues of mouse CKIε mediate binding of its protein substrates (Dahlberg et al., 2009). It is thus likely that Glu37 is important for protein–protein interactions requiring a negatively charged surface side chain. The effect of ckiA102 (Val295Phe) is more difficult to predict, as it maps to a residue included in motif conserved in the Pezizomycotina, but not in other fungi. Thus it is tempting to speculate that in Pezizomycotina such region is involved in specifically regulating, directly or indirectly, the delivery of YAT transporters to the plasma membrane. This region is immediately downstream from a conserved non-structured loop (see Fig. 3), and thus in a position suitable to engage in protein–protein interactions. The partners of these interactions and thus candidate CkiA substrates are currently unknown. Preliminary experiments using λ phosphatase digestion and comparison of AgtA–(HA)3 (Apostolaki et al., 2009) electrophoretic mobilities in wild-type and ckiA mutant backgrounds failed to provide any indication that AgtA could be a direct substrate of CkiA. It may be relevant that casein 1-δ modulates the traffic of the α sub-unit of the human epithelium sodium channel, while it does not phosphorylate directly this protein (Yan et al., 2007).
Involvement of the ascomycete casein kinases I in intracellular trafficking
Our results imply that partial deficiency of ckiA results in the inappropriate redirection of newly synthesized AgtA and PrnB (and by implication of other YATs of A. nidulans) to the vacuole under physiological conditions which should lead to their delivery to the plasma membrane. While palmitoylated Yck proteins of S. cerevisiæ are involved in the localization of membrane proteins (Decottignies et al., 1999; Hicke, 1999; Feng and Davis, 2000; Marchal et al., 2000; 2002; Gadura et al., 2006), non-palmitoylated casein kinases belonging to the HRR25/hhp1/hhp2 clade (Figs 3 and 4), play roles in DNA repair (Dhillon and Hoekstra, 1994; Ho et al., 1997), ribosome biogenesis (Ray et al., 2008), calcineurin signalling (Kafadar et al., 2003), meiosis (Petronczki et al., 2006) and circadian rhythms (Görl et al., 2001; He et al., 2006; Fan et al., 2009 and references therein), all of which are unrelated to intracellular traffic.
However, a role of S. cerevisiae Hrr25p in the ER-Golgi interface was suggested by the finding that an hrr25 partial loss-of-function mutation suppresses the thermo-sensitive phenotype of sec12-4, the latter mapping in the Sar1p guanine nucleotide exchange factor. The mutation (T176I; Fig. 3A) acts as a specific suppressor, as a null mutant does not suppress sec12-4 (Murakami et al., 1999). In the modelled structure of Hrr25p, Thr176 is a surface residue located in the strictly conserved region that hinges the β-sheet and α-helix moieties (Fig. 3C). It is thus likely that this residue is involved in a protein/protein interaction. Two recent articles lend support to a role of Hrr25p in cell trafficking. Gao and Kaiser (2006) reported that Ltv1p is a component of an endosomal GTPase complex that is necessary for Gap1 delivery to the plasma membrane. While the involvement of Hrr25p was not addressed in that study, Ltv1p was shown to be an Hrr25p substrate in a different context (Schäfer et al., 2006). More importantly, Lord et al. (2011) established that Hrr25p localizes also to the Golgi and specifically phosphorylates Sec23p. It is proposed that this phosphorylation establishes the directionality of the ER-Golgi traffic and prevents the back fusion of the COPII vesicles with the ER.
While these data point out to a previously unsuspected role for casein kinases I-δ in cell trafficking in S. cerevisiæ, our work establishes that in A. nidulans a specific function of CkiA is necessary for the delivery of amino acid transporters to the plasma membrane, a finding that is unprecedented in the literature.
Under physiological conditions under which the uptake of their substrates is no longer needed, transporters en route to the plasma membrane can be diverted to the endosomal system (and thus to the vacuole) from the Golgi. The best-studied example is that of the general amino acid permease Gap1p, whose sorting to endosomes involves ubiquitination (Rubio-Texeira and Kaiser, 2006). CkiA activity could inhibit, directly or indirectly a similar sorting step, so that re-routing to endosomes would occur when its activity is defective. It is noteworthy than in S. pombe, the amino acid permease Aat1p is retained in the Golgi in conditions of nitrogen sufficiency, and sorted to the plasma membrane in conditions of nitrogen starvation (Nakase et al., 2012) which illustrates the possible variations in intracellular traffic within the ascomycetes.
The fact that Hrr25p plays a role in determining the directionality of COPII-mediated ER-to-Golgi transport through phosphorylation of the coat component Sec23p (Lord et al., 2011) would also be consistent with the possibility that, when CkiA activity is defective, transporters incorporated into ER-derived vesicles are delivered to the vacuoles without passing through the Golgi. For example, these transporters could use the autophagy pathway, as the ER provides membranes for the biogenesis of autophagosomes (Mizushima et al., 2011). YAT transporters would next gain access to the vacuolar lumen after fusion of autophagosomes with vacuoles. If this were the case and assuming at least a partial conservation of function of the CkiA/Hrr25p orthologues, one implication of our studies is that Hrr25p could have a role involving anterograde trafficking of YAT transporters across the ER-Golgi interphase, a role which has not been investigated. Thus, it would be most interesting to know the fate of a secretory cargo, dependent on COPII-mediated ER-Golgi transport (such as Gap1p Malkus et al., 2002), in a yeast strain inactivated or depleted for Hrr25p, as well as to investigate whether CkiA is able to phosphorylate A. nidulans Sec23 (Pantazopoulou and Peñalva, 2009).
