Abstract
The phosphorylation-specific peptidyl-prolyl isomerase Pin1 catalyzes the isomerization of the peptide bond preceding a proline residue between cis and trans isomers. To best understand the mechanisms of Pin1 regulation, rigorous enzymatic assays of isomerization are required. However, most measures of isomerase activity require significant constraints on substrate sequence and only yield rate constants for the cis isomer, and apparent Michaelis constants, . By contrast, NMR lineshape analysis is a powerful tool for determining microscopic rates and populations of each state in a complex binding scheme. The isolated catalytic domain of Pin1 was employed as a first step towards elucidating the reaction scheme of the full-length enzyme. A 24-residue phosphopeptide derived from the amyloid precurser protein intracellular domain (AICD) phosphorylated at Thr668 served as a biologically-relevant Pin1 substrate. Specific 13C labeling at the Pin1-targeted proline residue provided multiple reporters sensitive to individual isomer binding and on-enzyme catalysis. We have performed titration experiments and employed lineshape analysis of phosphopeptide 13C-1H constant time HSQC spectra to determine , , , and for the catalytic domain of Pin1 acting on this AICD substrate. The on-enzyme equilibrium value of [E·trans]/[E·cis] = 3.9 suggests that the catalytic domain of Pin1 is optimized to operate on this substrate near equilibrium in the cellular context. This highlights the power of lineshape analysis for determining the microscopic parameters of enzyme catalysis, and demonstrates the feasibility of future studies of Pin1-PPIase mutants to gain insights on the catalytic mechanism of this important enzyme.
Keywords: Proyl isomerase, Pin1, Lineshape Analysis, isomerization, proline, APP
Introduction
Peptidyl-prolyl isomerases, or PPIases, are enzymes that catalyze the isomerization of the peptide bond preceding a proline residue (Fischer and Aumüller, 2003). The peptide bond between the amide nitrogen and the carbonyl carbon of the preceding residue is typically in the trans (ω = 180) configuration (Fanghanel, 2003). However, in the case of the peptide bond preceding a proline residue, the cis isomer (ω = 0) often exists in equilibrium with trans (Fanghanel, 2003). In the absence of stable tertiary structure, such as in short peptides, the population of the cis isomer can be between approximately 5% and 40%, depending on sequence (Reimer et al., 1998). Structure or protein-protein interactions can stabilize one isomer preferentially, often leading to nearly either 0% (Ramelot et al., 2000) or 100% cis (Sarkar et al., 2011). Prolyl isomerases can be important for facilitating catalysis by isomer-specific enzymes, and are thought to play an important role in facilitating binding by proteins which recognize only one of the isomers. The importance of this activity is highlighted in the example of dephosphorylation of the Ser(P)5-Pro6 motif in the heptad repeats of the C-terminal domain (CTD) of RNA polymerase II (Meinhart and Cramer, 2004; Xiang et al., 2010). This dephosphorylation is performed by the Proline-directed phosphatase Ssu72 which binds specifically to the lower populated cis isomer of the substrate(Werner-Allen et al., 2011; Xiang et al., 2010). Because phosphorylation and dephosphorylation of the CTD occurs on the same timescale as transcription (milliseconds to seconds)(Skinner et al., 2004), isomerization by the PPIase Pin1 is necessary to allow Ssu72-mediated dephosphorylation to proceed to completion on the proper timescale. This principle applies similarly to trans-specific phosphatases such as PP2A (Zhou et al., 2000), though the impact of Pin1 activity is presumably most pronounced if there is a substantial population of cis isomer to be converted.
Pin1 is the only PPIase in humans that is specific for pSer/pThr-Pro motifs (Lu et al., 1996; Ranganathan et al., 1997). While uncatalyzed inter-conversion between cis and trans isomers occurs naturally with a half-life on the order of 100 s (Grathwohl and Wüthrich, 1981), the presence of Pin1 can speed this exchange by orders of magnitude into the more biologically relevant timescale of seconds. Notably, aberrant function of Pin1 is implicated in cancer (Lu et al., 2006), asthma (Anderson, 2005), and Alzheimer's disease (AD) (Pastorino et al., 2006). Although Pin1 is a much-studied enzyme, we lack a detailed understanding of the actual significance of its isomerase activity towards its most-studied substrates, such as the microtubule-associated protein Tau or the amyloid precursor protein (APP), which are involved in Alzheimer's disease. In the case of APP, Pin1 knockout shifts the proteolytic processing fate of APP towards the amyloidogenic pathway in cell culture and mouse models (Pastorino et al., 2006). This may contribute to Pin1's established protective role against neurodegeneration (Meraz-Rios, 2010; Pastorino et al., 2006). Pin1 binds to and specifically catalyzes isomerization of the pThr668-Pro669 motif in APP in vitro (Pastorino et al., 2006), suggesting that the mechanism of Pin1's action on APP processing in vivo involves isomerization at this motif (Iijima et al., 2000; Lee et al., 2003). Pin1 is comprised of two domains with similar specificity, a catalytic PPIase domain (hereafter referred to as Pin1-PPIase) and a type IV WW binding domain (Verdecia et al., 2000). While essential for Pin1 function in vivo (Lu et al., 2002), the WW domain does not improve catalytic function in vitro on phosphopeptide substrates (Eckerdt et al., 2005; Peng et al., 2009; Rippmann et al., 2000). Hence, as an important step toward understanding a key molecular switch involved in the development of AD, focus on the isolated Pin1-PPIase domain avoids substrate competition with the WW domain and provides the opportunity to obtain a complete description of its catalytic reaction scheme.
Assays of isomerase activity (Fischer et al., 1984; Fischer et al., 1983; Lin and Brandts, 1985) commonly employ an isomer-specific protease such as chymotrypsin, which only cleaves after aromatic residues following a residue preceded by a trans peptide bond (Xaa-trans-Xaa-Yaa-Xaa, where Yaa is aromatic and Xaa is any amino acid). By coupling the reversible isomerization reaction with the essentially irreversible cleavage of the peptide bond, assays can indirectly measure conversion of cis to trans by a PPIase by monitoring the cleavage reaction. This is accomplished by incorporating the chromogenic compound p-nitroanilide (pNA) after the cleavage site (most commonly Xaa-Pro-Phe-pNA) and monitoring the change in fluorescence upon release of the pNA. This approach therefore does not allow for much variation in the sequence of the substrate C-terminal to the isomerized proline residue. Furthermore, it can only report on the cis-to-trans rate constant, , and an “apparent” Michaelis constant, , which is a composite of the binding constants for the trans and cis conformers. Other problems with this type of assay include the tendency of proteases to degrade the isomerase, and the likely possibility that cleavage of the substrate affects the value of . Other approaches have employed solvent jumps to perturb the equilibrium of cis and trans populations of substrates and monitor the return to equilibrium via UV absorption, exploiting the small differences in UV absorption between the cis and trans isomers (Garcia-Echeverria et al., 1992; Garcia-Echeverria et al., 1993). This, however, imposes similar restriction on substrate sequence because a UV-absorbing residue such as pNA must be located close to the isomerized peptide bond.
NMR provides direct observation of cis and trans isomers, which due to their slow timescale of interconversion can be identified as distinct resonances. NMR is therefore another popular tool used for the study of prolyl isomerization, and had in fact been used to measure intrinsic uncatalyzed isomerization in short peptides prior to the discovery of prolyl isomerases (Grathwohl and Wüthrich, 1981). Unlike most other approaches, NMR experiments have the ability to measure isomerization at chemical equilibrium, removing the need to perturb the cis-trans equilibrium and take time-resolved datasets. CPMG (Korzhnev and Kay, 2008) or R1ρ (Massi et al., 2004) experiments can measure the catalyzed rate of exchange as well as the rates and populations of on-enzyme motions. EXSY (Landrieu et al., 2000) and ROESY (Pastorino et al., 2006) experiments have been used to measure the rate of interconversion between two slowly-exchanging states. However, even in routine HSQC (Mulder et al., 1996) spectra, the supreme sensitivity of NMR line shapes to the kinetics and equilibria of individual reaction steps in a complex multi-state scheme is apparent. For complex systems in which multiple states are at equilibrium, NMR lineshape analysis provides a powerful tool with which to determine the microscopic kinetic rates and equilibrium constants connecting the different states in the reaction scheme, complementing these techniques.
