Abstract
The CCAAT motif is found in the promoters of many eukaryotic genes. In yeast a single complex of three proteins, termed HAP2, HAP3, and HAP5, binds to this sequence, and in mammals the three components of the equivalent complex (called variously NF-Y, CBF, or CP1) are also represented by single genes. Here we report the presence of multiple genes for each of the components of the CCAAT-binding complex, HAP2,3,5, from Arabidopsis. Three independent Arabidopsis HAP subunit 2 (AtHAP2) cDNAs were cloned by functional complementation of a yeast hap2 mutant, and two independent forms each of AtHAP3 and AtHAP5 cDNAs were detected in the expressed sequence tag database. Additional homologs (two of AtHAP3 and one of AtHAP5) have been identified from available Arabidopsis genomic sequences. Northern-blot analysis indicated ubiquitous expression for each AtHAP2 and AtHAP5 cDNA in a range of tissues, whereas expression of each AtHAP3 cDNA was under developmental and/or environmental regulation. The unexpected presence of multiple forms of each HAP homolog in Arabidopsis, compared with the single genes in yeast and vertebrates, suggests that the HAP2,3,5 complex may play diverse roles in gene transcription in higher plants.
The regulation of transcription of most eukaryotic genes is coordinated through sequence-specific binding of proteins to the promoter region located upstream of the gene. Many of these protein-binding sequences have been conserved during evolution and are found in a wide variety of organisms. One such feature is the CCAAT-box element (Gelinas et al., 1985). This motif is found between 80 and 300 bp 5′ from the transcription start site and may operate in either orientation, with possible cooperative interactions with multiple boxes (Tasanen et al., 1992) or other conserved motifs (Muro et al., 1992; Rieping and Schöffl, 1992).
Proteins that bind to the CCAAT motif were first characterized in the yeast Saccharomyces cerevisiae through analysis of mutants with reduced levels of expression of the CYC1 gene (encoding iso-1-Cyt c) (Guarente et al., 1984; Hahn et al., 1988). The CYC1 promoter comprises two UAS, one of which (UAS2) contains an inverted CCAAT motif that is required for UAS2-directed transcription. Activation of transcription from UAS2 requires HAP2, HAP3, and HAP5 (Pinkham and Guarente, 1985; Pinkham et al., 1987; Hahn et al., 1988; McNabb et al., 1995), which form a heterotrimeric CCAAT-box-binding complex. The yeast HAP complex recruits a fourth polypeptide, HAP4 (Forsburg and Guarente, 1989), which does not bind to DNA but associates with the HAP2,3,5 complex and activates transcription through an acidic domain. The HAP complex appears to control expression of genes important for mitochondrial biogenesis (de Winde and Grivell, 1993), demonstrated by the fact that yeast hap mutants show identical pleiotropic phenotypes, with a general reduction in cytochromes and reduced growth on nonfermentable carbon sources.
CCAAT-box-related motifs have also been identified in the promoters of a variety of vertebrate genes. A range of transcription factors has been shown to bind to different CCAAT boxes, with varying levels of specificity (Dorn et al., 1987; Raymondjean et al., 1988), and each is thought to play a distinct role in gene expression or DNA replication (Santoro et al., 1988). Direct homologs of the yeast HAP complex (called NF-Y, CP1, or CBF) have been identified in vertebrates (Maity et al., 1990; Becker et al., 1991; Li et al., 1992; Sinha et al., 1995). The individual vertebrate HAP subunits showed a remarkable similarity to the yeast homologs over short domains (Maity et al., 1990; Vuorio et al., 1990), which is sufficient to enable formation of a functional heterologous complex between the human HAP2 homolog and yeast HAP3 and HAP5 (Becker et al., 1991). However, outside of the highly conserved core protein motifs associated with DNA binding and subunit interactions, there is considerable divergence. Furthermore, there is no HAP4 homolog. Instead, the vertebrate HAP complex probably interacts with other classes of transcription factors to influence the level of transcription (Bellorini et al., 1997).
Based on their presence in other eukaryotes and sequence conservation between related plant gene promoters, putative CCAAT-box motifs have been identified for several plant genes (Rieping and Schöffl, 1992; Kehoe et al., 1994; Ito et al., 1995). As with vertebrates, there is no unifying expression pattern for plant genes containing putative CCAAT-promoter elements, indicating that they may play a complex role in regulating plant gene transcription, with greater similarity to the vertebrate model than to the yeast system. A homolog with sequence similarity to HAP3 has been isolated from maize (Li et al., 1992), and recently, a HAP2 homolog was characterized from Brassica napus (Albani and Robert, 1995).