Experimental procedures
Media growth conditions and genetic methodology
Minimal (MM) and complete (CM) media as well as growth conditions for A. nidulans were described by Cove (1966). Auxotrophies were supplemented at the concentrations given in http://www.fgsc.net/Aspergillus/gene_list/supplement.html, or as specifically detailed in Supplementary Experimental procedures. For growth tests, conidiospores were inoculated on minimal media supplemented with the appropriate nitrogen source and incubated at 37°C for 48 h unless otherwise indicated. Mutations were selected spontaneously or after chemical mutagenesis as described in Supplementary Experimental procedures. Crosses between A. nidulans strains and construction of diploids were described by Pontecorvo et al. (1953). Haploidization was as described by Hastie (1970).
Strains, plasmids and genomic libraries
Auxotrophic and morphological mutations of A. nidulans strains are compiled by A.J. Clutterbuck (http://www.gla.ac.uk/acad/ibls/molgen/aspergillus/index.html), where gene symbols are described. The strains used and constructed during this work are listed in Table 1. All strains are veA1.
Table 1.
Aspergillus nidulans strains used and constructed during this study.
| Strain | Genotype | Description/references |
|---|---|---|
| CS2498 | pabaA1 | Wild-type reference strain |
| LH59 | yA2; pantoB100; riboB2 | Wild-type reference strain |
| CS1957 | pantoB100; wA3 | Wild-type reference strain |
| CS1958 | pyrG89; biA1; pyroA4; fwA1 | Apostolaki (2003) |
| CS2374 | pantoB100; biA1; argB2 | Apostolaki (2003) |
| TNO2A7 | nkuAΔ::argB; pyrG89; pyroA4; riboB2 | Nayak et al. (2006) |
| TNO2A25 | nkuAΔ::argB; pyrG89; pabaB22; riboB2; argB2 | Nayak et al. (2006) |
| CS2290 | prnB377; pabaA1; riboB2; yA2 | prnB deleted strain (Tazebay et al., 1995) |
| CAM45 | prnB377; pabaA1; yA2 | Tavoularis et al. (2001) |
| TpA4 | prnB::gfp; pantoB100; yA2 | Tavoularis et al. (2001) |
| CS1945 | agtAΔ::riboB; biA; pyrG89; wA3; pyroA4; riboB2 | agtA deleted strain (Apostolaki et al., 2009) |
| CS3095 | areA600; sb43; biA1 | areA null mutant (Al Taho et al., 1984; Kudla et al., 1990) |
| C357 | sC12; ivoA1; galA1; yA2 | Glasgow collection |
| C358 | sC12; alX4; palG21; dilA1; pabaA1 | Glasgow collection |
| C190 | cnxH5; anA1; pyrF11; luA1; yA2 | Glasgow collection |
| LH61 | yA; pantoB100; riboB+; biA1; argB2/yA2; pantoB100; riboB2; biA+; argB+ | Diploid, this study |
| CS1924 | ckiA102; prnB6; pabaA9 | prnB6: loss of function mutation in prnB (Arst and MacDonald, 1975; Arst et al., 1981; Sharma, 1984) |
| CS1904 | ckiA102; biA1 | This study |
| CS1947 | ckiA102; pabaA1; yA2 | This study |
| CS1903 | ckiA102; prnB377; pabaA1; yA2 | This study |
| CS1912 | ckiA102; prnB377; pabaA1; riboB2; yA2 | This study |
| CS1901 | ckiA102; pyrG89; pabaA1; pyroA4; yA2 | This study |
| CS1902 | ckiA102; riboB2; biA1 | This study |
| VIE178 | ckiA102; prnB::gfp; pantoB100; biA1 | This study |
| VIE173 | ckiA2; puA2; biA1; fwA1 | This study |
| CS1959 | ckiA2; yA2; puA2 | This study |
| VIE174 | ckiA2; pyroA4; yA2 | This study |
| VIE175 | ckiA2; pabaA1 | This study |
| VIE181 | ckiA1919; prnB377; pabaA1; riboB2; yA2 | This study |
| VIE179 | ckiA1919; pyrG89; pabaA1 | This study |
| VIE180 | ckiA1919; pabaA1; pantoB100; yA2 | This study |
| VIE112 | fbaA1013; pabaA1; prnB377; riboB2; yA2 | Roumelioti et al. (2010) |
| KR1 | fbaA1013; pyrG89; pabaA1; pantoB100; yA2 | Roumelioti et al. (2010) |
| VIE116 | fbaA1013; pabaA1; pyrG89; pantoB100; yA2 | Roumelioti et al. (2010) |
| VIE111 | fbaA1013; pabaA1; riboB2; yA2 | This study |
| VIE107 | ckiA102; fbaA1013; pabaA1; yA2 | This study |
| VIE108 | ckiA102; fbaA1013; riboB2 | This study |
| VIE113 | ckiA2; fbaA1013; pabaA1; pantoB100 | This study |
| VIE114 | ckiA1919; fbaA1013; pantoB100; yA2 | This study |
| VIE115 | ckiA1919; fbaA1013; riboB2; yA2 | This study |
| LH121 | agtA::HA::AFpyrG; pyroA4 | Apostolaki et al. (2009) |
| CAM13 | ckiA102; agtA::HA::AFpyrG; riboB2; nkuAΔ::argB; pyrG89 | This study |