The detailed analysis of NMR line shapes is a well-established but underutilized tool for examining protein interactions and motions that occur within the microsecond to millisecond timescale (Gunther and Schaffhausen, 2002; Kern et al., 1995; Kovrigin and Loria, 2006). The processes of ligand binding and release, conformational exchange, and enzymatic turnover can all occur on this timescale (Mittermaier and Kay, 2009a). While classically applied to 1D NMR spectra, lineshape analysis is easily applied to 2D spectra by peak slice extraction. NMR lineshape broadening and changes in chemical shift in both dimensions of a series of HSQC spectra can yield information about kinetics, populations, and unobservable chemical shifts on a residue-specific basis (Gunther and Schaffhausen, 2002). Because lineshape analysis uses data taken at multiple concentrations of interacting proteins, it is especially applicable to complex models with three, four, or more states (Gunther and Schaffhausen, 2002). Software has been developed to fit lineshape data to multiple, arbitrarily complex models, allowing easy comparisons between models (http://lineshapekin.net/, http://biophysicslab.net/). Simultaneously fitting data from multiple peaks can allow a global understanding of complex physical processes to emerge. In addition to ligand binding and protein dynamics (Craven et al., 1996; Johnson et al., 1998), lineshape analysis is applicable to the study of catalyic mechanisms and enzyme function, such as that of PPIases (Kern et al., 1995). The advantages of NMR lineshape analysis over standard biochemical isomerase assays lie in the ability to use the natural substrate sequence and the convenience of performing experiments at chemical equilibrium.
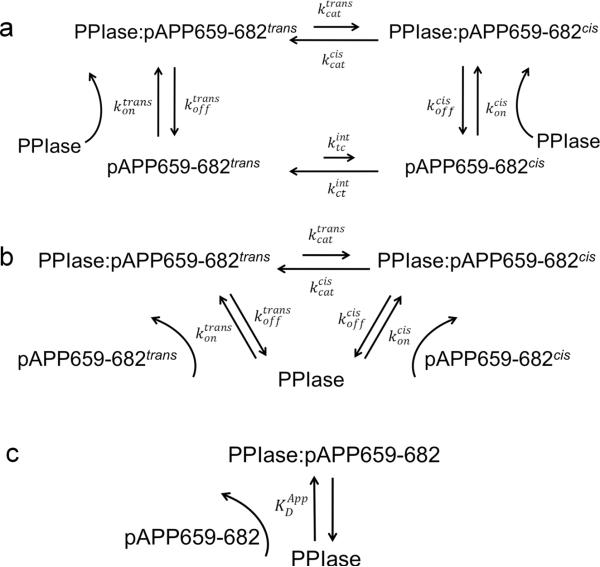
Here we present the complete characterization of the reaction scheme of Pin1-PPIase acting on the pT668-P669 motif in APP using NMR lineshape analysis. A specifically isotopically labeled (13C- and 15N) APP-derived phopshopeptide (pAPP659-682, corresponding to residues 659–682 of APP phosphorylated at T668) was titrated with unlabeled Pin1-PPIase, and the induced line broadening was fit to a four-state model (Fig. 1a). NMR titration experiments using 15N-labeled Pin1-PPIase and unlabeled pAPP659-682 allowed the measurement of the apparent dissociation constant for the binding reaction, which was used as a constraint in the lineshape analysis. This was in close agreement with that measured for full-length Pin1, indicating that the isolated Pin1-PPIase domain accurately represents the function of the intact enzyme under these conditions. Using lineshape analysis, the microscopic rates and of catalysis as well as isomer-specific dissociation constants and were determined. Moreover, 1H-1H ROESY exchange experiments were employed to measure the overall rate of exchange kex for both Pin1-PPIase and full-length Pin1, providing independent validation of the lineshape-derived fitted parameters and further supporting the accurate representation of Pin1 catalytic function by the isolated PPIase domain. Interestingly, the on-enzyme equilibrium of cis and trans substrate was found to be skewed slightly in favor of trans, suggesting that Pin1 may be optimized to operate on this substrate slightly off-equilibrium.
Fig. 1. Models used to interpret NMR data.
(a) Four state model for lineshape fitting of 13C-pAPP659-682 peptide isomerization data. This model describes four states: the cis and trans isomers of the peptide in the free state, and the cis and trans isomers bound to the enzyme. (b) Three state model of PPIase catalysis for PPIase-perspective titration data. This model describes Pin1-PPIase catalysis as a three-state process in which the PPIase can bind to either the cis or trans isomer of the pAPP659-682 peptide. (c) Two state approximation for PPIase-perspective titration data. Interpretation of peak positions with this model requires the approximation that exchange between the two bound states in the three-state model occurs on the fast chemical shift timescale. In this model the relative populations are described by a single parameter, the apparent dissociation constant .
Materials and Methods
Protein expression and purification
A pET vector [GE Health Sciences] imparting kanamycin resistance was used for recombinant Pin1-PPIase expression. This vector encodes a protein consisting of an N-terminal His tag and a TEV cut site followed by the PPIase domain of Pin1 (residues 46–163). Protein was expressed in BL21 star E. Coli [Invitrogen] cells grown in 1 L of either LB [BD] or M9 minimal media (using 19 mM 15NH4Cl [Isotec] with 50 μg/mL kanamycin [IBI Scientific]). Cultures were grown at 37°C to an O.D.600 ~0.6, and expression was induced with 1 mM IPTG [CalBioChem] at 15°C for ~20 hours. Cells were then pelleted and resuspended in lysis buffer (50 mM Tris-Hcl [Mallinckrodt Baker], 200 mM NaCl [Mallinckrodt Baker], 10 mM Imidazole [Alfa Aesar], pH 8.0, plus 100 μL protease inhibitor [Sigma], 1 mM DTT [Gold Biotechnology]). Cells were then lysed by freezing and thawing on ice, adding 1 mg/ml lysozyme [EMD], and sonicating (10 cycles). Cells were then centrifuged at 23,000 g and the supernatant was filtered with a 0.8-μm syringe filter [Corning]. Filtrate was then applied to a nickel-NTA column [Qiagen] and washed with 20 bed volumes wash buffer (50 mM Tris-Hcl, 200 mM NaCl, 18 mM Imidazole, pH 8.0). Protein was then eluted with elution buffer (50 mM Tris-Hcl, 150 mM NaCl, 300 mM Imidazole, pH 8.0) and dialyzed into cleavage buffer (50 mM Tris-Hcl, pH 8.0). Recombinant His-tagged TEV protease was then added and the Pin1-PPIase was cleaved from the His-tag overnight at 4°C. The His-tagged TEV was then removed by putting the cleavage reaction through a second Nickel-NTA column, and the flow-through containing pure Pin1-PPIase was dialyzed into NMR buffer (10 mM HEPES [Fisher, sodium salt], 1 mM DTT, pH 6.9). NMR samples also included 5 mM NaN3 [Fisher], 1 mM TCEP [Thermo Scientific], and 7% D2O [Cambridge Isotope Laboratories]. Protein was concentrated with VivaSpin-20 5000 MWC centrifugal concentrators [GE]. The concentration of Pin1-PPIase was measured by the UV absorbance spectrum, using the theoretical extinction coefficient at 280 nm of 7210 cm−1M−1. Protein purity was verified by SDS-PAGE. The pH of buffers and samples was adjusted using HCl [Mallinckrodt Baker] and NaOH [Sigma]. All peptides were synthesized using solid-phase F-moc chemistry at the Tufts University Core Facility, purified, and delivered as a lyophilized powder. Peptides were dissolved in NMR buffer (plus NaN3, TCEP and D2O), de-salted using a PD MiniTrap gravity column [GE], and their pH adjusted with NaOH before use.
NMR experiments/Data processing
NMR experiments were performed at 25°C on a Varian Inova 600 MHz spectrometer equipped with a {H,C,N} Z-axis gradient probe. Spectra were processed and analyzed using the software tools nmrPipe (Delaglio et al., 1995), nmrDraw (Delaglio et al., 1995), and Sparky (Goddard, 2008). Free induction decays were apodized using an exponential function to preserve the natural lorentzian lineshape, except for the ROESY data for which a shifted sine bell function was used, and zero filled prior to Fourier transformation. Peak heights and positions were measured using the peak detection modules of nmrDraw or Sparky, respectively.
In titration experiments to measure , 12 mM pAPP659-682 was added to 0.25 mM 15N-labeled Pin1-PPIase, and the peptide was reverse titrated from ~12 mM to ~0.8 mM by diluting with 15N-labeled Pin1-PPIase. Titration points were taken at peptide concentrations of 12, 7, 4, 2, 0.8, and 0 mM. The concentration of peptide was measured by UV absorbance based on the theoretical extinction coefficient of 1,209 cm−1M−1 at 280 nm. At each concentration, a two-dimensional 15N-1H fast HSQC spectrum (Mulder et al., 1996) was taken with a spectral width of 1.7 kHz (8 kHz) in t1 (t2), and a total of 512 (2048) complex data points. A total of 40 scans were acquired per FID with a 1.0 s delay, for a total time per spectrum of 7 hrs.