To characterize the role of the CCAAT motif in plants, we have isolated and characterized plant homologs of the HAP/CBF/NF-Y class of CCAAT-binding transcription factors from Arabidopsis. In contrast to the situation in yeast and in animals, in which single representations of each subunit are present, we show that multiple genes exist for each of the HAP2,3,5 subunits in Arabidopsis, providing the potential for multiple alternative forms of HAP complexes in plants.
MATERIALS AND METHODS
Arabidopsis ecotype Columbia seeds were grown in compost, Murashige and Skoog solution (0.46% Murashige and Skoog mixture, 2% Suc, pH 5.9), or solid agar (Murashige and Skoog solution, 0.8% agarose) at 25°C with a 16-h photoperiod.
Yeast Growth and Transformation
The Saccharomyces cerevisiae strains used were gifts from L. Guarente (Massachusetts Institute of Technology, Cambridge). The strains were BWG 1-7a (MATa leu2-3, 112 his4-519 ade1-100 ura3-52; Olesen and Guarente, 1990), which has a wild-type HAP complex, or isogenic derivatives in which the individual HAP genes had been disrupted: JO1–1a (Δhap2; Pinkham and Guarente, 1985), SHY40 (Δhap3; Hahn et al., 1988), SLF401 (Δhap4; Forsburg and Guarente, 1989), DMY110 (Δhap5; McNabb et al., 1995), and JO2–1 (Δhap2 Δhap3; Olesen and Guarente, 1990). All strains were grown at 30°C on yeast peptone dextrose medium (1% yeast extract, 1% peptone, and 2% Glc, pH 6.5). Primary transformants were selected on minimal medium without uracil (0.76% yeast nitrogen base, 2% Glc, His and Trp at 0.01 mg/mL, Leu at 0.12 mg/mL, pH 6.0). Functional complementation was selected for by growth on lactate medium (1% yeast extract, 1% peptone, and 2% lactate, pH 4.8). Solid medium contained 2% bactoagar. Yeast transformation was done with lithium acetate according to the method of Schiestl and Gietz (1989) and Gietz et al. (1992).
DNA- and RNA-Blot Analysis
Genomic DNA was extracted from soil-grown Arabidopsis leaves (Dellaporta et al., 1983). RNA was extracted from various Arabidopsis tissues using the acid-phenol method (Chomczynski and Sacchi, 1987). Genomic DNA was digested with different restriction enzymes, fractionated on gels of 0.8% agarose in Tris-acetate-EDTA buffer (Sambrook et al., 1989), transferred onto GeneScreen Plus (New England Nuclear) filters, and hybridized with 32P-labeled DNA probes, as described previously (Church and Gilbert, 1984). The filters were then washed in 40 mm sodium phosphate buffer, pH 7.2, 1% SDS for 30 min at 65°C, followed by several 5-min washes in the same buffer until the background radioactivity was undetectable. Total RNA was fractionated in formaldehyde denaturing agarose gels, transferred onto GeneScreen Plus filters, and hybridized with 32P-labeled DNA probes (Sambrook et al., 1989). The filters were then washed in 2× SSC, 1% SDS for 30 min at 70°C, followed by several 5-min washes in 1× SSC, 1% SDS until the background radioactivity was undetectable.
DNA Sequencing and Analysis
DNA sequencing was carried out using a dideoxy termination kit and an ABI 373A sequencer (Applied Biosystems) at the Protein and Nucleic Acid Chemistry Facility (Department of Biochemistry, University of Cambridge, UK). DNA-sequence analysis and comparisons of DNA and protein sequences were made using ClustalW and Genetics Computer Group facilities (Madison, WI) (Devereux et al., 1984).
