Abstract
Crohn’s disease (CD) and ulcerative colitis (UC), the two common forms of inflammatory bowel disease (IBD), affect over 2.5 million people of European ancestry with rising prevalence in other populations1. Genome-wide association studies (GWAS) and subsequent meta-analyses of CD and UC2,3 as separate phenotypes implicated previously unsuspected mechanisms, such as autophagy4, in pathogenesis and showed that some IBD loci are shared with other inflammatory diseases5. Here we expand knowledge of relevant pathways by undertaking a meta-analysis of CD and UC genome-wide association scans, with validation of significant findings in more than 75,000 cases and controls. We identify 71 new associations, for a total of 163 IBD loci that meet genome-wide significance thresholds. Most loci contribute to both phenotypes, and both directional and balancing selection effects are evident. Many IBD loci are also implicated in other immune-mediated disorders, most notably with ankylosing spondylitis and psoriasis. We also observe striking overlap between susceptibility loci for IBD and mycobacterial infection. Gene co-expression network analysis emphasizes this relationship, with pathways shared between host responses to mycobacteria and those predisposing to IBD.
We conducted an imputation-based association analysis using autosomal genotype level data from 15 GWAS of CD and/or UC (Supplementary Table 1, Supplementary Figure 1). We imputed 1.23 million SNPs from the HapMap3 reference set (Supplementary Methods), resulting in a high quality dataset with reduced genome-wide inflation (Supplementary Figures 2, 3) compared with previous meta-analyses of subsets of these data2,3. The imputed GWAS data identified 25,075 SNPs that had association p < 0.01 in at least one of the CD, UC or all IBD analyses. A meta-analysis of GWAS data with Immunochip6 validation genotypes from an independent, newly-genotyped set of 14,763 CD cases, 10,920 UC cases, and 15,977 controls was performed (Supplementary Table 1, Supplementary Figure 1). Principal components analysis resolved geographic stratification, as well as Jewish and non-Jewish ancestry (Supplementary Figure 4), and significantly reduced inflation to a level consistent with residual polygenic risk, rather than other confounding effects (from λGC = 2.00 to λGC = 1.23 when analyzing all IBD samples, Supplementary Methods, Supplementary Figure 5).
Our meta-analysis of the GWAS and Immunochip data identified 193 statistically independent signals of association at genome-wide significance (p < 5×10−8) in at least one of the three analyses (CD, UC, IBD). Since some of these signals (Supplementary Figure 6) probably represent associations to the same underlying functional unit, we merged these signals (Supplementary Methods) into 163 regions, of which 71 are reported here for the first time (Table 1, Supplementary Table 2). Figure 1A shows the relative contributions of each locus to the total variance explained in UC and CD. We have increased the total disease variance explained (variance being subject to fewer assumptions than heritability7) from 8.2% to 13.6% in CD and from 4.1% to 7.5% in UC (Supplementary Methods). Consistent with previous studies, our IBD risk loci seem to act independently, with no significant evidence of deviation from an additive combination of log odds ratios.
Table 1.