| AMC264 | ckiA2; agtA::HA::AFpyrG; pyroA4 | This study |
| LH127 | ckiA1919; agtA::HA::AFpyrG; pabaA1 | This study |
| LH115 | agtA::gfp::AFpyrG; pabaA1; pyroA4; pyrG89; yA2 | Apostolaki et al. (2009) |
| AMC129 | cki102; agtA::gfp::AFpyrG; yA2; pabaA1; pyrG89 | This study |
| AMC314 | ckiA2;agtA::GFP::AFpyrG; pabaA1; yA2 | This study |
| CAM51 | cki1919; agtA::GFP::AFpyrG; pyroA4 | This study |
| VIE050 | thiAp:ckiA; agtA::gfp::AFpyrG; pabaA1; yA2 | This study |
| VIE051 | thiAp::FLAG::ckiA; agtA::gfp::AFpyrG;pabaA1; yA2 | This study |
| VIE047 | thiAp::gfp::ckiA::AFriboB; pyrG89 nkuAΔ::argB; pyroA4; riboB2 | This study |
| VIE172 | ckiA::gfp::AFpyrG; pabaB22; riboB2 | This study |
Escherichia coli strains
The E. coli strains used were JM109b and DH10B.
Vectors and plasmids
Those are listed in Supplementary Experimental procedures, Tables S1–S3.
Transformation methods
Transformation of E. coli was carried out as described by Sambrook and Rusell (2001) and Dower et al. (1988). Transformation of A. nidulans is described by Tilburn et al. (1983).
DNA manipulations
Plasmid and cosmid preparation from E. coli strains was carried out as described by Sambrook and Rusell (2001) or by using the Qiagen Plasmid Mini and Plasmid Midi kit according to the manufacturer's instructions. Cosmid pool preparation is described in the Supplementary Experimental procedures. DNA digestion and cloning strategies were carried out as described by Sambrook and Rusell (2001). Genomic DNA extraction from A. nidulans is described by Lockington et al. (1985). Southern blot analysis was carried out according to Sambrook and Rusell (2001), and details of specific procedures are given in the Supplementary Experimental procedures. High-fidelity PCR reactions were carried out using the kit Expand™ Long Template PCR System (Roche). Conventional PCR reactions were carried out using Taq polymerase (Promega) and the REDTaq® ReadyMix™ (Sigma Aldrich). TA cloning was carried out using the pGEM™-T easy vector system (Promega). DNA bands were purified from agarose gels using the Wizard PCR preps DNA purification system (Promega) and the MinElute Gel Extraction Kit (Qiagen). Cloning and amplification of the replacement cassettes were carried out using Phusion® Flash High-Fidelity PCR Master Mix (New England Biolabs). The 32Pα-dCTP labelled DNA molecules, which were used as gene-specific probes, were prepared using the Megaprime™ DNA labelling systems kit (Amersham LIFE SCIENCE) or the Random Hexanucleotide Primer kit and purified on MicroSpin™ S-200 HR columns, following the supplier's instructions (Roche Applied Science). DNA sequences were determined in MWG AG Biotech and GENOME Express. The primers used are detailed in Table S4.
ckiA cloning
This was carried out by complementing appropriate ckiA102 strains with the minimal ordered compressed cosmid library of the third chromosome of A. nidulans constructed in the pWE15 and pLORIST2 cosmids (Prade et al., 1997) or the AMA-NotI genomic library constructed in the self-replicating pRG3NotI plasmid (Osherov and May, 2000), which was re-amplified as described in Apostolaki (2003). Further details are given in the Supplementary Experimental procedures.
Construction of ckiA deletions
These are detailed in Supplementary Experimental procedures.
Construction of strains containing in-locus ckiA transcriptional and translational fusions
A cassette containing ckiA::sgfp::AFpyrG was used to transform TNO2A25. Three cassettes containing thiAp::ckiA, thiaAp::FLAG–ckiA and AFriboB–thiAp::gfp::ckiA fusions were constructed. The first two were used to transform the AMC314 strain. Selection of transformants with fully restored ckiA function was carried out on MM with glutamate as sole nitrogen source. The third cassette was used to transform TNO2A7 and selection was carried out on MM media with urea as sole nitrogen source, in the absence of riboflavin. The intact single copy in-locus replacements were confirmed by Southern blot analysis. Further details are given in Supplementary Experimental procedures.
