Abstract
Background.Entamoeba moshkovskii is prevalent in developing countries and morphologically indistinguishable from pathogenic Entamoeba histolytica and nonpathogenic Entamoeba dispar. It is not known if E. moshkovskii is pathogenic.
Methods.Mice were intracecally challenged with the trophozoites of each Entamoeba spp. to test the ability to cause diarrhea, and infants in Bangladesh were prospectively observed to see if newly acquired E. moshkovskii infection was associated with diarrhea.
Results.E. moshkovskii and E. histolytica caused diarrhea and weight loss in susceptible mice. E. dispar infected none of the mouse strains tested. In Mirpur, Dhaka, Bangladesh, E. moshkovskii, E. histolytica, and E. dispar were identified in 42 (2.95%), 66 (4.63%), and 5 (0.35%), respectively, of 1426 diarrheal episodes in 385 children followed prospectively from birth to one year of age. Diarrhea occurred temporally with acquisition of a new E. moshkovskii infection: in the 2 months preceding E. moshkvskii-associated diarrhea, 86% (36 of 42) of monthly surveillance stool samples were negative for E. moshkovskii.
Conclusions.E. moshkovskii was found to be pathogenic in mice. In children, the acquisition of E. moshkovskii infection was associated with diarrhea. These data are consistent with E. moshkovskii causing disease, indicating that it is important to reexamine its pathogenicity.
Entamoeba histolytica causes extensive mortality and morbidity worldwide through diarrheal disease and abscess formation in parenchymal tissues such as liver, lung, and brain. In contrast, other amoebae that infect humans include Entamoeba dispar, Entamoeba moshkovskii, Entamoeba coli, Entamoeba hartmanni, and Endolimax nana, which have been considered nonpathogenic commensals of the human gut [1–3]. Dientamoeba fragilis and Entamoeba polecki have been associated with diarrhea and Entamoeba gingivalis with periodontal disease [4, 5].
E. moshkovskii is genetically related to E. histolytica and E. dispar and is microscopically indistinguishable from them in its cyst and trophozoite forms [6]. This species of Entamoeba was first identified in sewage in Moscow by Tshalaria in 1941 [7] and was initially thought to be a free-living common protozoan species in anoxic sediments and in environments such as brackish coastal pools. The first human isolate was obtained from a resident of Laredo, Texas, who suffered from diarrhea, weight loss, and epigastric pain in 1961 [8]. This finding would seem to suggest and/or support that E. moshkovskii can be pathogenic. At first, this isolate was named E. histolytica Laredo strain and shared biological features with E. moshkovskii. Both the Laredo strain and E. moshkovskii grow at room temperature and were resistant to osmotic shock and to drugs used in the chemotherapy of amoebiasis such as emetine [9]. Subsequent molecular studies revealed that E. histolytica Laredo is identical with E. moshkovskii [10].
E. moshkovskii is a common Entamoeba infection in humans in some settings. It is composed of anywhere from as little as 1% to as high as 50% of the E. histolytica/E. dispar/E. moshkovskii complex parasites detected in fecal samples in limited studies from Australia, Bangladesh, India, Iran, Tanzania, and Turkey [6, 11–16]. These studies for the most part tested stool samples submitted to clinical microbiology laboratories from patients with gastrointestinal symptoms, suggesting that E. moshkovskii could cause disease. However, in HIV-1–infected individuals in northern Tanzania, E. moshkovskii was not associated with enteric symptoms nor immune status [17]. Thus, the ability of E. moshkovskii to cause disease in humans remains unclear.
Here we tested the ability of E. moshkovskii to cause colitis and diarrhea in a murine model system, in which intracaecal inoculation with E. histolytica trophozoites into CBA/J, C3H/HeN, and C3H/HeJ mice leads to amebic colitis [18–20]. In addition, we tested in a longitudinal study of children in Bangladesh not only if E. moshkovskii was present in stool samples from infants with diarrhea, but whether the E. moshkovskii infection was newly acquired at the time of the diarrheal illness.
MATERIALS AND METHODS
Mice
Male CBA/J, C57BL6/J, BALB/c, C3H/HeN, and C3H/HeJ mice were purchased from the Jackson Laboratory. Animals were maintained under specific pathogen-free conditions at Animal Research Center for Tropical Infectious Diseases, Nagasaki University, and were challenged when they were 5–8 weeks old.
Cultivation of Entamoeba spp.
Trophozoites of the E. moshkovskii Laredo strain were a gift from Dr Seiki Kobayashi, Keio University, School of Medicine (originally from the late Professor Louis S. Diamond, National Institutes of Health, Bethesda, Maryland). Trophozoites of E. histolytica, originally laboratory strain HM1:IMSS (American Type Culture Collection, Manassas, Virginia), were from Professor Eric Houpt, University of Virginia, which were sequentially passaged in vivo through the mouse cecum [18]. Cecal contents were cultured at 25° and 37°C, respectively, in BIS-33 medium supplemented with heat-inactivated 10% adult bovine serum, 25 U/mL penicillin, and 25 mg/mL streptomycin [21]. Trophozoites of E. dispar AS16IR were also provided by Dr Seiki Kobayashi and cultured in YIMDHA-S media at 37°C. Trophozoites under log phase of growth were used in the experiments.