Crohn’s disease-specific, ulcerative colitis-specific and IBD general loci
| Chr | Position (hg19 (Mb)) | SNP | Key Genes (+N additional in locus) |
|---|---|---|---|
| Crohn’s Disease | |||
|
| |||
| 1 | 78.62 | rs17391694 | (5) |
| 1 | 114.3 | rs6679677§ | PTPN22||,(8) |
| 1 | 120.45 | rs3897478 | ADAM30,(5) |
| 1 | 172.85 | rs9286879 | FASLG,TNFSF18,(0) |
| 2 | 27.63 | rs1728918 | UCN,(23) |
| 2 | 62.55 | rs10865331 | (3) |
| 2 | 231.09 | rs6716753 | SP140,(5) |
| 2 | 234.15 | rs12994997 | ATG16L1||, (8) |
| 4 | 48.36 | rs6837335 | (6) |
| 4 | 102.86 | rs13126505 | (1) |
| 5 | 55.43 | rs10065637 | IL6ST,IL31RA,(1) |
| 5 | 72.54 | rs7702331 | (4) |
| 5 | 173.34 | rs17695092 | CPEB4,(2) |
| 6 | 21.42 | rs12663356 | (3) |
| 6 | 31.27 | rs9264942 | (22) |
| 6 | 127.45 | rs9491697 | (3) |
| 6 | 128.24 | rs13204742 | (2) |
| 6 | 159.49 | rs212388 | TAGAP,(5) |
| 7 | 26.88‡ | rs10486483 | (2) |
| 7 | 28.17 | rs864745 | CREB5,JAZF1,(1) |
| 8 | 90.87 | rs7015630 | RIPK2,(4) |
| 8 | 129.56 | rs6651252 | 0 |
| 13 | 44.45 | rs3764147 | LACC1,(3) |
| 15 | 38.89 | rs16967103 | RASGRP1,SPRED1,(2) |
| 16 | 50.66** | rs2066847§ | NOD2||, (6) |
| 17 | 25.84 | rs2945412 | LGALS9,NOS2,(3) |
| 19 | 1.12 | rs2024092 | GPX4,HMHA1,(20) |
| 19 | 46.85‡ | rs4802307 | (9) |
| 19 | 49.2 | rs516246 | FUT2, (25) |
| 21 | 34.77 | rs2284553 | IFNGR2,IFNAR1, (10) |
|
| |||
| Ulcerative Colitis | |||
|
| |||
| 1 | 2.5 | rs10797432 | TNFRSF14, (10) |
| 1 | 20.15** | rs6426833 | (9) |
| 1 | 200.09 | rs2816958 | (3) |
| 2 | 198.65 | rs1016883 | RFTN2,PLCL1,(7) |
| 2 | 199.70* | rs17229285 | 0 |
| 3 | 53.05 | rs9847710 | PRKCD,ITIH4,(8) |
| 4 | 103.51 | rs3774959 | NFKB1,MANBA,(2) |
| 5 | 0.59 | rs11739663 | SLC9A3,(8) |
| 5 | 134.44 | rs254560 | (6) |
| 6 | 32.595 | rs6927022 | (15) |
| 7 | 2.78 | rs798502 | CARD11, GNA12, (5) |
| 7 | 27.22‡ | rs4722672 | (14) |
| 7 | 107.45* | rs4380874 | DLD,(9) |
| 7 | 128.57 | rs4728142 | IRF5||, (13) |
| 11 | 96.02 | rs483905 | JRKL,MAML2,(2) |
| 11 | 114.38 | rs561722 | FAM55A,FAM55D,(5) |
| 15 | 41.55 | rs28374715 | (11) |
| 16 | 30.47 | rs11150589 | ITGAL,(20) |
| 16 | 68.58 | rs1728785 | ZFP90,(6) |
| 17 | 70.64 | rs7210086 | (3) |
| 19 | 47.12‡ | rs1126510 | CALM3,(14) |
| 20 | 33.8 | rs6088765 | (11) |
| 20 | 43.06 | rs6017342 | ADA,HNF4A,(9) |
|
| |||
| Inflammatory Bowel Disease | |||
|
| |||
| 1 | 1.24 | rs12103 | TNFRSF18,TNFRSF4,(30) |
| 1 | 8.02 | rs35675666 | TNFRSF9,(6) |
| 1 | 22.7 | rs12568930† | (3) |
| 1 | 67.68** | rs11209026† | IL23R||, (5) |
| 1 | 70.99 | rs2651244† | (3) |
| 1 | 151.79 | rs4845604† | RORC,(14) |
| 1 | 155.67 | rs670523† | (31) |
| 1 | 160.85 | rs4656958† | CD48, (15) |
| 1 | 161.47 | rs1801274† | FCGR2A/B, FCGR3A, (13) |
| 1 | 197.6 | rs2488389 | C1orf53,(2) |
| 1 | 200.87 | rs7554511 | KIF21B,(6) |
| 1 | 206.93 | rs3024505† | IL10, (10) |
| 2 | 25.12 | rs6545800† | ADCY3,(6) |
| 2 | 28.61 | rs925255† | FOSL2,BRE,(1) |
| 2 | 43.81 | rs10495903† | (5) |
| 2 | 61.2 | rs7608910 | REL, (9) |
| 2 | 65.67 | rs6740462 | SPRED2,(1) |
| 2 | 102.86* | rs917997† | IL18RAP, IL1R1, (7) |
| 2 | 163.1 | rs2111485 | IFIH1,(5) |
| 2 | 191.92 | rs1517352 | STAT1,STAT4,(2) |
| 2 | 219.14 | rs2382817 | (15) |
| 2 | 241.57* | rs3749171† | GPR35,(12) |
| 3 | 18.76 | rs4256159† | 0 |
| 3 | 48.96** | rs3197999 | MST1, PFKB4, (63) |
| 4 | 74.85 | rs2472649† | (11) |
| 4 | 123.22 | rs7657746 | IL2,IL21,(2) |
| 5 | 10.69 | rs2930047 | DAP,(2) |