RNA manipulations
Total RNA extraction from A. nidulans was carried out using the RNAPLUS™ (Q-BIOgene) or TRIzol® Reagent (Invitrogen) according to the corresponding manufacturer's instructions and also as described by Chomczynski and Sacchi (1987) and Lockington et al. (1987). RNA was separated on glyoxal agarose gels as described by Sambrook and Rusell (2001). The hybridization technique was described by Church and Gilbert (1984). To monitor RNA loading, the γ-actin gene of A. nidulans and the radish 18S rRNA gene were used as probes.
Protein manipulations
Both total proteins and membrane-bound proteins were analysed in Western blots as described respectively by Vangelatos et al. (2010) and Calcagno-Pizarelli et al. (2007). Further details are given in Supplementary Experimental procedures.
Amino acid uptake assays
3H-labeled amino acid uptake was measured in germinating conidia and was carried out following the procedures of Robinson et al. (1973) and Tazebay et al. (1995). Details are given in the Supplementary Experimental procedures.
Fluorescence microscopy
Germlings of strains AMC132, CAM51, AMC314 and AMC129 were grown for 16 h at 25°C on supplemented watch medium (WMM) containing GABA as sole nitrogen source (Peñalva, 2005). agtA expression was modulated as described in Apostolaki et al. (2009). Germlings of strains TpA4, VIE047, VIE050 and VIE172 were obtained after growth on supplemented MM at 25°C for 14–16 h in the presence of ammonium or GABA as nitrogen sources and then shifted to supplemented MM containing thiamine to a final concentration of 10 µM and GABA or urea as nitrogen sources and incubated for additional 2–6 h. Details of image acquisition are described in Supplementary Experimental procedures.
Bioinformatic tools and databases
See Supplementary Experimental procedures.
Acknowledgments
We thank Maria Billini for providing her cDNA sequence before submission to the database, Katerina Roumelioti with help in microscopy and Panagiotis Maniadis for help with plotting the uptake results. C.S. thanks Robert Lucking for helpful discussion on the phylogeny of the ascomycetes. S.A. thanks Joseph Strauss and Thorsten Schinko for helpful discussion and experimental facilities. We thank anonymous referees for a useful experimental suggestion and pointing out important references. A.A. was supported by European Union Contract EUROFUNG N° QLK3-CT1999-00729. L.H. was partially supported by the Direction des Rélations Internacionales (Université Paris-Sud) and by the Agencia Española de Cooperación Internacional (Spain). I.V. was supported by postgraduate research grants from NCSR Demokritos (Greece). Work in Orsay was supported by the above EUROFUNG contract, the Université Paris-Sud, the CNRS and the Institut Universitaire de France. Work in Madrid was supported DGCYT Grant BIO2009-BIO2009-7281 and Comunidad de Madrid Grant S2006/SAL-0246 to M.A.P. H.N.A. is grateful to the Royal Society for the Smithson Research Fellowship and the Science Research Council through a grant to Professor J.M. Thoday for support in Cambridge and the Wellcome Trust for grant 084660/Z/08/Z (to H.N.A. and Joan Tilburn) for support in London.
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abenza JF, Galindo A, Pantazopoulou A, Gil C, de los Rios V, Peñalva MA. Aspergillus RabB Rab5 integrates acquisition of degradative identity with the long distance movement of early endosomes. Mol Biol Cell. 2010;21:2756–2769. doi: 10.1091/mbc.E10-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Taho N, Sealy-Lewis HN, Scazzocchio C. Suppressible alleles in a positive control gene in Aspergillus nidulans. Curr Genet. 1984;8:245–251. doi: 10.1007/BF00419720. [DOI] [PubMed] [Google Scholar]
- André B. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast. 1995;11:1575–1611. doi: 10.1002/yea.320111605. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Apostolaki A. 2003. Topogenése des transporteurs d'acides aminés chez le champignon filamenteux Aspergillus nidulans. PhD Thesis, Université Paris-Sud.