Intracecal Inoculation of Entamoeba spp.
Trophozoites were harvested from culture tubes of E. histolytica HM1:IMSS, E. moshkovskii Laredo and E. dispar AS16IR strains by incubating the tubes on ice for 5–10 minutes. Then, the trophozites were collected, and the number of trophozoites was determined. We anesthetized mice with domitor (medetomidine hydrochloride: 0.1 mg/kg) and dormicum (midazolam: 0.1 mg/kg), shaved their abdomens to incise the skin and exteriorized each cecum from the peritoneum, and injected 150 µL of 1 × 106 each trophozoites into the proximal, middle, and apical sites of cecum. Then the cecum was blotted and the peritoneum and the skin were sutured. Mice were kept on warming blankets at 37°C throughout. Survival rates were ≥85% in all strains. The study was approved by the animal ethical review board of Nagasaki University.
PCR Amplification for Diagnosis of Entamoeba spp. Infection in Mice
For isolation of Entamoeba DNA from mouse stools, QIAamp DNA Stool kits (QIAGEN, Valencia, California) were used according to manufacturer's instructions. The primer sequences used for polymerase chain reaction (PCR) were described elsewhere [22].
Pathology of Murine Amoebic Colitis
At the indicated days after intracecal challenge, mice were killed, the ceca fixed in phosphate buffered 10% formalin, and then cut into 4–6 equal cross-sections and embedded in paraffin, and 4 µm slides were stained with H&E.
Child Study Area and Population
The study was conducted in Mirpur, an urban slum in Dhaka. Infants were enrolled in the first week after birth and followed until one year of age, beginning in January 2008. Field research assistants (FRAs) visited each study house every other day and collected information related to child morbidity, especially for diarrheal illness, through a structured questionnaire. If the FRA found any child with an acute illness, then she referred the child to the study clinic for further management by the medical officer. Parents or guardians were also encouraged to visit the study clinic for medical assistance if the study child became sick. FRAs collected nondiarrheal monthly stool specimens as well as diarrheal stool specimens from the home or in the study field clinic. All stool specimens were transported from the field to the clinic using a cold box. In the field clinic an aliquot of the diarrheal stool specimens was placed into Carry-Blair medium. All specimens were transported from the field clinic to the ICDDR,B Parasitology laboratory within 3 hours of collection, with a cold chain maintained. Diarrhea was defined as having ≥3 unformed or abnormal stools (as per the mother's perception) in a 24-hour period. A diarrheal episode was defined as being separated from another episode by at least 3 diarrhea-free days.
The study was approved by the Institutional Review Board of the University of Virginia, and the Ethical Review Committee of the International Centre for Diarrhoeal Disease Research, Bangladesh. Informed written consent was obtained from the parents or guardians for the participation of their child in the study.
Detection of Enteropathogens
Stool samples were cultured for enteric pathogens including Vibrio cholerae O1/O139, Salmonella spp., Shigella spp., and Campylobacter jejuni. Enzyme-linked immunosorbent assay (ELISA) methods were used to detect LT and ST producing enterotoxigenic E. coli (ETEC) [23]. Entamoeba histolytica, Cryptosporidium, and Giardia were identified by real-time PCR as described elsewhere [24]. Rotavirus, astrovirus, and adenovirus were detected by ELISA using commercial kits (ProSpectT Rotavirus Catalog R240396, ProSpectT Astrovirus Catalog R240196, and ProSpectT Adenovirus Catalog R240096, respectively). Multiplex (RT-)PCR and probe-based detection with Luminex beads for conceivable diarrhea-causative microbes was performed as described elsewhere in the literature [25–27].
The DNA was extracted using a slightly modified QIAamp DNA Stool Mini Kit protocol (Qiagen Inc, Valencia, California) [24]. The RNA was extracted using the QuickGene RNA tissue kit SII [25, 26]. For the (RT-)PCR-Luminex assay, either the forward or the reverse primer per target was labeled with biotin-TEG at 5′ ends. After (RT-)PCR was performed with the conditions described elsewhere, samples were analyzed on the BioPlex-200 system using bead on which coupling and hybridization were performed according to published protocols [28].
Amplification of ArgTCT Gene Fragment and Sequencing
The E. moshkovskii–specific primer pair, EmR-1 and EmR-2, was used to specifically amplify the E. moshkovskii ArgTCT gene fragment [13]. Amplification was performed using the high-fidelity Sahara DNA polymerase (Bio-Line, US). Sequencing was performed on an Applied Biosystems 377 Prism DNA Sequencer, using the BigDye terminator chemistry and EmR-1 or EmR-2 primer.
Statistical Analysis
The χ2 test and Mann–Whitney U test were used where they were applicable.