RESULTS
Cloning of Arabidopsis HAP2 Homologs by Functional Complementation
Functional complementation has been used previously to identify a human HAP2 homolog. We therefore used heterologous functional complementation of a yeast hap2 mutant to isolate cDNAs encoding Arabidopsis HAP2 homologs. The hap2 yeast mutant JP1-1C (Pinkham and Guarente, 1985), which is unable to grow on nonfermentable carbon sources such as lactate, was transformed with an Arabidopsis cDNA library in the yeast expression vector pFL61 (Minet et al., 1992). Five-hundred-thousand primary transformants were selected by growth on minimal medium containing Glc but no uracil to select for uptake of library plasmids. Colonies were replica plated to rich medium with lactate as the sole carbon source. After 5 to 14 d of growth at 30°C, six independent colonies (P1–P6) were observed growing on the lactate medium, suggesting that the plasmids they contained were able to rescue the growth defect. The extent of growth varied between rescued colonies on both lactate and glycerol media, suggesting that they might not all contain identical plasmids. Plasmids were transformed into an Escherichia coli host by electroporation and classified into three groups, with four of the six plasmids (P1, P2, P4, and P6) having an identical restriction pattern. The growth phenotypes on lactate of the yeast hap2 mutant rescued with each of the three types of cDNA are shown in Figure 1. It is clear that none of them grows as well as the parental strain with the wild-type HAP2 gene (MY68), and also that P3 is less effective at rescue than either P1 or P5.
Figure 1.
Complementation of yeast hap2 with AtHAP2 cDNAs. S. cerevisiae strain JO1-1a (MY69) was transformed with three independent Arabidopsis HAP2 cDNAs (P1, P3, and P5 for AtHAP2a, AtHAP2b, and AtHAP2c, respectively) in the yeast expression vector pFL61. After primary selection on medium without uracil, two different dilutions of transformants were plated onto lactate medium along with Δhap2 itself (MY69) and the parental strain with a wild-type HAP2 gene (MY68). A reduced level of complementation was observed for AtHAP2b (P3).
The three classes of clones were used to transform yeast hap2, hap3, hap4, and hap5 mutants in turn, along with the double-mutant hap2,3, followed by selection for complementation on lactate medium. All three clones were capable of recomplementing the hap2 mutant, although transformants from each clone showed different extents of growth on lactate medium, as observed previously. In contrast, as expected, the three clones were unable to rescue any of the other yeast mutants, indicating that they were not general transcriptional activators, which are able to act downstream of the HAP complex. Consequently, the clones P1, P3, and P5 were termed AtHAP2a, AtHAP2b, and AtHAP2c, respectively.
Arabidopsis Contains Multiple, Distinct HAP2 Genes
The sequence of each of the three characterized cDNA clones was determined on both strands; the EMBL accession numbers for the sequences are Y13720, Y13721, and Y13722 for AtHAP2a, AtHAP2b, and AtHAP2c, respectively. AtHAP2a is 1388 bp long and encodes a single open reading frame of 271 amino acids; AtHAP2b (1385 bp) encodes a protein of 295 amino acids; and AtHAP2c (1516 bp) encodes a protein of 340 amino acids. Both AtHAP2a and AtHAP2c cDNAs possesses unusually long 5′ untranslated sequences (279 and 255 bp, respectively) before the first ATG codon, whereas AtHAP2b had multiple ATG start codons 5′ of the predicted coding region, followed by in-frame stop codons.
Comparisons between the predicted AtHAP2 protein sequences and those from other organisms identified conservation of two domains within each of the sequences (Fig. 2) that have been shown to be HAP2 specific and required for HAP2 function (Olesen and Guarente, 1990). However, outside of these regions, as has been observed for HAP2 homologs from other organisms, the three Arabidopsis proteins were quite distinct both in sequence and in length (data not shown). Furthermore, the spacing between the subunit-association and DNA-binding domains was four amino acids longer in AtHAP2b than in AtHAP2a or AtHAP2c. This is unusual because the length of the spacer region has been conserved between HAP2 homologs from different organisms, except for Schizosaccharomyces pombe, which has an additional amino acid. This increased spacer length correlates with the reduced ability of AtHAP2b (P3) to rescue the yeast hap2 mutant (Fig. 1).
Figure 2.
Protein sequence alignment of the subunit association and DNA-binding domains of HAP2 from a range of organisms. Numbering relates to AtHAP2b. Sequences were identified in the EMBL database (accession numbers are given in the right column) and were aligned using the Lineup program (Genetics Computer Group). Dots indicate gaps in the sequence; dashes indicate identity with AtHAP2a. At, Arabidopsis; Bn, B. napus; Sc, S. cerevisiae; Kl, Kluyveromyces lactis; Sp, S. pombe; Hm, human; Ms, mouse; Rt, rat; Sm, Schistosoma mansoni; and Su, sea urchin.