- Apostolaki A, Erpapazoglou Z, Harispe L, Billini M, Kafasla P, Kizis D, et al. AgtA, the dicarboxylic amino acid transporter of Aspergillus nidulans, is concertedly down-regulated by exquisite sensitivity to nitrogen metabolite repression and ammonium-elicited endocytosis. Eukaryot Cell. 2009;8:339–352. doi: 10.1128/EC.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arst HN, MacDonald DW. A gene cluster in Aspergillus nidulans with an internally located cis-acting regulatory region. Nature. 1975;254:26–31. doi: 10.1038/254026a0. [DOI] [PubMed] [Google Scholar]
- Arst HN, Jones SA, Bailey CR. A method for the selection of deletion mutations in the L-proline catabolism gene cluster of Aspergillus nidulans. Genet Res. 1981;38:171–195. doi: 10.1017/s0016672300020516. [DOI] [PubMed] [Google Scholar]
- Berbee ML, Taylor JW. Rhynie chert: a window into a lost world of complex plant-fungus interactions. New Phytol. 2007;174:475–479. doi: 10.1111/j.1469-8137.2007.02080.x. [DOI] [PubMed] [Google Scholar]
- Bimbó A, Jia Y, Poh SL, Karuturi RK, den Elzen N, Peng X, et al. Systematic deletion analysis of fission yeast protein kinases. Eukaryot Cell. 2005;4:799–813. doi: 10.1128/EC.4.4.799-813.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcagno-Pizarelli AM, Negrete-Urtasun S, Denison SH, Rudnicka JD, Bussink HJ, Munera-Huertas T, et al. Establishment of the ambient pH signaling complex in Aspergillus nidulans: PalI assists plasma membrane localization of PalH. Eukaryot Cell. 2007;6:2365–2375. doi: 10.1128/EC.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcagno-Pizarelli AM, Hervás-Aguilar A, Galindo A, Abenza JF, Peñalva MA, Arst HN., Jr Rescue of Aspergillus nidulans severely debilitating null mutations in ESCRT-0, I, II and III genes by inactivation of a salt-tolerance pathway allows examination of ESCRT gene roles in pH signalling. J Cell Sci. 2011;124:4064–4076. doi: 10.1242/jcs.088344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove DJ. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;113:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- Dahlberg CL, Nguyen EZ, Goodlett D, Kimelman D. Interactions between Casein kinase I epsilon (CKI epsilon) and two substrates from disparate signaling pathways reveal mechanisms for substrate-kinase specificity. PLoS ONE. 2009;4:e4766. doi: 10.1371/journal.pone.0004766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson G, Wu W, Shen J, Bilic U, Fenger P, Stannek A, et al. Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature. 2005;438:867–872. doi: 10.1038/nature04170. [DOI] [PubMed] [Google Scholar]
- Decottignies A, Owsianik G, Ghislain M. Casein kinase I-dependent phosphorylation and stability of the yeast multidrug transporter Pdr5p. J Biol Chem. 1999;274:37139–37146. doi: 10.1074/jbc.274.52.37139. [DOI] [PubMed] [Google Scholar]
- Dhillon N, Hoekstra MF. Characterization of two protein kinases from Schizosaccharomyces pombe involved in the regulation of DNA repair. EMBO J. 1994;13:2777–2788. doi: 10.1002/j.1460-2075.1994.tb06571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erpapazoglou Z, Kafasla P, Sophianopoulou V. The product of the SHR3 orthologue of Aspergillus nidulans has restricted range of amino acid transporter targets. Fungal Genet Biol. 2006;43:222–233. doi: 10.1016/j.fgb.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Fan JY, Preuss F, Muskus MJ, Bjes ES, Price JL. Drosophila and vertebrate casein kinase I delta exhibits evolutionary conservation of circadian function. Genetics. 2009;181:139–152. doi: 10.1534/genetics.108.094805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Davis NG. Feedback phosphorylation of the yeast a-factor receptor requires activation of the downstream signaling pathway from G protein through mitogen-activated protein kinase. Mol Cell Biol. 2000;20:563–574. doi: 10.1128/mcb.20.2.563-574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadura N, Robinson LC, Michels CA. Glc7-Reg1 phosphatase signals to Yck 1,2 casein kinase 1 to regulate transport activity and glucose-induced inactivation of Saccharomyces maltose permease. Genetics. 2006;172:1427–1439. doi: 10.1534/genetics.105.051698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol. 2006;8:657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- Gilstring CF, Ljungdahl PO. A method for determining the in vivo topology of yeast polytopic membrane proteins demonstrates that Gap1p fully integrates into the membrane independently of Shr3p. J Biol Chem. 2000;275:31488–31495. doi: 10.1074/jbc.M005047200. [DOI] [PubMed] [Google Scholar]
- Görl M, Merrow M, Huttner B, Johnson J, Roenneberg T, Brunner M. A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. EMBO J. 2001;20:7074–7084. doi: 10.1093/emboj/20.24.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gournas C, Amillis S, Vlanti A, Diallinas G. Transport-dependent endocytosis and turnover of a uric acid-xanthine permease. Mol Microbiol. 2010;75:246–260. doi: 10.1111/j.1365-2958.2009.06997.x. [DOI] [PubMed] [Google Scholar]