| 5 | 40.38** | rs11742570† | PTGER4,(1) |
| 5 | 96.24 | rs1363907 | ERAP2, ERAP1, (3) |
| 5 | 130.01 | rs4836519† | (1) |
| 5 | 131.19* | rs2188962† | IBD5 locus, (18) |
| 5 | 141.51 | rs6863411† | SPRY4,NDFIP1,(5) |
| 5 | 150.27 | rs11741861† | IRGM||, (10) |
| 5 | 158.8** | rs6871626† | IL12B,(3) |
| 5 | 176.79 | rs12654812 | DOK3,(17) |
| 6 | 14.71 | rs17119 | 0 |
| 6 | 20.77* | rs9358372† | (2) |
| 6 | 90.96 | rs1847472 | (1) |
| 6 | 106.43 | rs6568421† | (2) |
| 6 | 111.82 | rs3851228 | TRAF3IP2, (4) |
| 6 | 138 | rs6920220† | TNFAIP3,(1) |
| 6 | 143.9 | rs12199775 | PHACTR2,(5) |
| 6 | 167.37 | rs1819333† | CCR6,RPS6KA2,(4) |
| 7 | 50.245* | rs1456896 | ZPBP,IKZF1,(4) |
| 7 | 98.75 | rs9297145 | SMURF1,(6) |
| 7 | 100.34 | rs1734907† | EPO,(21) |
| 7 | 116.89 | rs38904† | (6) |
| 8 | 126.53 | rs921720† | TRIB1,(1) |
| 8 | 130.62 | rs1991866 | (2) |
| 9 | 4.98 | rs10758669 | JAK2,(4) |
| 9 | 93.92 | rs4743820† | NFIL3,(2) |
| 9 | 117.60** | rs4246905 | TNFSF15, (4) |
| 9 | 139.32* | rs10781499† | CARD9, (22) |
| 10 | 6.08 | rs12722515† | IL2RA,IL15RA,(6) |
| 10 | 30.72 | rs1042058† | MAP3K8,(3) |
|
| |||
| Inflammatory Bowel Disease | |||
|
| |||
| 10 | 35.3 | rs11010067† | CREM,(3) |
| 10 | 59.99 | rs2790216 | CISD1,IPMK,(2) |
| 10 | 64.51** | rs10761659† | (3) |
| 10 | 75.67 | rs2227564† | (13) |
| 10 | 81.03 | rs1250546† | (5) |
| 10 | 82.25 | rs6586030† | TSPAN14,C10orf58,(4) |
| 10 | 94.43 | rs7911264 | (4) |
| 10 | 101.28 | rs4409764 | NKX2-3,(6) |
| 11 | 1.87 | rs907611 | TNNI2,LSP1,(17) |
| 11 | 58.33 | rs10896794 | CNTF,LPXN,(8) |
| 11 | 60.77 | rs11230563 | CD6, (14) |
| 11 | 61.56 | rs4246215† | (15) |
| 11 | 64.12 | rs559928 | CCDC88B,(23) |
| 11 | 65.65 | rs2231884† | RELA, (25) |
| 11 | 76.29 | rs2155219† | (5) |
| 11 | 87.12 | rs6592362 | (1) |
| 11 | 118.74 | rs630923† | CXCR5,(17) |
| 12 | 12.65 | rs11612508† | LOH12CR1,(8) |
| 12 | 40.77* | rs11564258† | MUC19,(1) |
| 12 | 48.2 | rs11168249† | VDR,(8) |
| 12 | 68.49 | rs7134599† | IFNG, (3) |
| 13 | 27.52 | rs17085007† | (2) |
| 13 | 40.86** | rs941823† | (3) |
| 13 | 99.95 | rs9557195 | GPR183,GPR18,(6) |
| 14 | 69.27 | rs194749† | ZFP36L1,(4) |
| 14 | 75.7 | rs4899554† | FOS,MLH3,(6) |
| 14 | 88.47 | rs8005161 | GPR65,GALC,(1) |
| 15 | 67.43 | rs17293632† | SMAD3,(2) |
| 15 | 91.17 | rs7495132 | CRTC3,(3) |
| 16 | 11.54* | rs529866† | SOCS1,LITAF, (11) |
| 16 | 23.86 | rs7404095 | PRKCB,(5) |
| 16 | 28.6 | rs26528† | IL27, (14) |
| 16 | 86 | rs10521318† | IRF8,(4) |
| 17 | 32.59 | rs3091316† | CCL13,CCL2, (5) |
| 17 | 37.91 | rs12946510 | ORMDL3, (16) |
| 17 | 40.53 | rs12942547† | STAT3, (15) |
| 17 | 57.96 | rs1292053† | TUBD1,RPS6KB1,(9) |
| 18 | 12.8 | rs1893217† | (6) |
| 18 | 46.39 | rs7240004† | SMAD7,(2) |
| 18 | 67.53 | rs727088 | CD226,(2) |
| 19 | 10.49* | rs11879191 | TYK2, (27) |
| 19 | 33.73 | rs17694108 | CEBPG,(8) |
| 19 | 55.38 | rs11672983 | (19) |
| 20 | 30.75 | rs6142618† | HCK,(10) |
| 20 | 31.37 | rs4911259 | DNMT3B,(8) |
| 20 | 44.74 | rs1569723† | CD40, (13) |
| 20 | 48.95 | rs913678 | CEBPB,(5) |
| 20 | 57.82 | rs259964 | ZNF831,CTSZ,(5) |
| 20 | 62.34 | rs6062504 | TNFRSF6B, (26) |
| 21 | 16.81 | rs2823286† | 0 |
| 21 | 40.46 | rs2836878† | (3) |
| 21 | 45.62 | rs7282490 | ICOSLG,(9) |
| 22 | 21.92 | rs2266959 | (13) |
| 22 | 30.43 | rs2412970 | LIF, OSM, (9) |
| 22 | 39.69* | rs2413583† | (19) |
The position given is the middle of the locus window.