RESULTS
E. moshkovskii Established the Infection in Mice
We previously showed that C3H/HeN, C3H/HeJ, and CBA/J mice allowed the establishment of E. histolytica infection, whereas many strains of mice including C57BL/6 and BALB/c mice did not, indicating that susceptibility to E. histolytica infection depended on the genetic background of the host [18–20]. Trophozoites of either E. histolytica, E. moshkovskii, or E. dispar were intracecally injected into congenic strains of mice. E. histolytica successfully infected the ceca of C3H/HeN, C3H/HeJ, and CBA/J mice. E. moshkovskii infected the ceca of CBA/J mice in approximately 68% (51 of 75) of mice at 4 days after challenge, as determined by both culture and PCR of intracecal contents. Likewise, C3H/HeN and C3H/HeJ mice were infected with E. moshkovskii in 60% and 40% of cases at 4 days, respectively, whereas infection rates of C57BL/6 and BALB/c mice were 5.6% and 0.0% at 4 days, respectively (Table 1). Nonpathogenic E. dispar did not infect any mouse strain tested. These data demonstrated that in contrast to nonpathogenic E. dispar that did not infect, E. moshkovskii had a similar host genetic susceptibility to infection in the murine model as did pathogenic E. histolytica.
Table 1.
Susceptibility of Congenic Strains of Mice to Entamoeba histolytica, Entamoeba moshkovskii, or Entamoeba dispar Infection
| E. histolytica (%) | E. moshkovskii (%) | E. dispar (%) | |
|---|---|---|---|
| BALB/c | 1/15 (6) | 0/10 (0) | 0/10 (0) |
| C57BL/6 | 2/20 (10) | 1/18 (6) | 0/15 (0) |
| C3H/HeJ | 8/15 (53) | 4/10 (40) | 0/10 (0)a |
| C3H/HeN | 7/15 (47) | 6/10 (60) | 0/10 (0)a |
| CBA/J | 61/90 (68) | 51/75 (68) | 0/20 (0)a |
a P < .05 compared to E. histolytica or E. moshkovskii (χ2 test).
E. moshkovskii Induced Intestinal Symptoms in Mice
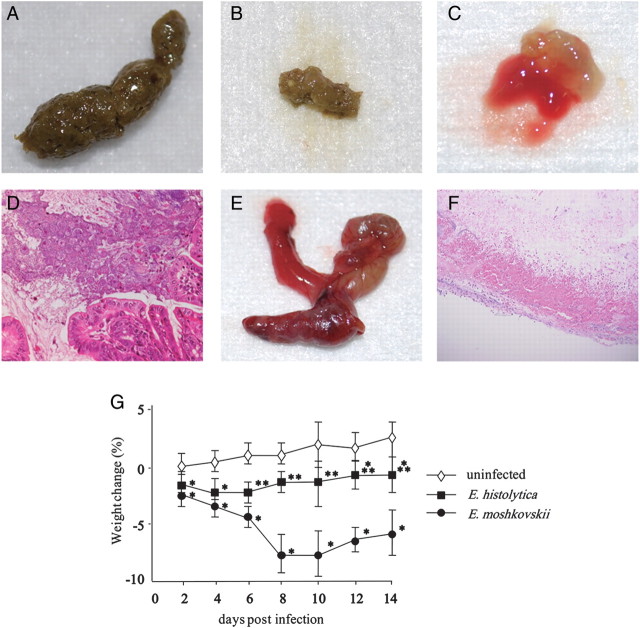
Intestinal symptoms and body weight were monitored after challenging CBA/J mice with E. moshkovskii. A total of 71% (51/72) of CBA/J mice inoculated with E. moshkovskii were infected by 3 days after challenge. Diarrhea was observed in 39% (20/51) and dysentery in 6% (3/51) (Figure 1A–C). In successfully infected mice, amoebae were observed in the lumen of the ceca (Figure 1D). Mice with bloody diarrhea exhibited a thickened and contracted ceca (Figure 1E). Histopathological examination of ceca from these mice revealed epithelial ulceration, hemorrhagic changes, and tissue destruction (Figure 1F). Furthermore, obvious weight loss was observed during the course of E. moshkovskii infection in CBA/J mice, which was more severe than that observed during pathogenic E. histolytica infection, both of which were significant compared to control sham-operated mice (Figure 1H). Together these data indicated that E. moshkovskii was virulent in mice.
Figure 1.
Entamoeba moshkovskii induced intestinal symptoms and weight loss in CBA/J mice. CBA/J mice were intracecally inoculated with 1 × 106 trophozoites of E. moshkovskii. After infection, diarrhea, colitis, and weight loss were monitored. Normal (A), loose (B), and bloody feces (C) were observed as was indicated in the results. Amoebae were observed in the lumen of the ceca in successfully infected mice (D). Macroscopic and histopathological observations of ceca in mice exhibited bloody diarrhea were shown in panels E and F. Changes in body weight were monitored in successfully infected 15 mice per group (G), in which CBA/J mice were intracecally inoculated with 1 × 106 trophozoites of Entamoeba histolytica (solid squares), E. moshkovskii (solid circles), or medium alone (open diamonds). The study was repeated 3 times with similar results. *P < 1.0 × 10−6, **P < 1.0 × 10−5 and ***P < 1.0 × 10−4 compared with sham-operated mice (Mann–Whitney U test).