- Gross SD, Anderson RA. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal. 1998;10:699–711. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hamari Z, Amillis S, Drevet C, Apostolaki A, Vagvolgyi C, Diallinas G, Scazzocchio C. Convergent evolution and orphan genes in the Fur4p-like family and characterization of a general nucleoside transporter in Aspergillus nidulans. Mol Microbiol. 2009;73:43–57. doi: 10.1111/j.1365-2958.2009.06738.x. [DOI] [PubMed] [Google Scholar]
- Hastie AC. Benlate-induced instability of Aspergillus diploids. Nature. 1970;226:771. doi: 10.1038/226771a0. [DOI] [PubMed] [Google Scholar]
- He Q, Cha J, He Q, Lee HC, Yang Y, Liu Y. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 2006;20:2552–2565. doi: 10.1101/gad.1463506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB. Molecular evidence for the early colonization of land by fungi and plants. Science. 2001;293:1129–1133. doi: 10.1126/science.1061457. [DOI] [PubMed] [Google Scholar]
- Herman C, Clutterbuck AJ. A method for selection of auxotrophs by means of ‘spidery’ growth. Aspergillus Newsl. 1966;7:13–14. [Google Scholar]
- Hicke L. Gettin' down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 1999;9:107–112. doi: 10.1016/s0962-8924(98)01491-3. [DOI] [PubMed] [Google Scholar]
- Ho Y, Mason S, Kobayashi R, Hoekstra M, Andrews B. Role of the casein kinase I isoform, Hrr25, and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:581–586. doi: 10.1073/pnas.94.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra MF, Liskay RM, Ou AC, DeMaggio AJ, Burbee DG, Heffron F. HRR25, a putative protein kinase from budding yeast: association with repair of damaged DNA. Science. 1991;253:1031–1034. doi: 10.1126/science.1887218. [DOI] [PubMed] [Google Scholar]
- Hutchings H, Stahmann KP, Roels S, Espeso EA, Timberlake WE, Arst HN, Jr, Tilburn J. The multiply-regulated gabA gene encoding the GABA permease of Aspergillus nidulans: a score of exons. Mol Microbiol. 1999;32:557–568. doi: 10.1046/j.1365-2958.1999.01371.x. [DOI] [PubMed] [Google Scholar]
- Jack DL, Paulsen IT, Saier MH. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology. 2000;146:1797–1814. doi: 10.1099/00221287-146-8-1797. [DOI] [PubMed] [Google Scholar]
- Kafadar KA, Zhu H, Snyder M, Cyert MS. Negative regulation of calcineurin signaling by Hrr25p, a yeast homolog of casein kinase I. Genes Dev. 2003;17:2698–2708. doi: 10.1101/gad.1140603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinghorn JR, Pateman JA. Mutations which affect amino acid transport in Aspergillus nidulans. J Gen Microbiol. 1975;86:174–184. doi: 10.1099/00221287-86-1-174. [DOI] [PubMed] [Google Scholar]
- Kota J, Gilstring CF, Ljungdahl PO. Membrane chaperone Shr3 assists in folding amino acid permeases preventing precocious ERAD. J Cell Sci. 2007;176:617–628. doi: 10.1083/jcb.200612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla B, Caddick MX, Langdon T, Martinez-Rossi NM, Bennett CF, Sibley S, et al. The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 1990;9:1355–1364. doi: 10.1002/j.1460-2075.1990.tb08250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Destri Nicosia MG, Brocard-Masson C, Demais S, Hua Van A, Daboussi MJ, Scazzocchio C. Heterologous transposition in Aspergillus nidulans. Mol Microbiol. 2001;39:1330–1344. [PubMed] [Google Scholar]
- Ljungdahl PO, Gimeno CJ, Styles CA, Fink GR. SHR3: a novel component of the secretory pathway specifically required for localization of amino acid permeases in yeast. Cell. 1992;71:463–478. doi: 10.1016/0092-8674(92)90515-e. [DOI] [PubMed] [Google Scholar]
- Lockington R, Scazzocchio C, Sequeval D, Mathieu M, Felenbok B. Regulation of alcR, the positive regulatory gene of the ethanol utilization regulon of Aspergillus nidulans. Mol Microbiol. 1987;1:275–281. doi: 10.1111/j.1365-2958.1987.tb01933.x. [DOI] [PubMed] [Google Scholar]
- Lockington RA, Sealy-Lewis HM, Scazzocchio C, Davies RW. Cloning and characterization of the ethanol utilization regulon in Aspergillus nidulans. Gene. 1985;33:137–149. doi: 10.1016/0378-1119(85)90088-5. [DOI] [PubMed] [Google Scholar]
- Longenecker KL, Roach PJ, Hurley TD. Three-dimensional structure of mammalian casein kinase I: molecular basis for phosphate recognition. J Mol Biol. 1996;257:618–631. doi: 10.1006/jmbi.1996.0189. [DOI] [PubMed] [Google Scholar]
- Lord C, Bhandari D, Menon S, Ghassemian M, Nycz D, Hay J, et al. Sequential interactions with Sec23 control the direction of vesicle traffic. Nature. 2011;473:181–186. doi: 10.1038/nature09969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucking R, Huhndorf S, Pfister DH, Plata ER, Lumbsch HT. Fungi evolved right on track. Mycologia. 2009;101:810–822. doi: 10.3852/09-016. [DOI] [PubMed] [Google Scholar]
- Malkus P, Jiang F, Schekman R. Concentrative sorting of secretory cargo proteins into COPII-coated vesicles. J Cell Sci. 2002;159:915–921. doi: 10.1083/jcb.200208074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal C, Haguenauer-Tsapis R, Urban-Grimal D. Casein kinase I-dependent phosphorylation within a PEST sequence and ubiquitination at nearby lysines signal endocytosis of yeast uracil permease. J Biol Chem. 2000;275:23608–23614. doi: 10.1074/jbc.M001735200. [DOI] [PubMed] [Google Scholar]
- Marchal C, Dupre S, Urban-Grimal D. Casein kinase I controls a late step in the endocytic trafficking of yeast uracil permease. J Cell Sci. 2002;115:217–226. doi: 10.1242/jcs.115.1.217. [DOI] [PubMed] [Google Scholar]