= additional genome-wide significant associated SNP in the region.
= two or more additional genome-wide significant SNPs in the region.
= These regions have overlapping but distinct UC and CD signals.
= heterogeneity of odds ratios.
= CD risk allele is significantly protective in UC.
= gene for which functional studies of associated alleles have been reported. Newly discovered loci. Bolded rs numbers indicate SNPs with p-values less than 10−13. Listed are genes implicated by one or more candidate genes approaches. Bolded genes have been implicated by two or more candidate gene approaches. For each locus, the top two candidate genes are listed. A complete listing of gene prioritization is provided in Supplementary Table 2.
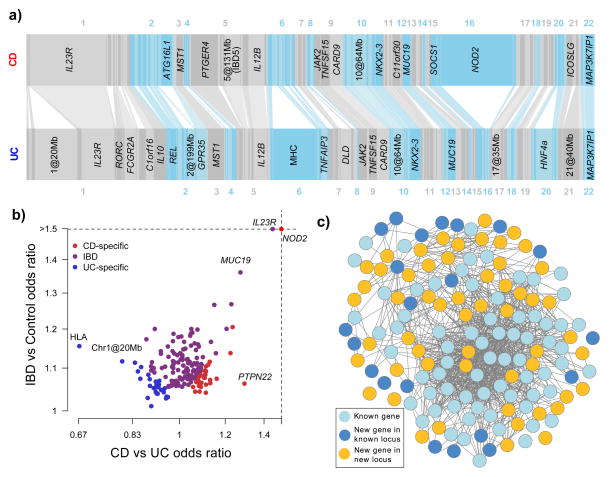
Figure 1. The IBD genome.
A) Variance explained by the 163 IBD loci. Each bar, ordered by genomic position, represents an independent locus. The width of the bar is proportional to the variance explained by that locus in CD and UC. Bars are connected together if they are identified as being associated with both CD and UC. Loci are labeled if they explain more than 1% of the total variance explained by all loci for that phenotype. B) The 193 independent signals, plotted by total IBD odds ratio and phenotype specificity (measured by the odds ratio of CD relative to UC), and colored by their IBD phenotype classification from Table 1. Note that many loci (e.g. IL23R) show very different effects in CD and UC despite being strongly associated to both. C) GRAIL network for all genes with GRAIL p < 0.05. Genes included in our previous GRAIL networks in CD and UC are shown in light blue, newly connected genes in previously identified loci in dark blue, and genes from newly associated loci in gold. The gold genes reinforce the previous network (light blue) and expand it to include dark blue genes.
Our combined genome-wide analysis of CD and UC enables a more comprehensive analysis of disease specificity than was previously possible. A model selection analysis (Supplementary Methods 1d) showed that 110/163 loci are associated with both disease phenotypes; 50 of these have an indistinguishable effect size in UC and CD, while 60 show evidence of heterogeneous effects (Table 1). Of the remaining loci, 30 are classified as CD-specific and 23 as UC-specific. However, 43 of these 53 show the same direction of effect in the non-associated disease (Figure 1B, overall p=2.8×10−6). Risk alleles at two CD loci, PTPN22 and NOD2, show significant (p < 0.005) protective effects in UC, exceptions that may reflect biological differences between the two diseases. This degree of sharing of genetic risk suggests that nearly all the biological mechanisms involved in one disease play some role in the other.
The large number of IBD associations, far more than reported for any other complex disease, increases the power of network-based analyses to prioritize genes within loci. We investigated the IBD loci using functional annotation and empirical gene network tools (Supplementary Table 2). Compared with previous analyses which identified candidate genes in 35% of loci2,3 our updated GRAIL8 -connectivity network identifies candidates in 53% of loci, including increased statistical significance for 58 of the 73 candidates from previous analyses. The new candidates come not only from genes within newly identified loci, but also integrate additional genes from previously established loci (Figure 1C). Only 29 IBD-associated SNPs are in strong linkage disequilibrium (r2 > 0.8) with a missense variant in the 1000 Genomes Project data, which reinforces previous evidence that a large fraction of risk for complex disease is driven by non-coding variation. In contrast, 64 IBD-associated SNPs are in linkage disequilibrium with variants known to regulate gene expression (Supplementary Table 2). Overall, we highlighted a total of 300 candidate genes in 125 loci, of which 39 contained a single gene supported by two or more methods.