E. moshkovskii Was Expelled Within 14 Days After Challenge, Whereas E. histolytica Chronically Infected in the Ceca of Mice
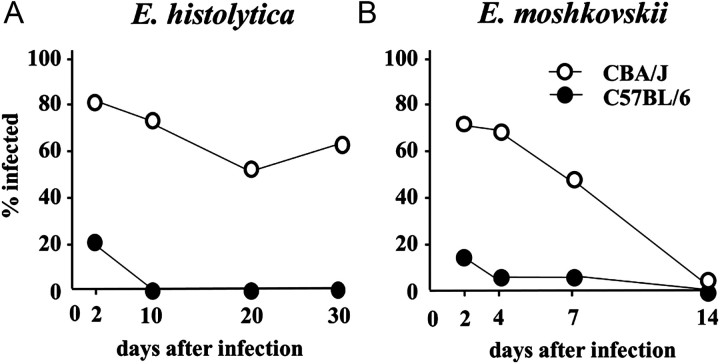
The time course of each Entamoeba spp. infection in susceptible strains of mice was observed. As was reported [20], E. histolytica established chronic infection in not only CBA/J but also C3H/HeJ and C3H/HeN mice, whereas neither C57BL/6 nor BALB/c allowed establishment of E. histolytica infection (Figure 2). In contrast, E. moshkovskii did not cause chronic infection, being expelled by approximately 2 weeks after challenge in CBA/J mice (Figure 2). A similar time to clearance was seen in C3H/HeN and C3H/HeJ mice.
Figure 2.
Entamoeba moshkovskii was expelled within 2 weeks in CBA/J Mice. CBA/J (open circles) and C57BL/6 (solid circles) mice were intracecally inoculated with 1 × 106 trophozoites of Entamoeba histolytica (A) or Entamoeba moshkovskii (B). Time course of each Entamoeba spp. infection was then monitored by detection of the parasites in stool by culture and polymerase chain reaction (PCR).
In Infants in Bangladesh, E. moshkovskii Was Detected in Diarrheal Samples With Similar Frequency to E. histolytica
The association between diarrheal episodes and infection with each Entamoeba spp. was tested in children in Mirpur, Dhaka, Bangladesh. These studies were part of a prospective cohort study on diarrheal diseases [29]. Newborn children were enrolled in the Mirpur community of Dhaka, Bangladesh, and prospectively followed for diarrheal illness by every other day home visits. A total of 1426 diarrheal episodes were recorded during the first 12 months of life in 385 children. PCR analyses of the diarrheal samples revealed that 66 episodes were positive for E. histolytica (4.63%), 42 were positive for E. moshkovskii (2.95%), and 5 episodes were positive for E. dispar (0.35%). As such, in diarrheal samples, the detection rates of either E. histolytica or E. moshkovskii were 13.2 and 8.4 times higher than that of nonpathogenic E. dispar. Two episodes were found to be mixed infections with E. histolytica and E. moshkovskii, but no other mixed infections of Entamoeba spp. were found.
E. moshkovskii Infection Was Newly Acquired in Children With Diarrhea
In order to attempt to discern if the E. moshkovskii detected in the diarrheal stool sample could be the cause of diarrhea, we tested if it was newly acquired at the time of diarrhea. The preceding 2 months of surveillance stool samples collected when the child did not have diarrhea were tested for the presence of E. moshkovskii. This study design therefore temporally controlled for E. moshkovskii infection in the 42 infants with diarrhea attributed to this parasite. In the 1 and 2 months preceding E. moshkvskii-associated diarrhea, 93% (39/42) and 86% (36/42) of monthly surveillance stool samples, respectively, were negative for E. moshkovskii (Table 2). This supported the hypothesis that temporal acquisition of a new E. moshkovskii infection led to diarrheal episodes in some proportion of these children.
Table 2.
Prevalence of Entamoeba moshkovskii Asymptomatic Infection Preceding E. moshkovskii-Associated Diarrhea in 42 Children
| Category | Preceding 1-Month Surveillance Stool | Preceding 2-Month Surveillance Stool |
|---|---|---|
| E. moshkovskii (+) | 3 | 6 |
| E. moshkovskii (−) | 39 | 36 |
| Total | 42 | 42 |
E. moshkovskii-Associated Diarrhea Was of Similar Severity to Other Causes of Diarrhea
The diarrheal severity score was comparable among episodes associated with E. histolytica, E. moshkovskii, and other causes: 4.89 ± 0.22, 4.71 ± 0.24, and 4.84 ± 0.05, respectively. The duration of diarrhea was also comparable among these episodes positive for E. histolytica, E. moshkovskii, and others: 4.44 ± 0.44, 4.74 ± 0.49, and 4.84 ± 0.10 days, respectively (Table 3). The mean age of the onset of diarrheal episodes associated with E. histolytica, E. moshkovskii, and others was found to be 7.72 ± 0.75, 9.12 ± 0.73, and 9.09 ± 0.18 months, respectively, without any significant differences. Thus, the diarrhea related to E. moshkovskii was indistinguishable from diarrhea related to E. histolytica in severity, duration, and age of onset (Table 3).
Table 3.