- Martínez P, Ljungdahl PO. The SHR3 homologue from S. pombe demonstrates a conserved function of ER packaging chaperones. J Cell Sci. 2000;113:4351–4362. doi: 10.1242/jcs.113.23.4351. [DOI] [PubMed] [Google Scholar]
- Martínez P, Ljungdahl PO. An ER packaging chaperone determines the amino acid uptake capacity and virulence of Candida albicans. Mol Microbiol. 2004;51:371–384. doi: 10.1046/j.1365-2958.2003.03845.x. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Murakami A, Kimura K, Nakano A. The inactive form of a yeast casein kinase I suppresses the secretory defect of the sec12 mutant. Implication of negative regulation by the Hrr25 kinase in the vesicle budding from the endoplasmic reticulum. J Biol Chem. 1999;274:3804–3810. doi: 10.1074/jbc.274.6.3804. [DOI] [PubMed] [Google Scholar]
- Nakase M, Nakase Y, Chardwiriyapreecha S, Kakinuma Y, Matsumoto T, Takegawa K. Intracellular trafficking and ubiquitination of the Schizosaccharomyces pombe amino acid permease Aat1p. Microbiology. 2012;158:659–673. doi: 10.1099/mic.0.053389-0. [DOI] [PubMed] [Google Scholar]
- Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, et al. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KL, Osmani AH, Osmani SA, Morris NR. bimA encodes a member of the tetratricopeptide repeat family of proteins and is required for the completion of mitosis in Aspergillus nidulans. J Cell Sci. 1991;99:711–719. doi: 10.1242/jcs.99.4.711. [DOI] [PubMed] [Google Scholar]
- Osherov N, May G. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics. 2000;155:647–656. doi: 10.1093/genetics/155.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani AH, Oakley BR, Osmani SA. Identification and analysis of essential Aspergillus nidulans genes using the heterokaryon rescue technique. Nat Protoc. 2006;1:2517–2526. doi: 10.1038/nprot.2006.406. [DOI] [PubMed] [Google Scholar]
- Padovan AC, Sanson GF, Brunstein A, Briones MR. Fungi evolution revisited: application of the penalized likelihood method to a Bayesian fungal phylogeny provides a new perspective on phylogenetic relationships and divergence dates of Ascomycota groups. J Mol Evol. 2005;60:726–735. doi: 10.1007/s00239-004-0164-y. [DOI] [PubMed] [Google Scholar]
- Pantazopoulou A, Peñalva MA. Organization and dynamics of the Aspergillus nidulans Golgi during apical extension and mitosis. Mol Biol Cell. 2009;20:4335–4347. doi: 10.1091/mbc.E09-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalva MA. Tracing the endocytic pathway of Aspergillus nidulans with FM4-64. Fungal Genet Biol. 2005;42:963–975. doi: 10.1016/j.fgb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, et al. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- Prade RA, Griffith J, Kochut K, Arnold J, Timberlake WE. In vitro reconstruction of the AspergillusEmericellanidulans genome. Proc Natl Acad Sci USA. 1997;94:14564–14569. doi: 10.1073/pnas.94.26.14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P, Basu U, Ray A, Majumdar R, Deng H, Maitra U. The Saccharomyces cerevisiae 60 S ribosome biogenesis factor Tif6p is regulated by Hrr25p-mediated phosphorylation. J Biol Chem. 2008;283:9681–9691. doi: 10.1074/jbc.M710294200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenberg B, During-Olsen L, Kielland-Brandt MC, Holmberg S. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr Genet. 1999;36:317–328. doi: 10.1007/s002940050506. [DOI] [PubMed] [Google Scholar]
- Risinger AL, Kaiser CA. Different ubiquitin signals act at the Golgi and plasma membrane to direct GAP1 trafficking. Mol Biol Cell. 2008;19:2962–2972. doi: 10.1091/mbc.E07-06-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JH, Anthony C, Drabble WT. The acidic amino-acid permease of Aspergillus nidulans. J Gen Microbiol. 1973;79:53–63. doi: 10.1099/00221287-79-1-53. [DOI] [PubMed] [Google Scholar]
- Roth AF, Feng Y, Chen L, Davis NG. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J Cell Sci. 2002;159:23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumelioti K, Vangelatos I, Sophianopoulou V. A cryptic role of a glycolytic-gluconeogenic enzyme (aldolase) in amino acid transporter turnover in Aspergillus nidulans. Fungal Genet Biol. 2010;47:254–267. doi: 10.1016/j.fgb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Rubio-Texeira M, Kaiser CA. Amino acids regulate retrieval of the yeast general amino acid permease from the vacuolar targeting pathway. Mol Biol Cell. 2006;17:3031–3050. doi: 10.1091/mbc.E05-07-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Rusell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schäfer T, Maco B, Petfalski E, Tollervey D, Bottcher B, Aebi U, Hurt E. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441:651–655. doi: 10.1038/nature04840. [DOI] [PubMed] [Google Scholar]
- Schreve JL, Sin JK, Garrett JM. The Saccharomyces cerevisiae YCC5 (YCL025c) gene encodes an amino acid permease, Agp1, which transports asparagine and glutamine. J Bacteriol. 1998;180:2556–2559. doi: 10.1128/jb.180.9.2556-2559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K. 1984. Proline catabolism in Aspergillus nidulans. PhD Thesis, University of Newcastle.