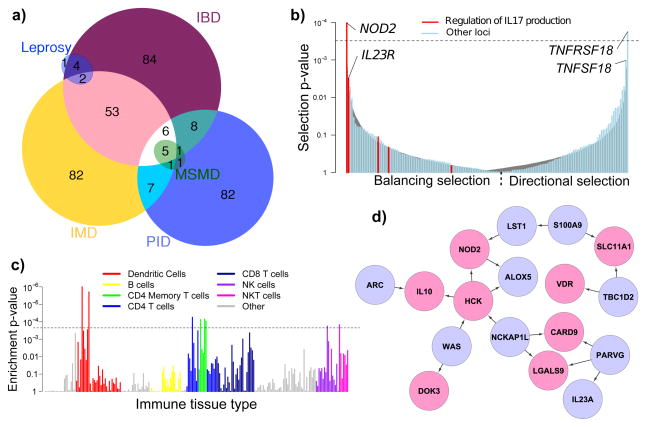
Seventy percent (113/163) of the IBD loci are shared with other complex diseases or traits, including 66 among the 154 loci previously associated with other immune-mediated diseases9, which is 8.6 times the number that would be expected by chance (Figure 2A, p < 10−16, Supplementary Figure 7). Such enrichment cannot be attributed to the immune-mediated focus of the Immunochip, (Supplementary Methods 4a(i), Supplementary Figure 8), since the analysis is based on our combined GWAS-Immunochip data. Comparing overlaps with specific diseases is confounded by the variable power in studies of different diseases. For instance, while type 1 diabetes (T1D) shares the largest number of loci (20/39, 10-fold enrichment) with IBD, this is partially driven by the large number of known T1D associations. Indeed, seven other immune-mediated diseases show stronger enrichment of overlap, with the largest being ankylosing spondylitis (8/11, 13-fold) and psoriasis (14/17, 14-fold).
Figure 2. Dissecting the biology of IBD.
A) Number of overlapping IBD loci with other immune-mediated diseases (IMD), leprosy, and Mendelian primary immunodeficiencies (PID). Within PID, we highlight Mendelian susceptibility to mycobacterial disease (MSMD). B) Signals of selection at IBD SNPs, from strongest balancing on the left to strongest directional on the right. The grey curve shows the 95% confidence interval for randomly chosen frequency-matched SNPs, illustrating our overall enrichment (p = 5.5 × 10-6), while the dashed line represents the Bonferroni significance threshold. SNPs highlighted in red are annotated as involved in regulation of IL17 production, a key IBD functional term related to bacterial defense, and are enriched for balancing selection. C) Evidence of enrichment in IBD loci of differentially expressed genes from various immune tissues. Each bar represents the empirical p-value in a single tissue, and the colours represent different cell type groupings. The dashed line is Bonferroni-corrected significance for the number of tissues tested. D) NOD2-focused cluster of the IBD causal subnetwork. Pink genes are in IBD associated loci, blue are not. Arrows indicate inferred causal direction of regulation of expression.
IBD loci are also markedly enriched (4.9-fold, p < 10−4) in genes involved in primary immunodeficiencies (PIDs, Figure 2A), which are characterized by a dysfunctional immune system resulting in severe infections10. Genes implicated in this overlap correlate with reduced levels of circulating T-cells (ADA, CD40, TAP1/2, NBS1, BLM, DNMT3B), or of specific subsets such as Th17 (STAT3), memory (SP110), or regulatory T-cells (STAT5B). The subset of PIDs genes leading to Mendelian susceptibility to mycobacterial disease (MSMD)10–12 is enriched still further; six of the eight known autosomal genes linked to MSMD are located within IBD loci (IL12B, IFNGR2, STAT1, IRF8, TYK2 and STAT3, 46-fold enrichment, p = 1.3 × 10−6), and a seventh, IFNGR1, narrowly missed genome-wide significance (p = 6 × 10−8). Overlap with IBD is also seen in complex mycobacterial disease; we find IBD associations in 7/8 loci identified by leprosy GWAS13, including 6 cases where the same SNP is implicated. Furthermore, genetic defects in STAT314–15 and CARD916, also within IBD loci, lead to PIDs involving skin infections with staphylococcus and candidiasis, respectively. The comparative effects of IBD and infectious disease susceptibility risk alleles on gene function and expression is summarized in Supplementary Table 3, and include both opposite (e.g. NOD2 and STAT3, Supplementary Figure 9) and similar (e.g., IFNGR2) directional effects.
To extend our understanding of the fundamental biology of IBD pathogenesis we conducted searches across the IBD locus list: (i) for enrichment of specific GeneOntology (GO) terms and canonical pathways, (ii) for evidence of selective pressure acting on specific variants and pathways, and (iii) for enrichment of differentially expressed genes across immune cell types. We tested the 300 prioritized genes (see above) for enrichment in GO terms (Supplementary Methods) and identified 286 GO terms and 56 pathways demonstrating significant enrichment in genes contained within IBD loci (Supplementary Table 4, Supplementary Figure 10,11). Excluding high-level GO categories such as “immune system processes” (p = 3.5 × 10−26), the most significantly enriched term is regulation of cytokine production (p=2.7×10−24), specifically IFNG-γ, IL-12, TNF-α, and IL-10 signalling. Lymphocyte activation was the next most significant (p=1.8 × 10−23), with activation of T-, B-, and NK-cells being the strongest contributors to this signal. Strong enrichment was also seen for response to molecules of bacterial origin (p=2.4 × 10−20), and for KEGG’s JAK-STAT signalling pathway (p = 4.8 × 10−15). We note that no enriched terms or pathways showed specific evidence of CD- or UC-specificity.