Severity and Duration of Diarrhea Associated With Entamoeba histolytica or Entamoeba moshkovskii
| Pathogens | Severity Score (mean ± SE) | Duration (days) (mean ± SE) | Age of Onset in Months (mean ± SE) |
|---|---|---|---|
| E. histolytica | 4.89 ± 0.22 | 4.44 ± 0.44 | 7.72 ± 0.75 |
| E. moshkovskii | 4.71 ± 0.24a | 4.74 ± 0.49a | 9.12 ± 0.73a |
| Others | 4.84 ± 0.05 | 4.84 ± 0.10 | 9.09 ± 0.18 |
a No significant difference in diarrheal severity score or duration for episodes associated with E. histolytica, E. moshkovskii, or other enteropathogens infection.
Additional Enteropathogens Were Identified in Stool Samples From E. moshkovskii Infected Children
As there are many microbes that can potentially induce diarrhea, the presence of other diarrheagenic microbes was tested in the 42 diarrheal samples that were associated with E. moshkovskii. The 42 samples were examined for other conceivable diarrhea-causative microbes infection using standard bacterial culture techniques, fecal antigen detection, and multiplex PCR combined with probe-based detection with Luminex beads (Table 4) [25, 26]. In the 42 diarrheal stool samples with E. moshkovskii, 12 samples (28.6%) contained >4 other pathogens, 13 (31.0%) had 3 pathogens, 14 (33.3%) had 2 pathogens, 1 (2.3%) had 1 pathogen, and 2 samples were positive solely for E. moshkovskii (Table 4). The application of these state-of-the-art diagnostic techniques in this cohort has on average identified a minimum of 2 different enteropathogens in every diarrheal stool sample (E. Houpt and M. Taniuchi, personal communication, 2011). It was therefore not surprising that the diarrheal episodes associated with E. moshkovskii were commonly coinfected.
Table 4.
Other Enteropathogens Detected in Entamoeba moshkovskii (+) Diarrheal Stool Samples
| Name of Organism | No. of Samples |
|---|---|
| Encephalitozoon intestinalis | 1 |
| Cyclospora cayetanensis | 1 |
| Cystoisospora belli | 1 |
| Enterocytozoon bieneusi | 6 |
| Adenovirus | 1 |
| Astrovirus | 4 |
| Sapovirus | 2 |
| Norovirus G1 | 0 |
| Norovirus G2 | 4 |
| Rotavirus | 1 |
| E. histolytica | 2 |
| Giardia intestinalis | 10 |
| Cryptosporidium spp. | 1 |
| Vibrio cholera/parahaemolyticus | 3 |
| EAEC | 15 |
| ETEC | 3 |
| EPEC | 5 |
| EHEC | 0 |
| EIEC/Shigella spp. | 23 |
| Salmonella (pan) | 2 |
| Aeromonas (pathogenic) | 14 |
| Yersinia (pan) | 0 |
| Campylobacter jejuni/coli | 23 |
E. moshkovskii Isolates Were Genetically Diverse in the Infants
In order to investigate the genetic diversity in E. moshkovskii strains detected in the infected children's stools, we used a tRNA-gene linked locus (R-R), which previously showed PCR size differences among E. moshkovskii strains from Bangladesh [12, 30]. Twenty-six E. moshkovskii-positive stool DNAs (6 from asymptomatic children and 20 from diarrheal children) were amplified using the E. moshkovskii specific nested PCR primers described elsewhere[12]. However, PCR did not reveal any obvious product size differences among these samples (data not shown). Because same size PCR products do not necessarily mean identical DNA sequences, we sequenced PCR products directly without cloning them into any vectors (in order to minimize the chances of any sequence selection) to detect sequence variation. Sequencing did reveal that the E. moshkovskii strains detected in this study were polymorphic in locus R-R; although unlike the E. histolytica and E. dispar sequences[31], no short tandem repeats could be detected in E. moshkovskii. Single-nucleotide polymorphisms (SNPs) were detected in 2 of the 6 asymptomatic children-derived sequences and in 6 of the 20 diarrheal children-derived sequences (Supplementary Figure 1). These SNPs could be used to divide them into 9 different genotypes—18 strains with identical locus R-R sequences and the remaining 8 strains containing ≥1 distinct SNPs (Supplementary Figure 1 and Table 5). Because we used a high-fidelity DNA polymerase (Bio-Line, US) during PCR amplification, it was unlikely that these SNPs were erroneously introduced by the DNA polymerase. The sequence alignment at locus R-R revealed that the E. moshkovskii strains of this study were comparatively more diverse than the reference E. moshkovskii Laredo strain, but closer to the only Bangladeshi strain (ID:MS15-3646) sequenced previously (labeled as Em-Laredo and Em-BANGLA, respectively, in Supplementary Figure 1). The SNPs detected in this study were distributed randomly across the locus R-R sequences, and as a result, these SNPs could not be used to differentiate asymptomatic and diarrheal strains of E. moshkovskii. However, we noticed from the sequence traces that the 2 asymptomatic strains (IDs:8056-CMS15 and 7086-CMS15) showed allelic variation in all 3 SNPs (T203W, T204Y, and A135R), whereas none of the 6 diarrheal strains showed any allelic variations in their respective SNPs (Table 5 and Supplementary Figure 2). The significance of this remains unknown at present.
Table 5.