For lineshape analysis titration experiments, ~1.0 mM unlabeled Pin1-PPIase was added to 0.7 mM labeled peptide and the protein was reverse titrated from 1.0 mM to 0.06 mM by diluting with labeled peptide. Titration points were taken at enzyme concentrations of 1, 0.5, 0.25, 0.1, 0.06, and 0 mM. At each concentration, a two-dimensional 13C-1H constant-time (constant time delay of 27 ms) HSQC spectrum (Vuister and Bax, 1992) was taken with a spectral width of 5.5 kHz (8 kHz) in t1 (t2), and a total of 256 (2048) complex data points. A total of 88 scans were acquired per FID with a with a repetition delay of 1.0 s, for a total time per spectrum of 7.5 hrs. Constant-time spectra were taken to prevent splitting in the carbon dimension. Wrapping the alpha and delta peaks in the 13C dimension allowed for a smaller window with still well-resolved peaks. The 13C time domain was extended to 512 points by mirror-image linear prediction prior to zero filling.
For rate measurements, 3 mM of peptide was dissolved in NMR buffer and a catalytic amount of Pin1-PPIase (50 μM) was added. The rate of interconversion between the cis and trans isomers was measured by taking a series of ROESY (Bax and Davis, 1985) spectra with varying mixing times. The rate measurement was taken with mixing times of 0, 0.01, 0.02, 0.04, 0.06 0.08, 0.10, 0.15, and 0.20 s. Spectral widths used were 6 kHz in both dimensions, using a total of 192 complex data points in t1 and 2048 in t2. A total of 52 scans per FID were acquired with a recycle delay of 1.0 s, for a total acquisition time of 64 hrs.
Data fitting
ROESY peak heights for autopeaks and crosspeaks corresponding to the amide proton of E670 were obtained using Sparky processing software after baseline correction using a Gaussian fit (Goddard, 2008). Peak heights were fit to the two-state solution to the Bloch-McConnell equations (Cavanagh, 2007; McConnell, 1958) by least-squares minimization using the Solver function in Microsoft Excel. Fixed parameters included the transverse relaxation constants for the cis and trans isomers, and determined independently by running the ROESY experiment without enzyme present, and the ratio of trans isomer to cis isomer, Keq, determined independently by comparing peak volumes of the two isomers. The cis autopeak was broadened significantly relative to the trans autopeak upon addition of Pin1-PPIase due to the high rate of isomerization. To accommodate this, the data was fit using an additional fitted parameter, , which scales the intensity of the trans-to-cis cross-peak. Other fitted parameters included the initial intensities of autopeaks, and , and the overall rate of isomerization, kex. Errors were determined by Monte Carlo analysis, using the noise of the spectrum as experimental error and fitting in Matlab 2010a.
Lineshape fitting was performed using the BiophysicsLab Matlab package (Kovrigin, E.L., manuscript in preparation, http://biophysicslab.net). BiophysicsLab fits data extracted as 1D slices from Sparky (Goddard, 2008) using a custom Python extension BiophysicsLab 1DNMR. 1H-dimension slices from cis and trans peaks of the 13C-labeled phosphopeptide were extracted separately and normalized by the area under each peak to correct for broadening in the 13C dimension. Cis and trans datasets were then added together (weighting appropriately) before lineshape fitting. Data was fit to the four-state reaction scheme in figure 1, which considers both the free and bound states of both isomers.
Fitted parameters included the on-enzyme equilibrium constant (), the on-enzyme catalysis rate from trans-to-cis (), and the chemical shifts of the proline resonances in the bound peptide ( and ). Constrained parameters included the apparent dissociation constant (), the equilibrium constant for the cis and trans isomers of the free peptide (), the intrinsic (uncatalyzed) rate from trans-to-cis () obtained by measuring isomer-specific binding kinetics (unpublished data), the chemical shifts ( and ) and linewidths () of the proline resonances in the free peptide, and the small 1H-1H J-couplings between proline ring protons . Because the parameters Keq, , and are intrinsic to the free peptide, and easily measured using only the apo spectrum, these parameters were fit once from the apo spectrum and held constant during lineshape fitting. The numerous couplings between proline protons, , are available in the literature (Aliev and Courtier-Murias, 2007) and thus held constant. The parameters and were not deemed to be reliably extractable parameters from the lineshape titration data, and so were obtained using independent experiments (enzyme-perspective titration, below, and binding kinetics, data not shown). Likewise, the contributions to the apparent transverse relaxation rates of resonances from the bound peptide not due to exchange and the reverse binding rate constants and were not deemed to be reliably extractable parameters, so rather than fitting for them they were constrained and systematically varied to assess the impact of their values on the results and quality of the fit, as discussed in results and discussion as well as supplementary information (table S-1).
BiophysicsLab fits titration lineshape data with the Bloch-McConnell equations in the following way: the population column vector P (equation 1a), which describes the populations of the four states (pi; free cis, free trans, bound cis and bound trans), is calculated for each spectrum using the thermodynamic parameters , , and , and the total enzyme/peptide concentrations at each titration point (supplementary information, equations S-1, S-2,S-3). The rate matrix, which describes the equilibrium rate equations for the four species, is then calculated for each spectrum using the calculated population of free Pin1-PPIase (PPIasefree) (equation 1b). The first-order rates for binding and isomerization are featured in the diagonal and off-diagonal elements of the rate matrix. The chemical shifts (νi) of each of the four states appear as the diagonal elements of a square matrix (equation 1c, term 2). Likewise, the apparent transverse relaxation rates of resonances not due to exchange appear in a second matrix (equation 1c, term 1), and the frequency variable ν appears in a third (equation 1d). The simulated spectrum is given by equation (1e) in the absence of three-bond proton coupling. To account for the small scalar coupling between proline ring protons, the final simulated spectrum was expressed as a sum of lorentzians that took the form of equation 1e with various small offsets in ν according to the scalar coupling constants reported in the literature (Aliev and Courtier-Murias, 2007) (supplementary information, equations S-4 and S-5 and table S-2). This classical approach is justified if scalar couplings are small compared to natural linewidths and exchange rates, as they are here (discussed in detail in supplementary information).
| (1a) |
| (1b) |
| (1c) |
| (1d) |
| (1e) |
Fitting was performed using the truncated Newton interior-point method (Kim et al., 2007) in Matlab 2010a. The quality of the fit was assessed by calculating the sum of squares of residuals, which are normalized by the square of the RMS noise of each spectrum. Fit results were insensitive to the starting values of fit parameters, indicating that in all cases a true minimum was reached. As errors we report the variations in the fitted parameters from varying within its standard deviation of measurement of 0.3 mM. Uncertainty in was deemed the largest single source of uncertainty in the fit, as varying other constrained parameters or the experimental data itself within their uncertainties did not impact the fit as much (supplementary information, table S-1).
Results and Discussion
Measurement of by NMR titration experiments
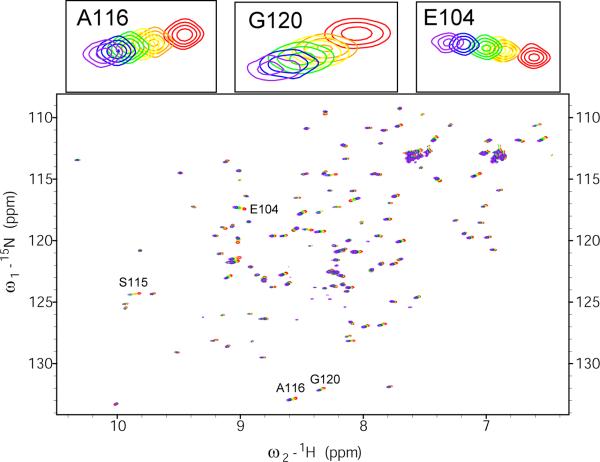
The Pin1-PPIase/pAPP659-682 interaction from the substrate perspective is represented by a four-state binding scheme (Fig. 1a) that is described by seven independent rate, equilibrium, or binding constants. In order to minimize the number of fitted parameters in the linshape analysis, the overall apparent binding constant of Pin1-PPIase and pAPP659-682 was measured using a chemical shift perturbation approach in an independent experiment. Since the binding of Pin1-PPIase to the pAPP659-682 peptide is very weak ( ~ 1 mM), it was not possible to reliably measure the binding constant from substrate-perspective titration data due to the limited solubility of Pin1-PPIase. Therefore, the apparent affinity was measured from the perspective of the 15N-labeled Pin1-PPIase domain. Pin1-PPIase exchanges between three states (free, cis-bound and trans-bound) during catalysis (Fig. 1b). However, approximation of the Pin1-PPIase interaction with the peptide substrate as a two-state (free and bound) exchange process with an equivalent (Fig. 1c) is reasonable if exchange between the bound cis and bound trans states is fast on the chemical shift timescale. A series of 15N-1H HSQC spectra of 0.25 mM 15N-labeled Pin1-PPIase was acquired while titrating with unlabeled pAPP659-682 peptide (soluble up to at least 10 mM). Backbone amide resonances moved progressively during the titration and followed a single trajectory rather than splitting into two peaks (Fig. 2), justifying the two-state approximation (Fig. 1c). The chemical shifts of selected residues were fit to the equation for fast exchange (kon + koff ⪢Δω) between two states:
| (2) |
where [P] refers to the PPIase enzyme concentration, [L] refers to the pAPP659-682 peptide concentration, and Δω is the composite chemical shift perturbation which is given by (Ayed et al., 2001; Mulder et al., 1999)
Fig. 2. Chemical Shift Perturbation of 15N-labeled Pin1-PPIase upon titration with unlabeled pAPP659-682 peptide.