As infectious organisms are known to be among the strongest agents of natural selection, we investigated whether the IBD-associated variants are subject to selective pressures (Supplementary Methods, Supplementary Table 5). Directional selection would imply that the balance between these forces shifted in one direction over the course of human history, whereas balancing selection would suggest an allele frequency dependent-scenario typified by host-microbe co-evolution, as can be observed with parasites. Two SNPs show Bonferroni-significant selection: the most significant signal, in NOD2, is under balancing selection (p = 5.2 × 10−5), and the second most significant, in the receptor TNFRSF18, showed directional selection (p = 8.9 × 10−5). The next most significant variants were in the ligand of that receptor, TNFSF18 (directional, p = 5.2 × 10−4), and IL23R (balancing, p = 1.5 × 10−3). As a group, the IBD variants show significant enrichment in selection (Figure 2B) of both types (p = 5.5 × 10−6). We discovered an enrichment of balancing selection (Figure 2B) in genes annotated with the GO term “regulation of interleukin-17 production” (p = 1.4 × 10−4). The important role of IL17 in both bacterial defense and autoimmunity suggests a key role for balancing selection in maintaining the genetic relationship between inflammation and infection, and this is reinforced by a nominal enrichment of balancing selection in loci annotated with the broader GO term “defense response to bacterium” (p = 0.007).
We tested for enrichment of cell-type expression specificity of genes in IBD loci in 223 distinct sets of sorted, mouse-derived immune cells from the Immunological Genome Consortium17. Dendritic cells showed the strongest enrichment, followed by weaker signals that support the GO analysis, including CD4+ T, NK and NKT cells (Figure 2C). Notably, several of these cell types express genes near our IBD associations much more specifically when stimulated; our strongest signal, a lung-derived dendritic cell, had p stimulated < 1×10−6 compared with p unstimulated = 0.0015, consistent with an important role for cell activation.
To further our goal of identifying likely causal genes within our susceptibility loci and to elucidate networks underlying IBD pathogenesis, we screened the associated genes against 211 co-expression modules identified from weighted gene co-expression network analyses18, conducted with large gene expression datasets from multiple tissues19–21. The most significantly enriched module comprised 523 genes from omental adipose tissue collected from morbidly obese patients19, which was found to be 2.9-fold enriched for genes in the IBD-associated loci (p = 1.1 × 10−13, Supplementary Table 6, Supplementary Figure 12). We constructed a probabilistic causal gene network using an integrative Bayesian network reconstruction algorithm22–24 which combines expression and genotype data to infer the direction of causality between genes with correlated expression. The intersection of this network and the genes in the IBD-enriched module defined a sub-network of genes enriched in bone marrow-derived macrophages (p < 10−16) and is suggestive of dynamic interactions relevant to IBD pathogenesis. In particular, this sub-network featured close proximity amongst genes connected to host interaction with bacteria, notably NOD2, IL10, and CARD9.
A NOD2-focused inspection of the sub-network prioritizes multiple additional candidate genes within IBD-associated regions. For example, a cluster near NOD2 (Figure 2D) contains multiple IBD genes implicated in M.tb response, including SLC11A1, VDR and LGALS9. Furthermore, both SLC11A1 (also known as NRAMP1) and VDR have been associated with M.tb infection by candidate gene studies25–26, and LGALS9 modulates mycobacteriosis27. Of interest, HCK (located in our new locus on chromosome 20 at 30.75Mb) is predicted to upregulate expression of both NOD2 and IL10, an anti-inflammatory cytokine associated with Mendelian28 and non-Mendelian IBD29. HCK has been linked to alternative, anti-inflammatory activation of monocytes (M2 macrophages)30; while not identified in our aforementioned analyses, these data implicate HCK as the causal gene in this new IBD locus.
We report one of the largest genetic experiments involving a complex disease undertaken to date. This has increased the number of confirmed IBD susceptibility loci to 163, most of which are associated with both CD and UC, and is substantially more than reported for any other complex disease. Even this large number of loci explains only a minority of the variance in disease risk, which suggests that other factors such as rarer genetic variation not captured by GWAS or environmental exposures make substantial contributions to pathogenesis. Most of the evidence relating to possible causal genes points to an essential role for host defence against infection in IBD. In this regard the current results focus ever closer attention on the interaction between the host mucosal immune system and microbes both at the epithelial cell surface and within the gut lumen. In particular, they raise the question, in the context of this burden of IBD susceptibility genes, as to what triggers components of the commensal microbiota to switch from a symbiotic to a pathogenic relationship with the host. Collectively, our findings have begun to shed light on these questions and provide a rich source of clues to the pathogenic mechanisms underlying this archetypal complex disease.