Identification of Arabidopsis HAP3 and HAP5 Homologs by Sequence Similarity
Screening of the EMBL nucleotide sequence database led to the identification of Arabidopsis ESTs, which showed considerable sequence similarity to HAP3 (ESTs T45165 and H76589) and HAP5 (ESTs T44300 and T43909) from other organisms. All cDNAs were obtained and sequenced on both strands.
Comparisons of predicted protein sequences with HAP3 and HAP5 homologs from other organisms identified extensive homology in domains that have been shown to be specific and required for HAP function (Fig. 3). The cDNAs identified here were given the names AtHAP3a, AtHAP3b, AtHAP5a, and AtHAP5b; their EMBL accession numbers are Y13723, Y13724, Y13726, and Y13725, respectively. In addition, distinct HAP3 and HAP5 genes were recently identified in the Arabidopsis genome sequence and are referred to as AtHAP3c (accession no. Z97336), AtHAP3d, and AtHAP5c (both on bacterial artificial chromosome F7G19). AtHAP3a (832 bp) and AtHAP3b (874 bp) encode open reading frames of 141 and 187 amino acids, whereas AtHAP5a (716 bp) and AtHAP5b (584 bp) encode open reading frames of 155 and 135 amino acids, respectively. The open reading frame in AtHAP3a is the only one that starts with a Met, suggesting that the ESTs encoding AtHAP3b, AtHAP5a, and AtHAP5b are truncated clones.
Figure 3.
Alignment of HAP3 and HAP5 core sequences from different organisms. Sequences were identified in the EMBL database (accession numbers are given in the right column). The symbols in the top line (/ / / / / / /********///////) denote the position of the predicted histone fold triple helix. Numbering refers to the position within AtHAP3a or S. cerevisiae HAP5. a, Alignment of HAP3 sequences. b, Alignment of HAP5 sequences. It is clear that AtHAP5a is incomplete because it is missing the first of the conserved helices. Dots indicate gaps in the sequence; dashes indicate sequence identity. At, Arabidopsis; Mz, maize; Sc, S. cerevisiae; Kl, K. lactis; Sp, S. pombe; En, Emericella nidulans; Hm, human; Ms, mouse; Rt, rat; Td, toad; Ch, chicken; Lm, lamprey; and An, Aspergillus nidulans.
Comparison between the Arabidopsis HAP3 or HAP5 homologs showed a high degree of sequence identity, with the level of sequence similarity being greatest within a central core domain. Sequences that encode HAP3 and HAP5 homologs from other organisms were retrieved from the EMBL database and aligned with their respective homologs. This confirmed the presence of highly conserved core domains within HAP3 and HAP5 (Fig. 3), flanked by sequences with a lower level of sequence identity and variable length, as was found for HAP2. The conserved domain lies within the predicted histone fold motif, a structural triple helix important for dimerization (Arents and Moudrianakis, 1995; Baxevanis et al., 1995).
Expression of Arabidopsis HAP Homologs
Northern-blot analysis was carried out on RNA from different Arabidopsis tissues using each AtHAP cDNA as a probe, under conditions in which the homologs do not cross-hybridize (Fig. 4). Five different tissue samples were chosen to identify potential tissue-specific or environmental regulation of gene expression. RNA was extracted from leaves (lane 1), from a mixture of flowers and siliques (lane 2), from roots (lane 3), from whole seedlings grown under a regular 16-h day (lane 4), and from whole seedlings grown for 48 h in the dark (lane 5). For each of the cDNAs analyzed, it was necessary to expose the autoradiographs for several days before a signal was detectable, indicating that the HAP genes are expressed at low levels.
Figure 4.
Expression of AtHAP cDNAs in various tissues from Arabidopsis. RNA was extracted from leaves (lane 1), flowers and siliques (lane 2), roots (lane 3), whole seedlings grown under a 16-h day/8-h night (lane 4), and whole seedlings grown in the dark for 48 h (lane 5). Ten micrograms of total RNA was loaded onto each lane, separated by electrophoresis, and blotted onto a nylon membrane. Membranes were probed with each radiolabeled AtHAP cDNA, followed by autoradiography. The bands were quantified by densitometry and normalized against the hybridization of an rDNA probe to the same filter. The relative values are shown in the histograms to the right of the autoradiographs. The sizes of the transcripts in kilobase pairs are indicated on the left.