Single-Nucleotide Polymorphisms (SNPs) in the E. moshkovskii Strains from Bangladesh at Locus R-R
| ID | Clinical Status | No. of SNPs | Position and SNP Type |
|---|---|---|---|
| 7040-CDS05 | Diarrhea | 6 | T204A, C205T, T206C, C208del, T209del, T210del |
| 7063-CDS02 | Diarrhea | 1 | T71C |
| 7161-CDS05 | Diarrhea | 1 | T141G |
| 7146-CDS02 | Diarrhea | 1 | A235T |
| 8119-CDS02 | Diarrhea | 2 | T83C, T137C |
| 8113-CDS04 | Diarrhea | 3 | T81C, C120T, G221A |
| 7086-CMS15 | Asymptomatic | 2 | T203W, T204Y |
| 8056-CMS15 | Asymptomatic | 1 | A135R |
All positions are based on the consensus sequence in the alignment. W = A/T; Y = C/T; R = A/G.
DISCUSSION
This work draws into question the paradigm that E. moshkovskii is avirulent. In the murine model of intestinal amebiasis, E. moshkovskii caused diarrhea, weight loss, and colitis. In this way, E. moshkovskii shared with E. histolytica, but not the nonpathogen E. dispar, the ability to cause disease. In children in Bangladesh, the new acquisition of E. moshkovskii infection was associated with diarrhea.
E. moshkovskii infected the ceca of C3H/HeN, C3H/HeJ, and CBA/J mice, but not C57BL/6 or BALB/c mice, which was consistent with the host range of pathogenic E. histolytica. In contrast, the nonpathogenic parasite E. dispar was unable to infect the intestine of any strains of mice tested. The finding that E. moshkovskii shared with E. histolytica the ability to infect mice indicates that they share virulence mechanisms, which are not present in E. dispar.
Mouse strain-dependent resistance to E. histolytica infection was mediated by nonhematopoietic cells [19]. Relatively few loci on C57BL/6 chromosomes 1 and 2 correlated with resistance to intestinal amebiasis [32]. In humans, one important means of innate resistance of intestinal epithelial cells to amebiasis is leptin, which acts via STAT3 signaling to protect intestinal epithelial cells from parasite killing [33, 34]. In this context, it will be interesting to examine whether this observation is also true in E. moshkovskii infection, because of the similar host range as E. histolytica. If the mechanism of resistance to E. histolytica and E. moshkovskii observed in many inbred strains of mice is shared with humans, identification of regional candidate genes in mice has implications for further understanding the human variability to amebic infection.
E. moshkovskii induced intestinal symptoms including diarrhea and bloody stool, typical symptoms of amebiasis, indicating that E. moshkovskii was pathogenically similar to E. histolytica at least in mice. Weight loss was also observed during the course of infection, which was more severe in mice infected with E. moshkovskii than with E. histolytica. The observation that E. moshkovskii induced severe intestinal symptoms accompanied by weight loss reemphasizes that it is potentially pathogenic.
However, it is unclear what kinds of differences among Entamoeba spp. result in the different outcomes of infection in the murine model. Entamoeba histolytica possesses molecules such as pathogen-associated molecular patterns (PAMPs) on its surface that stimulate proinflammatory cytokines production from antigen-presenting cells [35]. We are investigating whether parasite PAMPs, host MyD88 signaling, and the pattern of proinflammatory cytokines produced in response qualitatively differ between Entamoeba species, may provide clues to the different severity between CBA/J mice infected with E. histolytica, E. moshkovskii, and E. dispar.
Entamoeba moshkovskii isolates infecting children were genetically heterogeneous, as evidenced by PCR typing of tRNA locus R-R. It will be important in future studies of the potential pathogenicity of E. moshkovskii to take into account this heterogeneity; as for the case of E. histolytica, not every genotype is equally capable of causing disease [36].
The study subjects reported here differ from those of the previous study examining E. moshkovskii infection in preschool children in Dhaka, Bangladesh [13], which was not focused solely on diarrheal stool samples, but also included monthly stool samples from asymptomatic children. In addition, the current study reports on a novel birth cohort longitudinally followed from birth to 1 year of age. Therefore, it is important to discuss the association between diarrhea in infants and E. moshkovskii infection in the context of the cohort.
In conclusion, we found that E. moshkovskii caused diarrhea, colitis, and weight loss in mice and that in Bangladeshi children acquisition of a new E. moshkovskii infection occurred temporally with diarrhea. These data are consistent with E. moshkovskii causing diarrhea and indicate that it is important to reexamine its pathogenicity.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank M. Iwami-Sano and C. Matsuzaki-Moriya for technical assistance, F. Hara and M. Hayashida for animal husbandry, H. Hisaeda for comments, members of Department of Parasitology, Kyushu University and Institute of Tropical Medicine, Nagasaki University for helpful discussion, and members of Parasitology Laboratory, ICDDR, B., and all of field research assistants for helpful assistance in the field.