Overlay of 15N-1H HSQC spectra of 0.25 mM 15N-labeled Pin1-PPIase with varying amounts of unlabeled pAPP659-682 peptide, from 0 mM (red) to 12 mM (purple). Expanded regions (above) show example peak trajectories used in the determination of .
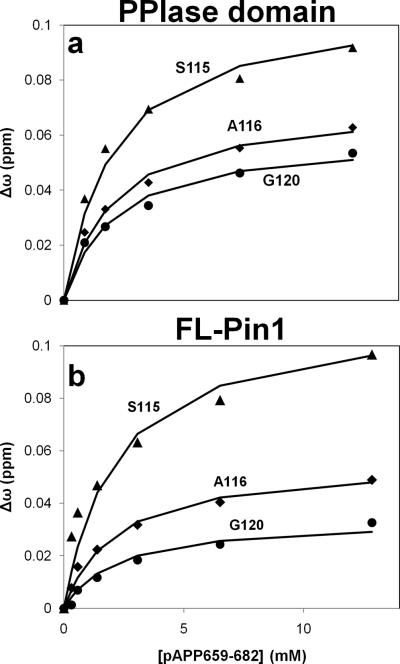
Residues that provided the best fits were in close agreement with each other, yielding a of 1.3 ± 0.3 mM, where error is the standard deviation across 10 individually fit peak walks (Fig. 3a). A global fit of the same 10 residues also yielded an identical of 1.3 mM. This dissociation constant represents a weighted sum of the binding affinities for each isomer, cis and trans, of the substrate peptide:
| (3) |
where Keq=13.6±0.7 is the ratio of free trans isomer to free cis isomer, as determined from the ratio of [trans peak/cis peak] volumes in a 13C-1H HSQC spectrum of pAPP659-682 (error is standard deviation across three sets of peaks). The of 1.3 mM provides a useful constraint on the fitted parameters and accessible from lineshape analysis as described below.
Fig. 3. Overall binding constant of Pin1 for pAPP659-682 is approximately independent of the presence of the WW domain.
To measure the apparent ligand binding affinity, , 15N-labeled PPIase domain (a) or full-length Pin1 (b) were titrated with the APP-derived phosphopeptide pAPP659-682 and a series of 2D 15N-1H HSQC spectra were acquired. The chemical shift perturbation,Δω (see main text), is plotted for a subset of residues that were used to determine for each protein.
In order to assess the pertinence of using the isolated PPIase domain as a model to study Pin1 activity, this analysis was repeated with labeled full-length Pin1 (Fig. 3b). In this case, individual fitting of 7 residues yielded a of 2.2 ± 1.3mM, while a global fit yielded . Because the corresponding values in full-length-Pin1 and isolated PPIase are within error of each other, as well as the close correspondence of the globally fit values, we concluded that Pin1-PPIase binding to pAPP659-682 is essentially independent of the presence of the WW domain under the experimental conditions used.
Lineshape analysis yields kinetic rates and equilibrium constant for Pin1-PPIase isomerization of pAPP659-682
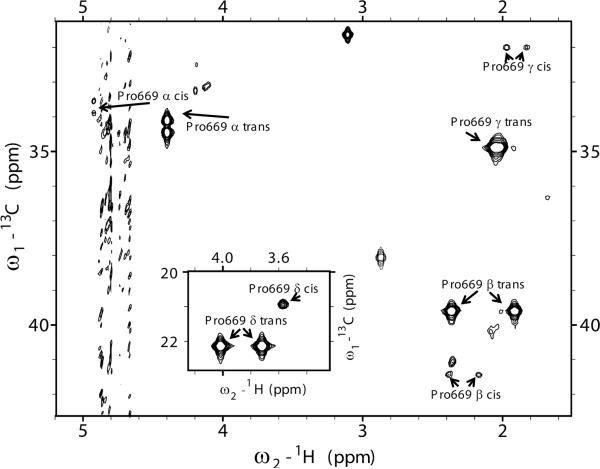
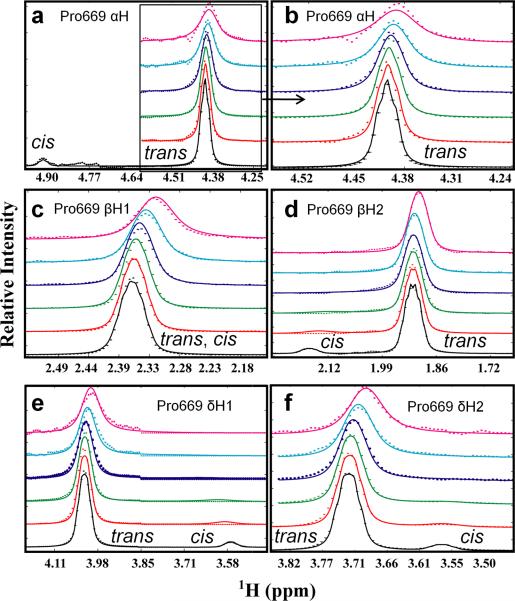
The kinetic rates and equilibrium constants describing catalysis by the Pin1 PPIase domain acting on the pT668-P669 motif were next extracted from the lineshapes of nuclei on the pAPP659-682 phosphopeptide substrate. Although measuring dynamics from the perspective of a saturated enzyme provides a wealth of information about enzyme function and mechanisms, the acquired data can be ambiguous to interpret. Enzyme-perspective dynamics can represent substrate turnover, but they can also represent numerous other exchange processes, related or not related to catalysis (Bosco et al., 2010). Furthermore, depending on the catalytic mechanism and the timescale of substrate binding, resonances from enzyme nuclei may not be sensitive to the actual process of substrate turnover. For these reasons, we sought to measure catalysis by the Pin1 PPIase domain primarily by considering the lineshapes of nuclei on the substrate, which exchange between four states (free cis, free trans, bound cis and bound trans, Fig. 1a). This approach provides the most direct measure of the actual catalytic activity, although it does not by itself provide information about the various residues and motions in the enzyme that contribute to catalysis as enzyme-perspective analysis can (Labeikovsky et al., 2007). We utilized a specifically labeled phosphopeptide, identical to pAPP659-682 but 13C and 15N labeled at Pro669 (13C-pAPP659-682). A constant-time 1H-13C HSQC of 13C-pAPP659-682 contains peaks corresponding to the alpha proton as well as the two beta, two gamma, and two delta protons of the proline ring (Fig. 4). Each proton yielded two peaks, a large peak corresponding to the trans isomer and a smaller peak corresponding to the cis isomer. With the exception of the gamma protons, which have overlap in their trans isomer peaks, and the delta protons, which have overlap in their cis isomer peaks, these peaks are well-resolved from each other. The generally large chemical shift differences between the cis and trans isomer resonances (e.g. ~0.5 ppm in the 1H dimension for the alpha proton) ensure that their linewidths are particularly sensitive to catalyzed exchange between the two states on the second-to-millisecond timescale. Furthermore, the presence of five sets of well-resolved peaks associated with the major trans population (Hα, Hβ1, Hβ2, Hδ1, Hδ2) allows for a global analysis, which can yield a much more reliable fit than analysis of a single pair of resonances.
Fig. 4. 13C-1H constant-time HSQC spectrum of specifically labeled 13C-pAPP659-682 peptide.
A 13C-1H constant-time HSQC (constant time delay of 27 ms) of the APP-derived phosphopeptide 13C-pAPP659-682, specifically 13C labeled on Proline 669, yields a spectrum with peaks corresponding to the proline sidechain protons in both the trans and cis conformations. Both negative (alpha, delta protons) and positive (gamma, beta protons) peaks are shown in black. The alpha proton peaks are split in the carbon dimension due to coupling with the 13C-labeled carbonyl carbon. The trans peaks of the gamma protons were found to overlap, and therefore not considered in the lineshape analysis. (inset) Delta proton peaks.