All three AtHAP2 genes appeared to be expressed ubiquitously in each of the tissues analyzed (Fig. 4). The sizes of the transcripts detected were comparable to those of the isolated cDNAs, suggesting that the clones are full length or nearly full length.
In contrast, a differential expression pattern was detected for both AtHAP3a and AtHAP3b (Fig. 4). AtHAP3a was predominantly expressed in the flower/silique sample (lane 2), with an estimated transcript size of approximately 700 nt, indicating that the cDNA encodes the full-length transcript and that its expression is developmentally regulated. Significant levels of AtHAP3b expression were detected in both leaf and flower/silique samples of soil-grown plants (lanes 1 and 2), with low levels of expression detected in the remaining samples. The whole seedlings (grown in liquid culture) contained a large proportion of leaf material, so the absence of expression in this sample (lane 4) may be contrasted to its presence in the leaves of soil-grown plants (lane 1).
AtHAP5a expression was detected at an equal level in all samples, with a transcript size of approximately 1400 nt, confirming that this cDNA clone does not encode the full- length transcript. Two transcripts, approximately 1400 and 600 nt long, were detected in each sample after hybridization with AtHAP5b cDNA (Fig. 4). The larger transcript was expressed equally in each sample, whereas the smaller transcript was more abundant in leaf and reproductive tissue. The presence of two transcripts was unexpected because each of the other HAP cDNAs detected only a single mRNA species. This correlates with the results found when genomic Southern blots were probed with AtHAP5b: a complex pattern was observed (Fig. 5, right), suggesting that more than one gene encodes this isoform. In contrast, each of the other HAP cDNAs appeared to be single-copy genes, as illustrated by AtHAP5a (Fig. 5, left).
Figure 5.
Southern-blot analysis of Arabidopsis genomic DNA. Ten micrograms of DNA was digested with EcoRI (lane 1), BamHI (lane 2), or HindIII (lane 3), separated by gel electrophoresis, and blotted onto a nylon membrane. This was probed with radiolabeled AtHAP5a or AtHAP5b cDNAs, followed by autoradiography. The sizes of the bands in kilobase pairs are indicated to the right of each blot.
From this analysis it can be concluded that the HAP2 and HAP5 genes are not expressed differently in different tissues, in contrast to both HAP3 isoforms. Furthermore, there is no effect of light on the expression of any of the HAP genes.
DISCUSSION
CCAAT boxes are a feature of gene promoters in many eukaryotes, and analysis of several plant gene promoters has indicated the presence of CCAAT-box motifs, which contribute to gene expression (Rieping and Schöffl, 1992; Kehoe et al., 1994; Ito et al., 1995). A protein complex conserved in both yeast and vertebrates has been shown to bind to the CCAAT motif. In yeast the complex is composed of the three proteins HAP2, HAP3, and HAP5, and the mammalian complex NF-Y (also known as CBF or CP1) has equivalent subunits sharing conserved domains with the yeast proteins. A homolog of yeast HAP2 has been reported from B. napus (Albani and Robert, 1995), as has a protein with similarity to HAP3 from maize (Li et al., 1992). However, there has been no systematic examination of HAP-related proteins in plants; in particular, plant HAP5 homologs have not been identified, nor have the three necessary components for HAP complex function been isolated from a single plant species. We have used both complementation and computational approaches to examine the presence and diversity of these genes in Arabidopsis, and show that Arabidopsis contains at least three isoforms of each of the HAP complex components.
Multiple Members of the HAP Family Are Present in Arabidopsis
Functional complementation of a yeast hap2 mutant with an Arabidopsis cDNA library led to the isolation of three independent AtHAP2 cDNAs. This method has the advantage of isolating only functional, usually full-length cDNAs, as well as the ability to isolate clones that have sequence divergence but maintain the same function (Murray and Smith, 1996). Sequencing, northern-blot analysis, and the presence of putative initiation ATG codons suggest that these are independent, full-length, functional Arabidopsis HAP2 homologs. A search of the EMBL nucleotide database led to the identification of seven more independent and distinct Arabidopsis HAP homologs, four as ESTs (AtHAP3a, AtHAP3b, AtHAP5a, and AtHAP5b) and three as genomic sequences (AtHAP3c, AtHAP3d, and AtHAP5c). The identification of multiple and distinct genes for each HAP homolog contrasts with the situation in yeast and vertebrates, in which only one form of each homolog has been identified. This raises the possibility that these factors adopt more complex roles in plants and suggests that this transcription-factor family may have particular significance for the regulation of plant gene expression.