METHODS SUMMARY
We conducted a meta-analysis of GWAS datasets after imputation to the HapMap3 reference set, and aimed to replicate in the Immunochip data any SNPs with p < 0.01. We compared likelihoods of different disease models to assess whether each locus was associated with CD, UC or both. We used databases of eQTL SNPs and coding SNPs in linkage disequilibrium with our hit SNPs, as well as the network tools GRAIL and DAPPLE, and a co-expression network analysis to prioritize candidate genes in our loci. Gene Ontology, ImmGen mouse immune cell expression resource, the TreeMix selection software, and a Bayesian causal network analysis were used to functionally annotate these genes.
Supplementary Material
Acknowledgments
We thank all the subjects who contributed samples and the physicians and nursing staff who helped with recruitment globally. UK case collections were supported by the National Association for Colitis and Crohn’s disease, Wellcome Trust grant 098051 (LJ, CAA, JCB), Medical Research Council UK, the Catherine McEwan Foundation, an NHS Research Scotland career fellowship (RKR), Peninsular College of Medicine and Dentistry, Exeter, the National Institute for Health Research, through the Comprehensive Local Research Network and through Biomedical Research Centre awards to Guy’s & St. Thomas’ National Health Service Trust, King’s College London, Addenbrooke’s Hospital, University of Cambridge School of Clinical Medicine and to the University of Manchester and Central Manchester Foundation Trust. The British 1958 Birth Cohort DNA collection was funded by Medical Research Council grant G0000934 and Wellcome Trust grant 068545/Z/02, and the UK National Blood Service controls by the Wellcome Trust. The Wellcome Trust Case Control Consortium projects were supported by Wellcome Trust grants 083948/Z/07/Z, 085475/B/08/Z and 085475/Z/08/Z. North American collections and data processing were supported by funds to the NIDDK IBD Genetics Consortium which is funded by the following grants: DK062431 (SRB), DK062422 (JHC), DK062420 (RHD), DK062432 (JDR), DK062423 (MSS), DK062413 (DPM), DK076984 (MJD), DK084554 (MJD and DPM) and DK062429 (JHC). Additional funds were provided by funding to JHC (DK062429-S1 and Crohn’s & Colitis Foundation of America, Senior Investigator Award (5-2229)), and RHD (CA141743). KYH is supported by the NIH MSTP TG T32GM07205 training award. Cedars-Sinai is supported by USPHS grant PO1DK046763 and the Cedars-Sinai F. Widjaja Inflammatory Bowel and Immunobiology Research Institute Research Funds, National Center for Research Resources (NCRR) grant M01-RR00425, UCLA/Cedars-Sinai/Harbor/Drew Clinical and Translational Science Institute (CTSI) Grant [UL1 TR000124-01], the Southern California Diabetes and Endocrinology Research Grant (DERC) [DK063491], The Helmsley Foundation (DPM) and the Crohn’s and Colitis Foundation of America (DPM). RJX and ANA are funded by DK83756, AI062773, DK043351 and the Helmsley Foundation. The Netherlands Organization for Scientific Research supported RKW with a clinical fellowship grant (90.700.281) and CW (VICI grant 918.66.620). CW is also supported by the Celiac Disease Consortium (BSIK03009). This study was also supported by the German Ministry of Education and Research through the National Genome Research Network, the Popgen biobank, through the Deutsche Forschungsgemeinschaft (DFG) cluster of excellence ‘Inflammation at Interfaces’ and DFG grant no. FR 2821/2-1. S Brand was supported by (DFG BR 1912/6-1) and the Else-Kröner-Fresenius-Stiftung (Else Kröner-Exzellenzstipendium 2010_EKES.32). Italian case collections were supported by the Italian Group for IBD and the Italian Society for Paediatric Gastroenterology, Hepatology and Nutrition and funded by the Italian Ministry of Health GR-2008-1144485. Activities in Sweden were supported by the Swedish Society of Medicine, Ihre Foundation, Örebro University Hospital Research Foundation, Karolinska Institutet, the Swedish National Program for IBD Genetics, the Swedish Organization for IBD, and the Swedish Medical Research Council. DF and SV are senior clinical investigators for the Funds for Scientific Research (FWO/FNRS) Belgium. We acknowledge a grant from Viborg Regional Hospital, Denmark. VA was supported by SHS Aabenraa, Denmark. We acknowledge funding provided by the Royal Brisbane and Women’s Hospital Foundation, National Health and Medical Research Council, Australia and by the European Community (5th PCRDT). We gratefully acknowledge the following groups who provided biological samples or data for this study: the Inflammatory Bowel in South Eastern Norway (IBSEN) study group, the Norwegian Bone Marrow Donor Registry (NMBDR), the Avon Longitudinal Study of Parents and Children, the Human Biological Data Interchange and Diabetes UK, and Banco Nacional de ADN, Salamanca. This research also utilizes resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the NIDDK, NIAID, NHGRI, NICHD, and JDRF and supported by U01 DK062418. The KORA study was initiated and financed by the Helmholtz Zentrum München – German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. KORA research was supported within the Munich Center of Health Sciences (MC Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ.