The four-state binding scheme for the Pin1-PPIase/pAPP659-682 interaction (Fig. 1a) is described by three independent equilibrium or binding constants (the ratio of the trans to cis isomers of the free peptide, , the on-enzyme equilibrium constant, , and the overall apparent binding constant , as defined in Equation 3) as well as four independent rate constants (the uncatalyzed and catalyzed conversion rates from cis-to-trans, and , and off rates for the cis and trans isomers, and ). In order to determine these microscopic rates and equilibria, lineshape analysis was performed on a series of constant-time 13C-1H HSQC spectra of 13C-pAPP659-682 with varying concentrations of unlabeled Pin1-PPIase. Resonances corresponding to the trans isomer typically showed a small upfield shift and modest, progressive broadening (Fig. 5). In all cases, the peaks corresponding to the cis isomer broadened substantially upon addition of even low (60 μM) concentrations of enzyme. This can be expected because, in the limit of slow exchange, exchange broadening preferentially affects the minor exchanging component as it has a shorter lifetime (faster rate constant) (Wang and Palmer Iii, 2003). Because of this, essentially only the apo peak positions for cis peaks impacted the fit. Constant-time HSQC spectra were utilized to eliminate splitting in the carbon dimension, and to provide resolution of peaks that would otherwise be overlapped in the 1H dimension. However, use of constant-time 13C evolution precluded lineshape analysis in this dimension, since this mode of evolution provides no encoding of exchange processes into the 13C lineshapes.
Fig. 5. NMR lineshape fitting of 13C-pAPP659-682 peptide titration data.
Individual 1H lineshapes for five well-resolved 13C-bonded protons of Pro669 (panels A-F) were globally fit using lineshape analysis. Stack plots show extracted 1D slices (frequency axis expressed in ppm) from 2D 13C-1H constant-time HSQC spectra of 0.7 mM 13C-pAPP659-682 peptide with varying concentrations of unlabeled Pin1-PPIase, from 0 mM (black dots) to 1 mM (magenta dots). The corresponding best-fit globally analyzed lineshapes are shown as solid lines in matching colors. Close proximity of cis-Pro669 αH to the water resonance detracted from its inclusion in the stack plot (A); lineshapes of trans-Pro669 αH are shown in more detail (B).
Proton dimension peak slices were extracted using the BiophysicsLab Sparky extension (Kovrigin, E.L., http://biophysicslab.net). To allow simultaneous visualization and analysis of data from both the trans and cis peaks, peak slices from each isomer of a given nucleus were added together in Matlab to generate a single 1D 1H spectrum containing both isomer peaks. The resulting 1D 1H spectra were then fit to a four-state model that considered the bound trans and bound cis states as well as apo trans and cis states (Fig. 1a). The on-enzyme apparent transverse relaxation rate in the absence of exchange () was constrained to be 20 Hz, the average value for aliphatic protons in the enzyme. The theoretical value of the transverse relaxation rate for a 13C-attached proton in a 16 kDa complex was also calculated to be 17 Hz using Stokes' law and Fig. 1.6 in (Cavanagh, 2007). Because varying this constrained parameter within a range of ± 4 Hz did not impact the result or quality of the fit substantially (table S-1), this approach was deemed valid. Because in general these 1H lineshapes exhibited small splitting due to J-coupling between the proline ring protons, each peak was modeled as a superposition of lorentzians, using the literature scalar coupling values between the ring protons of L-proline as reported by (Aliev and Courtier-Murias, 2007) (detailed in supplementary information). In order to obtain consistent results, it was necessary to simultaneously consider all five groups of peaks in a global fit. The obtained rate, equilibrium, and dissociation constants are presented in the top section of table 1, while constrained global parameters and nucleus-specific parameters are presented in the bottom section.
Table 1.
Summary of fitted and constrained parameters
| Fit parameters | on-enzyme trans/cis | (mM) | (mM) | on-enzyme kex (s−1) |
|---|---|---|---|---|
|
|
||||
| 3.9 ± 0.8 | 1.5 ± 0.4 | 0.4 ± 0.1 | 1500 ± 100 | |
| Constrained parameters | free trans/cisa | (mM)b | Uncatalyzed kex (s−1)c | (s−1)d | (s−1)d |
|---|---|---|---|---|---|
|
|
|||||
| 13.6 ± 0.7 | 1.3 ± 0.3 | 0.0085 ± 0.0008 | ≥ 4,000 | ≥ 20,000 | |
| Proline Protons | Δω trans (ppm) | Δω cis (ppm) | (Hz) | |
|---|---|---|---|---|
|
|
||||
| Nucleus-specific parameters | Hα | −0.075 | −0.197 | 4.4 |
| Hβ1 | −0.074 | −0.477 | 5.0 | |
| Hβ2 | −0.040 | −0.434 | 4.3 | |
| Hδ1 | −0.019 | 0.357 | 4.7 | |
| Hδ2 | −0.038 | −0.143 | 4.8 | |
Determined from peak volumes
Determined from NMR titration of enzyme
Determined from binding kinetics (data not shown)
Assuming a diffusion-limited on-rate of ≥ 107 M−1s−1
The value obtained for the on-enzyme rate of exchange, , was 1500 ± 100 s–1, in very close agreement with the rate of conformational exchange during catalysis of a model peptide (1200 ± 200 s−1) as detected by Labeikovsky et. al. (Labeikovsky et al., 2007). Furthermore, since that study showed similar motions in the free enzyme, this measurement seems to suggest that the rates of Pin1 catalysis are dictated by intrinsic motions that are largely independent of the actual substrate. The ratio of bound trans to bound cis was found to be 3.9 ± 0.8, which may indicate a role for the enzyme in restoring equilibrium to this substrate when it is skewed slightly towards the cis isomer, as discussed later. Determination of this ratio allows for the separation of the apparent into the isomer-specific binding constants and , using equation 3 and the definition . Errors were obtained by propagating the errors in and Keq.
The dissociation rates and were found to have only a small effect on the fit unless they were constrained to be below 1,000 s−1, which reduced the quality of the fit (supplementary information, table S-1). This observation suggests that and are sufficiently fast as to have no significant impact on lineshapes. This interpretation is consistent with the enzyme-perspective data, which indicated fast exchange. Assuming diffusion-limited on-rates of greater than 107 M−1s−1, we can only bracket and as greater than 4,000 s−1 and 20,000 s−1 respectively.
The best global fit of the data yielded rather large chemical shift changes for the bound cis populations of the beta protons and one of the delta protons, on the order of 0.4 ppm. While not unusual for chemical shift changes due to protein-ligand interactions, these were significantly larger than those for the trans peaks. This may reflect a distinct environment for the cis isomer in the active site of the enzyme, possibly involving closer interactions with the proline ring than for the trans isomer. It should be noted that in the fitting these three chemical shift changes diverged to even larger values when left unconstrained, and that the values presented reflect the upper or lower allowed limits in the fit. Loosening the limits, however, did not result in a substantially improved fit. Therefore, the reported bound chemical shifts for the cis isomer, while approximate, clearly indicate a significant change in chemical environment upon binding to the active site.
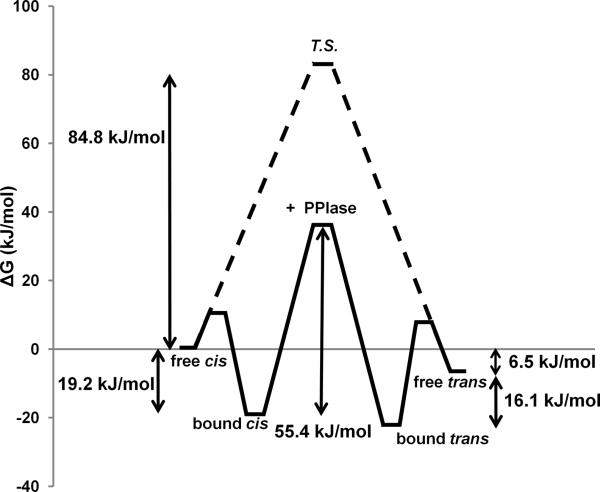
In order to construct the energy diagram of Pin1-PPIase catalysis, free energies for each of the four states in the model were calculated using the equations
| (4a) |
| (4b) |
where ΔGbind is the Gibbs free energy of binding for each isomer, kB is the Boltzmann constant, T is temperature, kD is the dissociation constant for each isomer, ΔGcis − ΔGtrans is the difference between the Gibbs free energies of the cis and trans isomers, and keq is the ratio of free trans and cis populations. Activation energies were calculated using the Eyring equation:
| (5) |
where ΔGi refers to the activation barrier (catalyzed or uncatalyzed) for either the cis (i=ct) or trans (i=tc) isomer, h is Planck's constant, and ki is the measured rate of exchange (catalyzed or uncatalyzed) between cis and trans (i=ct) or between trans and cis (i=tc). The values obtained for the peptide bond torsion barrier on (55.4 kJ/mol) and off (84.8 kJ/mol) the enzyme are in good agreement with published measurements and estimates for these values (Kofron et al., 1991; Park et al., 1992; Schutkowski et al., 1998).