Sequence Relationships of the Plant HAP Homologs
Comparative alignment of the conserved domains from a range of HAP2s identified residues that have previously been shown to be functionally important by mutational analysis. Although many of the residues for subunit association characterized in yeast mutation studies (Xing et al., 1994) have been conserved, there is a greater variation between the Arabidopsis homologs than between animal and yeast HAP2 (Fig. 6). This is exemplified by the various residues equivalent to position 147 in AtHAP2b (Fig. 2). In organisms other than plants, this relative position is occupied by an Arg residue. Mutation of this residue in the yeast HAP2 protein to Pro, Leu, or Gly leads to a greatly reduced ability to associate with the HAP3,5 dimer and to form a functional complex (Xing et al., 1994). Each AtHAP2 contains a short aliphatic residue at this position, either Gly or Ala, which by comparison with the mutated yeast protein should inhibit their ability to form a functional HAP complex and complement the yeast hap2 mutant. The ability of these AtHAP2s to complement the yeast hap2 mutant means that the inhibitory aspect of this amino acid change must presumably be compensated for by other changes in the sequence.
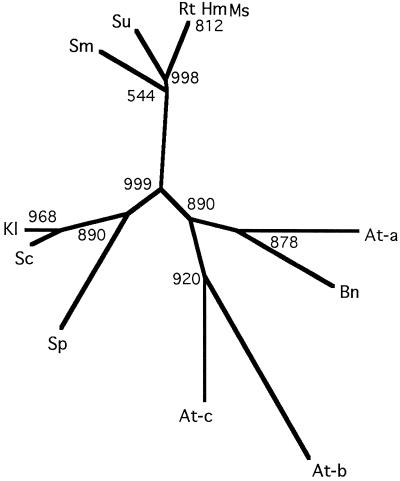
Figure 6.
Unrooted phylogenetic tree relating HAP2 homologs from different organisms. Multiple sequence alignment and generation of the tree was the program ClustalW (Genetics Computer Group), excluding gap positions and with correction for multiple substitutions. Bootstrap values are for 1000 replicates. Abbreviations are as in Figure 2.
The variation within the subunit-association domains of each AtHAP2 may allow specific recognition between each AtHAP2 and AtHAP3,5 dimer, although there is currently no evidence for such discrimination. Based on their functional complementation of the yeast hap2 mutant, each AtHAP2 is capable of forming a functional complex with the yeast HAP3,5 dimer, indicating that any such specificity must be subtle.
The predicted structures for AtHAP3 and AtHAP5 indicate the presence of putative TATA-box-binding protein association domains (Bellorini et al., 1997) and the histone fold motif (Arents and Moudrianakis, 1995; Baxevanis et al., 1995). The positions of residues within the histone fold motif can be directly compared with their homologous residues within the crystal structure of core histone proteins. The conservation of dimerization residues between species indicates that each AtHAP3 or AtHAP5 should recognize either of its respective partners and would be likely to interact with a dimerization partner from another species. This, and the ability of each AtHAP2 to rescue the hap2 yeast mutant, indicates that complex formation may occur through any combination of each subunit. However, the specificity of the subunit combinations may be determined by more subtle mechanisms, such as further protein-protein interactions or the availability of each of the components.
Although the core structure of the HAP complex has been highly conserved throughout evolution, the exact mechanism for transcriptional activation and the role of this complex in gene regulation seems to have evolved to suit the specific regulatory requirements of particular groups of organisms, with a greater complexity of HAP use and interactions in multicellular organisms than in yeast.
Expression and Potential Role of the HAP Complex in Plants
The expression patterns of the characterized AtHAP3 clones suggests a complex role in both the developmental and the environmental regulation of gene expression. In the absence of AtHAP3a or AtHAP3b, presumably, either another form such as AtHAP3c or AtHAP3d associates to produce a heterotrimer or no complex is formed. The expression of AtHAP3b in leaves from plants grown in soil but not in those from liquid culture may suggest environmental regulation of this gene (Fig. 4, compare lanes 1 and 4), perhaps in relation to osmotic stress. Northern-blot analysis of plants grown in tissue culture on different carbon sources (data not shown) indicates that there is no Glc repression of transcription, as observed for yeast HAP2 (de Winde and Grivell, 1993). Currently, the exact nature of this regulation remains elusive, and further research is required to understand the regulation of these factors and their role in developmental and environmental responses.