Financial support. This work was supported by NIH grant 5RO1 AI043596 (to W. A. P.), a Grant-in-Aid for Scientific Research on Priority Areas from MEXT (21022037 to S. H.), Grants-in-Aid for International Scientific Research (B) from JSPS (20406008, 23406009 to S. H.), a Health Labour Sciences Research Grant (H20-Shinkoh-Ippan-016, H23-Shinkoh-Ippan-014 to S. H.), the Takeda Foundation, the Uehara Foundation (to S. H.), and the Global COE Program, Nagasaki University, supported by MEXT (to S. H.).
Potential conflicts of interest. Dr Petri receives royalties from a licensing agreement with TechLab, Inc., for amebiasis diagnostics. These royalties are donated in their entirety to the American Society of Tropical Medicine and Hygiene without benefit to Dr Petri. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Irusen EM, Jackson TF, Simjee AE. Asymptomatic intestinal colonization by pathogenic Entamoeba histolytica in amebic liver abscess: prevalence, response to therapy, and pathogenic potential. Clin Infect Dis. 1992;14:889–93. doi: 10.1093/clinids/14.4.889. [DOI] [PubMed] [Google Scholar]
- 2.Blessmann J, Ali IK, Nu PA, et al. Longitudinal study of intestinal Entamoeba histolytica infections in asymptomatic adult carriers. J Clin Microbiol. 2003;41:4745–50. doi: 10.1128/JCM.41.10.4745-4750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gathiram V, Jackson TF. A longitudinal study of asymptomatic carriers of pathogenic zymodemes of Entamoeba histolytica. S Afr Med J. 1987;72:669–72. [PubMed] [Google Scholar]
- 4.Vandenberg O, Peek R, Souayah H, et al. Clinical and microbiological features of dientamoebiasis in patients suspected of suffering from a parasitic gastrointestinal illness: a comparison of Dientamoeba fragilis and Giardia lamblia infections. Int J Infect Dis. 2006;10:255–61. doi: 10.1016/j.ijid.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 5.Stark D, Beebe N, Marriott D, Ellis J, Harkness J. Prospective study of the prevalence, genotyping, and clinical relevance of Dientamoeba fragilis infections in an Australian population. J Clin Microbiol. 2005;43:2718–23. doi: 10.1128/JCM.43.6.2718-2723.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fotedar R, Stark D, Marriott D, Ellis J, Harkness J. Entamoeba moshkovskii infections in Sydney, Australia. Eur J Clin Microbiol Infect Dis. 2008;27:133–7. doi: 10.1007/s10096-007-0399-9. [DOI] [PubMed] [Google Scholar]
- 7.Scaglia M, Gatti S, Strosselli M, Grazioli V, Villa MR. Entamoeba moshkovskii (Tshalaia, 1941): morpho-biological characterization of new strains isolated from the environment, and a review of the literature. Ann Parasitol Hum Comp. 1983;58:413–22. doi: 10.1051/parasite/1983585413. [DOI] [PubMed] [Google Scholar]
- 8.Dreyer DA. Growth of a strain of Entamoeba histolytica at room temperature. Tex Rep Biol Med. 1961;19:393–6. [PubMed] [Google Scholar]
- 9.Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. Laboratory diagnostic techniques for Entamoeba species. Clin Microbiol Rev. 2007;20:511–32. doi: 10.1128/CMR.00004-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark CG, Diamond LS. The Laredo strain and other ‘Entamoeba histolytica-like’ amoebae are Entamoeba moshkovskii. Mol Biochem Parasitol. 1991;46:11–8. doi: 10.1016/0166-6851(91)90194-b. [DOI] [PubMed] [Google Scholar]
- 11.Tanyuksel M, Ulukanligil M, Guclu Z, Araz E, Koru O, Petri WA., Jr Two cases of rarely recognized infection with Entamoeba moshkovskii. Am J Trop Med Hyg. 2007;76:723–4. [PubMed] [Google Scholar]
- 12.Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. PCR detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii in stool samples from Sydney, Australia. J Clin Microbiol. 2007;45:1035–7. doi: 10.1128/JCM.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ali IK, Hossain MB, Roy S, et al. Entamoeba moshkovskii infections in children, Bangladesh. Emerg Infect Dis. 2003;9:580–4. doi: 10.3201/eid0905.020548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khairnar K, Parija SC, Palaniappan R. Diagnosis of intestinal amoebiasis by using nested polymerase chain reaction-restriction fragment length polymorphism assay. J Gastroenterol. 2007;42:631–40. doi: 10.1007/s00535-007-2080-6. [DOI] [PubMed] [Google Scholar]
- 15.Nazemalhosseini Mojarad E, Nochi Z, Sahebekhtiari N, et al. Discrimination of Entamoeba moshkovskii in patients with gastrointestinal disorders by single-round PCR. Jpn J Infect Dis. 2010;63:136–8. [PubMed] [Google Scholar]
- 16.Parija SC, Khairnar K. Mutation detection analysis of a region of 16S-like ribosomal RNA gene of Entamoeba histolytica, Entamoeba dispar and Entamoeba moshkovskii. BMC Infect Dis. 2008;8:131. doi: 10.1186/1471-2334-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck DL, Dogan N, Maro V, Sam NE, Shao J, Houpt ER. High prevalence of Entamoeba moshkovskii in a Tanzanian HIV population. Acta Trop. 2008;107:48–9. doi: 10.1016/j.actatropica.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamano S, Asgharpour A, Stroup SE, Wynn TA, Leiter EH, Houpt E. Resistance of C57BL/6 mice to amoebiasis is mediated by nonhemopoietic cells but requires hemopoietic IL-10 production. J Immunol. 2006;177:1208–13. doi: 10.4049/jimmunol.177.2.1208. [DOI] [PubMed] [Google Scholar]