- Sophianopoulou V, Scazzocchio C. The proline transport protein of Aspergillus nidulans is very similar to amino acid transporters of Saccharomyces cerevisiae. Mol Microbiol. 1989;3:705–714. doi: 10.1111/j.1365-2958.1989.tb00219.x. [DOI] [PubMed] [Google Scholar]
- Spatafora JW, Sung GH, Johnson D, Hesse C, O'Rourke B, Serdani M, et al. A five-gene phylogeny of Pezizomycotina. Mycologia. 2006;98:1018–1028. doi: 10.3852/mycologia.98.6.1018. [DOI] [PubMed] [Google Scholar]
- Sun B, Chen L, Cao W, Roth AF, Davis NG. The yeast casein kinase Yck3p is palmitoylated, then sorted to the vacuolar membrane with AP-3-dependent recognition of a YXXPhi adaptin sorting signal. Mol Biol Cell. 2004;15:1397–1406. doi: 10.1091/mbc.E03-09-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavoularis S, Scazzocchio C, Sophianopoulou V. Functional expression and cellular localization of a green fluorescent protein-tagged proline transporter in Aspergillus nidulans. Fungal Genet Biol. 2001;33:115–125. doi: 10.1006/fgbi.2001.1280. [DOI] [PubMed] [Google Scholar]
- Tavoularis SN, Tazebay UH, Diallinas G, Sideridou M, Rosa A, Scazzocchio C, Sophianopoulou V. Mutational analysis of the major proline transporter (PrnB) of Aspergillus nidulans. Mol Membr Biol. 2003;20:285–297. doi: 10.1080/0968768031000106339. [DOI] [PubMed] [Google Scholar]
- Tazebay UH, Sophianopoulou V, Cubero B, Scazzocchio C, Diallinas G. Post-transcriptional control and kinetic characterization of proline transport in germinating conidiospores of Aspergillus nidulans. FEMS Microbiol Lett. 1995;132:27–37. doi: 10.1111/j.1574-6968.1995.tb07806.x. [DOI] [PubMed] [Google Scholar]
- Tilburn J, Scazzocchio C, Taylor GG, Zabicky-Zissman JH, Lockington RA, Davies RW. Transformation by integration in Aspergillus nidulans. Gene. 1983;26:205–221. doi: 10.1016/0378-1119(83)90191-9. [DOI] [PubMed] [Google Scholar]
- Van Belle D, André B. A genomic view of yeast membrane transporters. Curr Opin Cell Biol. 2001;13:389–398. doi: 10.1016/s0955-0674(00)00226-x. [DOI] [PubMed] [Google Scholar]
- Vancura A, Sessler A, Leichus B, Kuret J. A prenylation motif is required for plasma membrane localization and biochemical function of casein kinase I in budding yeast. J Biol Chem. 1994;269:19271–19278. [PubMed] [Google Scholar]
- Vangelatos I, Vlachakis D, Sophianopoulou V, Diallinas G. Modelling and mutational evidence identify the substrate binding site and functional elements in APC amino acid transporters. Mol Membr Biol. 2009;26:356–370. doi: 10.1080/09687680903170546. [DOI] [PubMed] [Google Scholar]
- Vangelatos I, Roumelioti K, Gournas C, Suarez T, Scazzocchio C, Sophianopoulou V. Eisosome organization in the filamentous ascomycete Aspergillus nidulans. Eukaryot Cell. 2010;9:1441–1454. doi: 10.1128/EC.00087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hoekstra MF, DeMaggio AJ, Dhillon N, Vancura A, Kuret J, et al. Prenylated isoforms of yeast casein kinase I, including the novel Yck3p, suppress the gcs1 blockage of cell proliferation from stationary phase. Mol Cell Biol. 1996;16:5375–5385. doi: 10.1128/mcb.16.10.5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RM, Carmel G, Sweet RM, Kuret J, Cheng X. Crystal structure of casein kinase-1, a phosphate-directed protein kinase. EMBO J. 1995;14:1015–1023. doi: 10.1002/j.1460-2075.1995.tb07082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Spruce L, Rosenblatt MM, Kleyman TR, Rubenstein RC. Intracellular trafficking of a polymorphism in the COOH terminus of the alpha-subunit of the human epithelial sodium channel is modulated by casein kinase 1. Am J Physiol Renal Physiol. 2007;293:F868–F876. doi: 10.1152/ajprenal.00194.2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