The differential expression observed for AtHAP3a and AtHAP3b is in contrast to that for each AtHAP2 and AtHAP5 cDNA, in which there appears to be little variation in all of the RNA samples analyzed. The ubiquitous expression of AtHAP2 and AtHAP5 cDNAs in a wide range of tissues suggests either a general role for these forms or regulation at the posttranscriptional level. Furthermore, no evidence for differential splicing for the AtHAP2 was obtained, in contrast to that seen for B. napus HAP2 (Albani and Robert, 1995).
The presence of multiple forms of each AtHAP indicates that gene duplication followed by divergence may have increased the CCAAT-box-binding transcription factor repertoire in plants. The various combinations of subunits could, in principle, allow the combinatorial modulation of transcription. The lack of protein sequence similarity outside of the conserved HAP domains required for subunit association indicates that each homolog may interact with its own set of associating factors, leading to a specific, modulated response. This possibility is currently the focus of further analysis.
ACKNOWLEDGMENTS
We are very grateful to Dr. L. Guarente for supplying yeast strains, including the mutant JP1-1C, to Dr. F. Lacroute for the Arabidopsis cDNA library in pFL61, and to the Arabidopsis Biological Resource Center (Ohio State University, Columbus) for the EST clones.
Abbreviations:
- AtHAP
Arabidopsis HAP subunit
- EST
expressed sequence tag
- nt
nucleotide
- UAS
upstream activating sequence(s)
LITERATURE CITED
- Albani D, Robert LS. Cloning and characterization of a Brassica napus gene encoding a homologue of the B-subunit of a heteromeric CCAAT-binding factor. Gene. 1995;167:209–213. doi: 10.1016/0378-1119(95)00680-x. [DOI] [PubMed] [Google Scholar]
- Arents G, Moudrianakis EN. The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc Natl Acad Sci USA. 1995;92:11170–11174. doi: 10.1073/pnas.92.24.11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxevanis AD, Arents G, Moudrianakis EN, Landsman D. A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res. 1995;23:2685–2691. doi: 10.1093/nar/23.14.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker DM, Fikes JD, Guarente L. A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc Natl Acad Sci USA. 1991;88:1968–1972. doi: 10.1073/pnas.88.5.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellorini M, Kun Lee D, Dantonel JC, Zemozoumi K, Roeder RG, Tora L, Mantovani R. CCAAT binding NF-Y-TBP interactions: NF-YB and NF-YC require short domains adjacent to their histone fold motifs for association with TBP basic residues. Nucleic Acids Res. 1997;25:2174–2181. doi: 10.1093/nar/25.11.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Church G, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winde JH, Grivell LA. Global regulation of mitochondrial biogenesis in Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1993;46:51–91. doi: 10.1016/s0079-6603(08)61018-1. [DOI] [PubMed] [Google Scholar]
- Dorn A, Bollekens J, Staub A, Benoist C, Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987;50:863–872. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Guarente L. Identification and characterization of HAP4: a 3rd component of the CCAAT-bound HAP2 HAP3 heteromer. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- Gelinas R, Endlich B, Pfeiffer C, Yagi M, Stamatoyannopoulos G. G-substitution to A-substitution in the distal CCAAT box of the gamma-globin gene in Greek hereditary persistence of fetal hemoglobin. Nature. 1985;313:323–325. doi: 10.1038/313323a0. [DOI] [PubMed] [Google Scholar]
- Gietz D, Stjean A, Woods RA, Schiestl RH. Improved method for high-efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L, Lalonde B, Gifford P, Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984;36:503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Hahn S, Pinkham J, Wei R, Miller R, Guarente L. The HAP3 regulatory locus of Saccharomyces cerevisiae encodes divergent overlapping transcripts. Mol Cell Biol. 1988;8:655–663. doi: 10.1128/mcb.8.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Fujimoto Y, Nakayama T, Iwabuchi M. A far-upstream sequence of the wheat histone H3 promoter functions differently in rice and tobacco cultured cells. Plant Cell Physiol. 1995;36:1281–1289. [PubMed] [Google Scholar]