Footnotes
Author Contributions Conceived and designed study, managed study and funding: JHC, JCB, RKW, RHD, DPM, MDA, VA, AF, MP, SV. Manuscript preparation: JHC, JCB, LJ, SR, RKW, RHD, DPM, MJD, MP, CGM. Performed or supervised statistical and computational analyses: JHC, JCB, LJ, SR, RKW, KYH, CAA, JE, KN, SLS, SR, ZW, CA, AC, GB, MH, XH, BZ, CKZ, HZ, JDR, EES, MJD. Study subject recruitment and assembled phenotypic data: RKW, RHD, DPM, JCL, LPS, YS, PG, JPA, TA, LA, ANA, VA, JMA, LB, PAB, AB, SB, CB, SC, MDA, DDJ, KLD, MD, CE, LRF, DF, MG, RG, JG, AH, CH, THK, LK, SK, AL, DL, EL, ICL, CWL, ARM, CM, GM, JM, WN, OP, CYP, UP, NJP, MR, JIR, RKR, JDS, MS, JS, SS, LAS, JS, SRT, MT, HWV, MDV, CW, DCW, JW, RJX, SZ, MSS, VA, HH, SRB, JDR, GRS, CGM, AF, MP, SV, JHC. Established DNA collections, genotyping and data management: RKW, RHD, DPM, LPS, YS, MM, IC, ET, TB, DE, KF, TH, KDT, CGM, AF, MP, JHC. All authors read and approved the final manuscript before submission.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Data have been deposited in NCBI’s database of Genotypes and Phenotypes (dbGaP) through study accession numbers phs000130.v1.p1 and phs000345.v1.p1. Summary statistics for imputed GWAS are available at http://www.broadinstitute.org/mpg/ricopili/. Summary statistics for the meta-analysis markers are available at http://www.ibdgenetics.org/. The 523 causal gene network cytoscape file is available on request. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Molodecky NA, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khor BGA, Xavier RJ. Genetics pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho JH, Gregersen PK. Genomics and the multifactorial nature of human autoimmune disease. N Engl J Med. 2011;365:1612–1623. doi: 10.1056/NEJMra1100030. [DOI] [PubMed] [Google Scholar]
- 6.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci USA. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.International Union of Immunological Societies Expert Committee on Primary I et al. Primary immunodeficiencies: 2009 update. J Allergy Clin Immunol. 2009;124:1161–1178. doi: 10.1016/j.jaci.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bustamante J, Picard C, Boisson-Dupuis S, Abel L, Casanova JL. Genetic lessons learned from X-linked Mendelian susceptibility to mycobacterial diseases. Ann NY Acad Sci. 2011;1246:92–101. doi: 10.1111/j.1749-6632.2011.06273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel SY, Doffinger R, Barcenas-Morales G, Kumararatne DS. Genetically determined susceptibility to mycobacterial infection. J Clin Pathol. 2008;61:1006–1012. doi: 10.1136/jcp.2007.051201. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F, et al. Identification of two new loci at IL23R and RAB32 that influence susceptibility to leprosy. Nat Genet. 2011;43:1247–1251. doi: 10.1038/ng.973. [DOI] [PubMed] [Google Scholar]
- 14.Holland SM, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357:1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 15.Minegishi Y, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 16.Glocker EO, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361:1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu X, et al. Integrating autoimmune risk loci with gene-expression data identifies specific pathogenic immune cell subsets. Am J Hum Genet. 2011;89:496–506. doi: 10.1016/j.ajhg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 19.Greenawalt DM, et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 2011;21:1008–1016. doi: 10.1101/gr.112821.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 21.Schadt EE, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong H, et al. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 2010;6:e1000932. doi: 10.1371/journal.pgen.1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu J, et al. Increasing the power to detect causal associations by combining genotypic and expression data in segregating populations. PLoS Comput Biol. 2007;3:e69. doi: 10.1371/journal.pcbi.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis SJ, Baker I, Davey Smith G. Meta-analysis of vitamin D receptor polymorphisms and pulmonary tuberculosis risk. Int J Tuberc Lung Dis. 2005;9:1174–1177. [PubMed] [Google Scholar]
- 26.Li X, et al. SLC11A1 (NRAMP1) polymorphisms and tuberculosis susceptibility: updated systematic review and meta-analysis. PloS One. 2011;6:e15831. doi: 10.1371/journal.pone.0015831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar D, et al. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140:731–743. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 28.Glocker EO, et al. Infant colitis--it’s in the genes. Lancet. 2010;376:1272. doi: 10.1016/S0140-6736(10)61008-2. [DOI] [PubMed] [Google Scholar]
- 29.Franke A, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–1323. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 30.Bhattacharjee A, Pal S, Feldman GM, Cathcart MK. Hck is a key regulator of gene expression in alternatively activated human monocytes. J Biol Chem. 2011;286:36709–36723. doi: 10.1074/jbc.M111.291492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.