Measurement of overall cis/trans exchange by ROESY
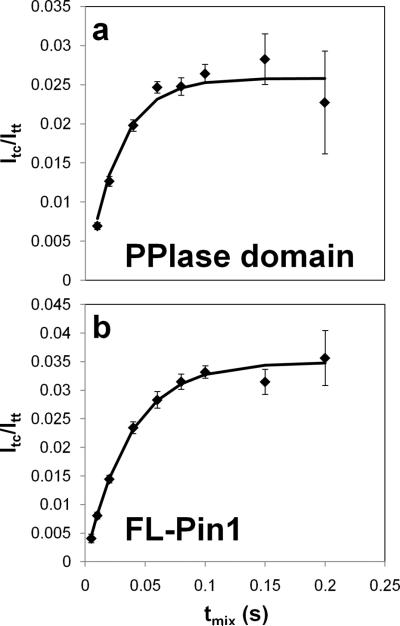
As a validation of the best-fit parameters, the catalyzed rate of exchange between the cis and trans isomers was measured by 1H-1H ROESY experiments. A catalytic amount of PPIase (50μ M) was added to a 3 mM sample of unlabeled pAPP659-682 and 1H-1H ROESY spectra were taken with a series of mixing times. In the absence of Pin1, the cis and trans conformations of the amide backbone resonance of E670 are resolved and do not detectably exchange in a ROESY spectrum. Upon addition of Pin1, chemical exchange crosspeaks appear and vary in intensity as a function of mixing time. The intensities of auto-peaks (cis and trans, Icc(t) and Itt (t)) and cross-peaks (trans-to-cis and cis-to-trans, Itc(t) and Ict (t)) were obtained and fit to the two-state solution of the Bloch-McConnell equations (Cavanagh, 2007; McConnell, 1958) to extract out the rate of isomerization (Fig. 6a). All four curves for cross- and auto-peaks were simultaneously fit to obtain rates. The value of kex = kcis→trans + ktrans→cis = 41 ± 1 s−1 was obtained and found to be in excellent agreement with the value of 47 s–1 predicted by the measured microscopic rates, using the reversible Michaelis-Menten equation . When this analysis was repeated with full-length Pin1 (Fig. 6b), a value of kex = 28 ± 1 s−1 was obtained. Although this is modestly lower than the value for the isolated PPIase domain, we concluded that there is no drastic difference in catalysis of pAPP659-682 by Pin1-PPIase and full-length Pin1 under the conditions of the experiment, validating our approach of focusing on the isolated catalytic domain.
Fig. 6.
In order to measure isomerization rates, a series of 1H-1H ROESY spectra of 3.0 mM pAPP659-682 were taken in the presence of 50 µM PPIase (A) or Full-length Pin1 (B). Shown are the build-up normalized cross-peak curves Itc / Itt as a function of mixing time tmix. The rate of isomerization was determined by fitting auto- and crosspeaks, yielding a kex = kcis→trans + ktrans→cis of 41 ± 1 s−1 for the PPIase domain and 28 ± 1 s−1 for full-length Pin1.
A complete description of the Pin1-PPIase thermodynamic cycle for catalysis of a biological substrate
Taken together, the results obtained here provide a complete kinetic and thermodynamic description of the Pin1-PPIase cycle for isomerization of a phosphopeptide corresponding to the Pin1-targeted motif in the intracellular domain of APP. The kinetic rates for each individual step and the corresponding equilibrium constants have been determined (Fig. 1a, table 1). A diagram mapping the corresponding relative free energies of each state (Fig. 7) illustrates that the modest binding energies for each isomer (19.2 kJ and 16.1 kJ for the cis and trans isomers, respectively), combined with the more favorable binding to the syn transition state (49 kJ/mol, or 11.6 kcal/mol) result in a lowering of the activation barrier by 29.4 kJ (7.0 kcal/mol) relative to the uncatalyzed reaction. This enzymatic reduction of the N–C' torsion barrier is comparable to that reported for other PPIase enzymes such as FKBP and CypA (Dugave and Demange, 2003; Fischer et al., 1993; Kofron et al., 1991; Lu et al., 2007; Park et al., 1992). Overall, these results provide the microscopic constants associated with the multi-state thermodynamic cycle of the Pin1 catalytic domain, and demonstrate that these parameters that quantitatively describe the activity of Pin1 on this important biological target are similar to those for other well-studied PPIase enzymes acting on their biological substrates.
Fig. 7.
Free energy diagram of Pin1-PPIase catalysis. The binding, equilibrium, and rate constants obtained by lineshape fitting (Table 1) were used to calculate the energy diagram of the PPIase-pAPP659-682 isomerization reaction. The determined equilibrium and binding constants were used to elucidate the thermodynamic cycle of PPIase binding to the cis and trans isomers of pAPP659-682 (Equations 4a, 4b). The measured rates of isomerization were used to determine the activation barriers between the two states either on or off the enzyme (Equation 5).
Implications of Kint tfor function of Pin1
The internal equilibrium constant, Kint, of an enzyme is defined as the ratio of bound substrate to bound product at equilibrium (Kint = ([ES]eq /[EP]eq). The catalytic efficiency of an enzyme is highly dependent on the relationship between the value of Kint and the conditions under which the enzyme operates. For an enzyme that operates near equilibrium of its substrates and products, the optimal value of Kint is 1 (Burbaum et al., 1989). However, for an enzyme that operates far from equilibrium, the optimal value of Kint is correspondingly skewed. Specifically, if the ratio of substrate to product is much lower than the equilibrium value ([S]/[P]⪡[S]eq /[P]eq) the optimal value of Kint is correspondingly increased (Kint ≫ 1). There is still a limited understanding of the role of Pin1-catalyzed isomerization of many of its substrates, such as the AICD of APP (Pastorino et al., 2006). An important factor when considering a mechanism of Pin1 function is the value of Kint. For example, if Pin1 exhibits a significantly skewed Kint for the pAPP659-682 it may indicate that Pin1 is optimized to operate far from equilibrium in its isomerization of this substrate. This in turn would have implications for the role of isomerization in the biological system, i.e. whether Pin1 serves to restore equilibrium to a population of substrates abnormally enriched for the cis isomer or the trans isomer. The measured value of Kint for the pAPP659-682/Pin1-PPIase complex is 3.9, which may suggest Pin1 is optimized to restore equilibrium to the AICD under conditions where it is slightly skewed towards the cis isomer. Pin1 has evolved under selective pressure from multiple substrates (Lu and Zhou, 2007), so it should not be assumed that Pin1 is optimized for its catalysis of the AICD; however, this result is suggestive because evidence exists that implicates the cis isomer of the pT668-Pro motif in the AICD in amyloidogenic processing of APP (Iijima et al., 2000; Lee et al., 2003; Ramelot and Nicholson, 2001), and Pin1 protects against this (Pastorino et al., 2006). Future work on other substrates such as the heptad motifs in the CTD of RNA Polymerase II (Buratowski, 2003) should provide insights into the significance of the value of Kint.
Concluding Remarks
During the past twenty years, the power of NMR spectroscopy for elucidating the detailed motions of proteins has dramatically increased, with decisive contributions from Lewis Kay and his research group (Grishaev et al., 2008; Hansen et al., 2008a; Hansen et al., 2008b; Religa et al., 2010; Sprangers and Kay, 2007; Vallurupalli et al., 2008). Particularly important is the development of methods to detect and characterize microsecond to millisecond timescale motions via CPMG (Hansen et al., 2008a) or R1ρ (Massi et al., 2004) relaxation experiments that yield dispersion profiles that can be fit to reveal rates, populations and chemical shifts associated with exchange processes. These methods have opened important windows into motions that occur on the timescale of as many biological processes, and have greatly enhanced our understanding of relationships between internal protein motions and function (Mittermaier and Kay, 2009b). While the interpretation of relaxation dispersion profiles has been extended to three-state (Sugase et al., 2007) and four state (Li et al., 2008) exchange processes, the complexity of the exchange reaction remains the major limitation of these methods. Here we have applied classic lineshape analysis as a complementary approach for measuring exchange processes (kex < 105 s−1) that offers the advantage of being based on the convenient and sensitive HSQC experiment (Mulder et al., 1996).