- 19.Houpt ER, Glembocki DJ, Obrig TG, et al. The mouse model of amebic colitis reveals mouse strain susceptibility to infection and exacerbation of disease by CD4+ T cells. J Immunol. 2002;169:4496–503. doi: 10.4049/jimmunol.169.8.4496. [DOI] [PubMed] [Google Scholar]
- 20.Asgharpour A, Gilchrist C, Baba D, Hamano S, Houpt E. Resistance to intestinal Entamoeba histolytica infection is conferred by innate immunity and Gr-1+ cells. Infect Immun. 2005;73:4522–9. doi: 10.1128/IAI.73.8.4522-4529.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diamond LS, Harlow DR, Cunnick CC. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans R Soc Trop Med Hyg. 1978;72:431–2. doi: 10.1016/0035-9203(78)90144-x. [DOI] [PubMed] [Google Scholar]
- 22.Hamzah Z, Petmitr S, Mungthin M, Leelayoova S, Chavalitshewinkoon-Petmitr P. Differential detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii by a single-round PCR assay. J Clin Microbiol. 2006;44:3196–200. doi: 10.1128/JCM.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qadri F, Saha A, Ahmed T, Al Tarique A, Begum YA, Svennerholm AM. Disease burden due to enterotoxigenic Escherichia coli in the first 2 years of life in an urban community in Bangladesh. Infect Immun. 2007;75:3961–8. doi: 10.1128/IAI.00459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haque R, Roy S, Siddique A, et al. Multiplex real-time PCR assay for detection of Entamoeba histolytica, Giardia intestinalis, and Cryptosporidium spp. Am J Trop Med Hyg. 2007;76:713–7. [PubMed] [Google Scholar]
- 25.Liu J, Kibiki G, Maro V, et al. Multiplex reverse transcription PCR Luminex assay for detection and quantitation of viral agents of gastroenteritis. J Clin Virol. 2011;50:308–13. doi: 10.1016/j.jcv.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taniuchi M, Verweij JJ, Noor Z, et al. High throughput multiplex PCR and probe-based detection with Luminex beads for seven intestinal parasites. Am J Trop Med Hyg. 2011;84:332–7. doi: 10.4269/ajtmh.2011.10-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taniuchi M, Verweij JJ, Sethabutr O, et al. Multiplex PCR method to detect Cyclospora, Cystoisospora, and Microsporidia in stool samples. Diagn Microbiol Infect Dis. 2011;71:386–90. doi: 10.1016/j.diagmicrobio.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baums IB, Goodwin KD, Kiesling T, Wanless D, Diaz MR, Fell JW. Luminex detection of fecal indicators in river samples, marine recreational water, and beach sand. Mar Pollut Bull. 2007;54:521–36. doi: 10.1016/j.marpolbul.2006.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mondal D, Minak J, Alam M, et al. Contribution of enteric infection, altered intestinal barrier function and maternal malnutrition to infant malnutrition in Bangladesh. Clin Infect Dis. 2011;54:185–92. doi: 10.1093/cid/cir807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ali IK, Mondal U, Roy S, Haque R, Petri WA, Jr, Clark CG. Evidence for a link between parasite genotype and outcome of infection with Entamoeba histolytica. J Clin Microbiol. 2007;45:285–9. doi: 10.1128/JCM.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali IK, Zaki M, Clark CG. Use of PCR amplification of tRNA gene-linked short tandem repeats for genotyping Entamoeba histolytica. J Clin Microbiol. 2005;43:5842–7. doi: 10.1128/JCM.43.12.5842-5847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamano S, Becker S, Asgharpour A, et al. Gender and genetic control of resistance to intestinal amebiasis in inbred mice. Genes Immun. 2008;9:452–61. doi: 10.1038/gene.2008.37. [DOI] [PubMed] [Google Scholar]
- 33.Duggal P, Guo X, Haque R, et al. A mutation in the leptin receptor is associated with Entamoeba histolytica infection in children. J Clin Invest. 2011;121:1191–8. doi: 10.1172/JCI45294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo X, Roberts MR, Becker SM, et al. Leptin signaling in intestinal epithelium mediates resistance to enteric infection by Entamoeba histolytica. Mucosal Immunol. 2011;4:294–303. doi: 10.1038/mi.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo X, Houpt E, Petri WA., Jr Crosstalk at the initial encounter: interplay between host defense and ameba survival strategies. Curr Opin Immunol. 2007;19:376–84. doi: 10.1016/j.coi.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ali IK, Solaymani-Mohammadi S, Akhter J, et al. Tissue invasion by Entamoeba histolytica: evidence of genetic selection and/or DNA reorganization events in organ tropism. PLoS Negl Trop Dis. 2008;2:e219. doi: 10.1371/journal.pntd.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.