- Kehoe DM, Degenhardt J, Winicov I, Tobin EM. Two 10-bp regions are critical for phytochrome regulation of a Lemna gibba LHCb-gene promoter. Plant Cell. 1994;6:1123–1134. doi: 10.1105/tpc.6.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Mantovani R, Vanhuijsduijnen RH, Andre I, Benoist C, Mathis D. Evolutionary variation of the CCAAT-binding transcription factor NF-Y. Nucleic Acids Res. 1992;20:1087–1091. doi: 10.1093/nar/20.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity SN, Vuorio T, Decrombrugghe B. The B-subunit of a rat heteromeric CCAAT-binding transcription factor shows a striking sequence identity with the yeast HAP2 transcription factor. Proc Natl Acad Sci USA. 1990;87:5378–5382. doi: 10.1073/pnas.87.14.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb DS, Xing YY, Guarente L. Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 1995;9:47–58. doi: 10.1101/gad.9.1.47. [DOI] [PubMed] [Google Scholar]
- Minet M, Dufour M, Lacroute F. Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 1992;2:417–422. doi: 10.1111/j.1365-313x.1992.00417.x. [DOI] [PubMed] [Google Scholar]
- Muro AF, Bernath VA, Kornblihtt AR. Interaction of the −170-cyclic AMP response element with the adjacent CCAAT box in the human fibronectin gene promoter. J Biol Chem. 1992;267:12767–12774. [PubMed] [Google Scholar]
- Murray JAH, Smith AG. Functional complementation in yeast and E. coli. In: Foster GD, Twell D, editors. Plant Gene Isolation: Principles and Practice. Chichester, UK: John Wiley & Sons; 1996. pp. 177–211. [Google Scholar]
- Olesen JT, Guarente L. The HAP2 subunit of yeast CCAAT transcriptional activator contains adjacent domains for subunit association and DNA recognition: model for the HAP2/3/4 complex. Genes Dev. 1990;4:1714–1729. doi: 10.1101/gad.4.10.1714. [DOI] [PubMed] [Google Scholar]
- Pinkham JL, Guarente L. Cloning and molecular analysis of the HAP2 locus: a global regulator of respiratory genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:3410–3416. doi: 10.1128/mcb.5.12.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkham JL, Olesen JT, Guarente LP. Sequence and nuclear localization of the Saccharomyces cerevisiae HAP2 protein, a transcriptional activator. Mol Cell Biol. 1987;7:578–585. doi: 10.1128/mcb.7.2.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymondjean M, Cereghini S, Yaniv M. Several distinct CCAAT box binding-proteins coexist in eukaryotic cells. Proc Natl Acad Sci USA. 1988;85:757–761. doi: 10.1073/pnas.85.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieping M, Schöffl F. Synergistic effect of upstream sequences, CCAAT box elements, and HSE sequences for enhanced expression of chimeric heat-shock genes in transgenic tobacco. Mol Gen Genet. 1992;231:226–232. doi: 10.1007/BF00279795. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Santoro C, Mermod N, Andrews PC, Tjian R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988;334:218–224. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High-efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Sinha S, Maity SN, Lu JF, Decrombrugghe B. Recombinant rat CBF-C, the 3rd subunit of CBF/N-FY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc Natl Acad Sci USA. 1995;92:1624–1628. doi: 10.1073/pnas.92.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasanen K, Oikarinen J, Kivirikko KI, Pihlajaniemi T. Promoter of the gene for the multifunctional protein disulfide isomerase polypeptide: functional significance of the 6 CCAAT boxes and other promoter elements. J Biol Chem. 1992;267:11513–11519. [PubMed] [Google Scholar]
- Vuorio T, Maity SN, Decrombrugghe B. Purification and molecular cloning of the A-chain of a rat heteromeric CCAAT-binding protein: sequence identity with the yeast HAP3 transcription factor. J Biol Chem. 1990;265:22480–22486. [PubMed] [Google Scholar]
- Xing YY, Zhang SU, Olesen JT, Rich A, Guarente L. Subunit interaction in the CCAAT-binding heteromeric complex is mediated by a very short alpha-helix in HAP2. Proc Natl Acad Sci USA. 1994;91:3009–3013. doi: 10.1073/pnas.91.8.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]