This study highlights the ability of NMR lineshape analysis to provide a quantitative description of functional motions during catalysis for the four-state reaction scheme of Pin1-PPIase acting on a biological substrate involved in the pathogenesis of Alzheimer's disease. Lineshape analysis of the 13C-Pro-labeled substrate yielded individual kinetic rates, equilibrium constants, and an overall catalytic rate that was independently verified using ROESY exchange spectroscopy. Importantly, since 13C-labeled proline yields a very characteristic spectrum with many well-resolved alpha, beta, delta, and gamma proton peaks, this labeling strategy coupled with lineshape analysis is applicable to the study of any prolyl isomerase and any peptide substrate. Our results support the interpretation of CPMG-derived chemical exchange occurring in and near the active site of Pin1-PPIase saturated with an optimized peptide substrate as on-enzyme isomerization of the substrate (Labeikovsky et al., 2007). These results further suggest that, since similar on-enzyme exchange rates were observed for distinctly different substrates, the intrinsic motions of Pin1-PPIase pre-determine the approximate timescale of this exchange and substrate-specific variations in overall catalytic rate are governed by their relative affinities. Of particular interest for future lineshape analysis studies is the characterization of specific Pin1-PPIase mutants to gain an in-depth understanding of the Pin1 catalytic mechanism.
Supplementary Material
Acknowledgements
Financial support was provided by the US National Institutes of Health (R01-AG029385).
References
- Aliev AE, Courtier-Murias D. Conformational analysis of L-prolines in water. J Phys Chem B. 2007;111:14034–14042. doi: 10.1021/jp076729c. [DOI] [PubMed] [Google Scholar]
- Anderson P. Pin1: a proline isomerase that makes you wheeze? Nature Immunology. 2005;6:1211–1212. doi: 10.1038/ni1205-1211. [DOI] [PubMed] [Google Scholar]
- Ayed A, Mulder FA, Yi GS, Lu Y, Kay LE, Arrowsmith CH. Latent and active p53 are identical in conformation. Nat Struct Biol. 2001;8:756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- Bax AD, Davis DG. Practical aspects of two-dimensional transverse NOE spectroscopy. J. Magn. Reson. 1985;63:207–213. [Google Scholar]
- Bosco DA, Eisenmesser EZ, Clarkson MW, Wolf-Watz M, Labeikovsky W, Millet O, Kern D. Dissecting the microscopic steps of the cyclophilin A enzymatic cycle on the biological HIV-1 capsid substrate by NMR. J Mol Biol. 2010;403:723–738. doi: 10.1016/j.jmb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- Burbaum JJ, Raines RT, Albery WJ, Knowles JR. Evolutionary optimization of the catalytic effectiveness of an enzyme. Biochemistry. 1989;28:9293–9305. doi: 10.1021/bi00450a009. [DOI] [PubMed] [Google Scholar]
- Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR spectroscopy: principles and practice. Second edn Academic Pr; 2007. [Google Scholar]
- Craven CJ, Whitehead B, Jones SK, Thulin E, Blackburn GM, Waltho JP. Complexes formed between calmodulin and the antagonists J-8 and TFP in solution. Biochemistry. 1996;35:10287–10299. doi: 10.1021/bi9605043. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dugave C, Demange L. Cis-trans isomerization of organic molecules and biomolecules: implications and applications. Chem Rev. 2003;103:2475–2532. doi: 10.1021/cr0104375. [DOI] [PubMed] [Google Scholar]
- Eckerdt F, Yuan J, Saxena K, Martin B, Kappel S, Lindenau C, Kramer A, Naumann S, Daum S, Fischer G. Polo-like kinase 1-mediated phosphorylation stabilizes Pin1 by inhibiting its ubiquitination in human cells. Journal of Biological Chemistry. 2005;280:36575. doi: 10.1074/jbc.M504548200. [DOI] [PubMed] [Google Scholar]
- Fanghanel J. Enzymatic catalysis of the peptidyl-prolyl bond rotation: are transition state formation and enzyme dynamics directly linked? Angew Chem Int Ed Engl. 2003;42:490–492. doi: 10.1002/anie.200390149. [DOI] [PubMed] [Google Scholar]
- Fischer G, Aumüller T. Regulation of peptide bond cis/trans isomerization by enzyme catalysis and its implication in physiological processes. Reviews of physiology, biochemistry and pharmacology. 2003:105–150. doi: 10.1007/s10254-003-0011-3. [DOI] [PubMed] [Google Scholar]
- Fischer G, Bang H, Berger E, Schellenberger A. Conformational specificity of chymotrypsin toward proline-containing substrates. Biochimica et Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology. 1984;791:87–97. doi: 10.1016/0167-4838(84)90285-1. [DOI] [PubMed] [Google Scholar]
- Fischer G, Heins J, Barth A. The conformation around the peptide bond between the P1-and P2-positions is important for catalytic activity of some proline-specific proteases. Biochimica et Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology. 1983;742:452–462. doi: 10.1016/0167-4838(83)90261-3. [DOI] [PubMed] [Google Scholar]
- Fischer S, Michnick S, Karplus M. A mechanism for rotamase catalysis by the FK506 binding protein (FKBP) Biochemistry. 1993;32:13830–13837. doi: 10.1021/bi00213a011. [DOI] [PubMed] [Google Scholar]
- Garcia-Echeverria C, Kofron JL, Kuzmic P, Kishore V, Rich DH. Continuous fluorimetric direct (uncoupled) assay for peptidyl prolyl cis-trans isomerases. Journal of the American Chemical Society. 1992;114:2758–2759. [Google Scholar]
- Garcia-Echeverria C, Kofron JL, Kuzmic P, Rich DH. A continuous spectrophotometric direct assay for peptidyl prolyl cis-trans isomerases. Biochemical and Biophysical Research Communications. 1993;191:70–75. doi: 10.1006/bbrc.1993.1185. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. Sparky 3. University of California; San Franscisco: 2008. [Google Scholar]
- Grathwohl C, Wüthrich K. NMR studies of the rates of proline cis-trans isomerization in oligopeptides. Biopolymers. 1981;20:2623–2633. [Google Scholar]
- Grishaev A, Tugarinov V, Kay LE, Trewhella J, Bax A. Refined solution structure of the 82-kDa enzyme malate synthase G from joint NMR and synchrotron SAXS restraints. Journal of Biomolecular NMR. 2008;40:95–106. doi: 10.1007/s10858-007-9211-5. [DOI] [PubMed] [Google Scholar]
- Gunther UL, Schaffhausen B. NMRKIN: simulating line shapes from two-dimensional spectra of proteins upon ligand binding. J Biomol NMR. 2002;22:201–209. doi: 10.1023/a:1014985726029. [DOI] [PubMed] [Google Scholar]
- Hansen DF, Vallurupalli P, Kay LE. An improved 15N relaxation dispersion experiment for the measurement of millisecond time-scale dynamics in proteins. The Journal of Physical Chemistry B. 2008a;112:5898–5904. doi: 10.1021/jp074793o. [DOI] [PubMed] [Google Scholar]
- Hansen DF, Vallurupalli P, Lundström P, Neudecker P, Kay LE. Probing Chemical Shifts of Invisible States of Proteins with Relaxation Dispersion NMR Spectroscopy: How Well Can We Do? Journal of the American Chemical Society. 2008b;130:2667–2675. doi: 10.1021/ja078337p. [DOI] [PubMed] [Google Scholar]
- Iijima K, Ando K, Takeda S, Satoh Y, Seki T, Itohara S, Greengard P, Kirino Y, Nairn AC, Suzuki T. Neuron Specific Phosphorylation of Alzheimer's Amyloid Precursor Protein by Cyclin Dependent Kinase 5. Journal of neurochemistry. 2000;75:1085–1091. doi: 10.1046/j.1471-4159.2000.0751085.x. [DOI] [PubMed] [Google Scholar]
- Johnson PE, Creagh AL, Brun E, Joe K, Tomme P, Haynes CA, McIntosh LP. Calcium binding by the N-terminal cellulose-binding domain from Cellulomonas fimi −1, 4-glucanase CenC. Biochemistry. 1998;37:12772–12781. doi: 10.1021/bi980978x. [DOI] [PubMed] [Google Scholar]
- Kern D, Kern G, Scherer G, Fischer G, Drakenberg T. Kinetic analysis of cyclophilin-catalyzed prolyl cis/trans isomerization by dynamic NMR spectroscopy. Biochemistry. 1995;34:13594–13602. doi: 10.1021/bi00041a039. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Koh K, Lustig M, Boyd S, Gorinevsky D. An interior-point method for large-scale l1-regularized least squares. Selected Topics in Signal Processing, IEEE Journal of. 2007;1:606–617. [Google Scholar]
- Kofron JL, Kuzmic P, Kishore V, Colon-Bonilla E, Rich DH. Determination of kinetic constants for peptidyl prolyl cis-trans isomerases by an improved spectrophotometric assay. Biochemistry. 1991;30:6127–6134. doi: 10.1021/bi00239a007. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Kay LE. Probing invisible, low-populated States of protein molecules by relaxation dispersion NMR spectroscopy: an application to protein folding. Acc Chem Res. 2008;41:442–451. doi: 10.1021/ar700189y. [DOI] [PubMed] [Google Scholar]
- Kovrigin EL, Loria JP. Enzyme dynamics along the reaction coordinate: critical role of a conserved residue. Biochemistry. 2006;45:2636–2647. doi: 10.1021/bi0525066. [DOI] [PubMed] [Google Scholar]
- Labeikovsky W, Eisenmesser EZ, Bosco DA, Kern D. Structure and dynamics of pin1 during catalysis by NMR. J Mol Biol. 2007;367:1370–1381. doi: 10.1016/j.jmb.2007.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrieu I, De Veylder L, Fruchart JS, Odaert B, Casteels P, Portetelle D, Van Montagu M, Inzé D, Lippens G. The Arabidopsis thaliana PIN1At Gene Encodes a Single-domain Phosphorylation-dependent Peptidyl Prolylcis/trans Isomerase. Journal of Biological Chemistry. 2000;275:10577. doi: 10.1074/jbc.275.14.10577. [DOI] [PubMed] [Google Scholar]
- Lee MS, Kao SC, Lemere CA, Xia W, Tseng HC, Zhou Y, Neve R, Ahlijanian MK, Tsai LH. APP processing is regulated by cytoplasmic phosphorylation. J Cell Biol. 2003;163:83–95. doi: 10.1083/jcb.200301115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Martins IR, Amarasinghe GK, Rosen MK. Internal dynamics control activation and activity of the autoinhibited Vav DH domain. Nat Struct Mol Biol. 2008;15:613–618. doi: 10.1038/nsmb.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LN, Brandts JF. Isomer-specific proteolysis of model substrates: influence that the location of the proline residue exerts on cis trans specificity. Biochemistry. 1985;24:6533–6538. doi: 10.1021/bi00344a034. [DOI] [PubMed] [Google Scholar]
- Lu KP, Finn G, Lee TH, Nicholson LK. Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol. 2007;3:619–629. doi: 10.1038/nchembio.2007.35. [DOI] [PubMed] [Google Scholar]
- Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- Lu KP, Suizu F, Zhou XZ, Finn G, Lam P, Wulf G. Targeting carcinogenesis: a role for the prolyl isomerase Pin1? Mol Carcinog. 2006;45:397–402. doi: 10.1002/mc.20216. [DOI] [PubMed] [Google Scholar]
- Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Zhou XZ, Liou YC, Noel JP, Lu KP. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. Journal of Biological Chemistry. 2002;277:2381. doi: 10.1074/jbc.C100228200. [DOI] [PubMed] [Google Scholar]
- Massi F, Johnson E, Wang C, Rance M, Palmer AG., 3rd NMR R1 rho rotating-frame relaxation with weak radio frequency fields. J Am Chem Soc. 2004;126:2247–2256. doi: 10.1021/ja038721w. [DOI] [PubMed] [Google Scholar]
- McConnell HM. Reaction rates by nuclear magnetic resonance. The Journal of Chemical Physics. 1958;28:430. [Google Scholar]
- Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3'-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- Meraz-Rios MAL-DL, K.I., Campos-Pena V, De Anda-Hernandez MA, Mena-Lopez R. Tau oligomers and aggregates in Alzheimer's disease. J Neurochemistry. 2010;112:1353–1367. doi: 10.1111/j.1471-4159.2009.06511.x. [DOI] [PubMed] [Google Scholar]
- Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci. 2009a;34:601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends in biochemical sciences. 2009b;34:601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Mulder FA, Schipper D, Bott R, Boelens R. Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J Mol Biol. 1999;292:111–123. doi: 10.1006/jmbi.1999.3034. [DOI] [PubMed] [Google Scholar]
- Mulder FAA, Spronk CAEM, Slijper M, Kaptein R, Boelens R. Improved HSQC experiments for the observation of exchange broadened signals. Journal of Biomolecular NMR. 1996;8:223–228. doi: 10.1007/BF00211169. [DOI] [PubMed] [Google Scholar]
- Park ST, Aldape RA, Futer O, DeCenzo MT, Livingston DJ. PPIase catalysis by human FK506-binding protein proceeds through a conformational twist mechanism. J Biol Chem. 1992;267:3316–3324. [PubMed] [Google Scholar]
- Pastorino L, Sun A, Lu PJ, Zhou XZ, Balastik M, Finn G, Wulf G, Lim J, Li SH, Li X, et al. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature. 2006;440:528–534. doi: 10.1038/nature04543. [DOI] [PubMed] [Google Scholar]
- Peng JW, Wilson BD, Namanja AT. Mapping the dynamics of ligand reorganization via 13 CH 3 and 13 CH 2 relaxation dispersion at natural abundance. Journal of Biomolecular NMR. 2009;45:171–183. doi: 10.1007/s10858-009-9349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramelot TA, Gentile LN, Nicholson LK. Transient structure of the amyloid precursor protein cytoplasmic tail indicates preordering of structure for binding to cytosolic factors. Biochemistry. 2000;39:2714–2725. doi: 10.1021/bi992580m. [DOI] [PubMed] [Google Scholar]
- Ramelot TA, Nicholson LK. Phosphorylation-induced structural changes in the amyloid precursor protein cytoplasmic tail detected by NMR. J Mol Biol. 2001;307:871–884. doi: 10.1006/jmbi.2001.4535. [DOI] [PubMed] [Google Scholar]
- Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- Reimer U, Scherer G, Drewello M, Kruber S, Schutkowski M, Fischer G. Side-chain effects on peptidyl-prolyl cis/trans isomerisation. J Mol Biol. 1998;279:449–460. doi: 10.1006/jmbi.1998.1770. [DOI] [PubMed] [Google Scholar]
- Religa TL, Sprangers R, Kay LE. Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science. 2010;328:98. doi: 10.1126/science.1184991. [DOI] [PubMed] [Google Scholar]
- Rippmann JF, Hobbie S, Daiber C, Guilliard B, Bauer M, Birk J, Nar H, Garin-Chesa P, Rettig WJ, Schnapp A. Phosphorylation-dependent proline isomerization catalyzed by Pin1 is essential for tumor cell survival and entry into mitosis. Cell growth & differentiation: the molecular biology journal of the American Association for Cancer Research. 2000;11:409. [PubMed] [Google Scholar]
- Sarkar P, Saleh T, Tzeng SR, Birge RB, Kalodimos CG. Structural basis for regulation of the Crk signaling protein by a proline switch. Nat Chem Biol. 2011;7:51–57. doi: 10.1038/nchembio.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutkowski M, Bernhardt A, Zhou XZ, Shen M, Reimer U, Rahfeld JU, Lu KP, Fischer G. Role of phosphorylation in determining the backbone dynamics of the serine/threonine-proline motif and Pin1 substrate recognition. Biochemistry. 1998;37:5566–5575. doi: 10.1021/bi973060z. [DOI] [PubMed] [Google Scholar]
- Skinner GM, Baumann CG, Quinn DM, Molloy JE, Hoggett JG. Promoter binding, initiation, and elongation by bacteriophage T7 RNA polymerase. A single-molecule view of the transcription cycle. J Biol Chem. 2004;279:3239–3244. doi: 10.1074/jbc.M310471200. [DOI] [PubMed] [Google Scholar]
- Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445:618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- Vallurupalli P, Hansen DF, Kay LE. Structures of invisible, excited protein states by relaxation dispersion NMR spectroscopy. Proceedings of the National Academy of Sciences. 2008;105:11766. doi: 10.1073/pnas.0804221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- Vuister GW, Bax A. Resolution enhancement and spectral editing of uniformy 13C-enriched proteins by homonuclear braodband 13C decoupling. Journal of Magnetic Resonance. 1992;98:428–435. [Google Scholar]
- Wang C, Palmer Iii AG. Solution NMR methods for quantitative identification of chemical exchange in 15N labeled proteins. Magnetic Resonance in Chemistry. 2003;41:866–876. [Google Scholar]
- Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P. cis-Proline-mediated Ser(P)5 Dephosphorylation by the RNA Polymerase II C-terminal Domain Phosphatase Ssu72. J Biol Chem. 2011;286:5717–5726. doi: 10.1074/jbc.M110.197129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, Tong L. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010;467:729–733. doi: 10.1038/nature09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XZ, Kops O, Werner A, Lu PJ, Shen M, Stoller G, Kullertz G, Stark M, Fischer G, Lu KP. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol Cell. 2000;6:873–883. doi: 10.1016/s1097-2765(05)00083-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.