SUMMARY
Trypanothione reductase is a key enzyme in the trypanothione-based redox metabolism of pathogenic trypanosomes. Since this system is absent in humans, being replaced with glutathione and glutathione reductase, it offers a target for selective inhibition. The rational design of potent inhibitors requires accurate structures of enzyme-inhibitor complexes, but this is lacking for trypanothione reductase. We therefore used quinacrine mustard, an alkylating derivative of the competitive inhibitor quinacrine, to probe the active site of this dimeric flavoprotein. Quinacrine mustard irreversibly inactivates Trypanosoma cruzi trypanothione reductase, but not human glutathione reductase, in a time-dependent manner with a stoichiometry of two inhibitors bound per monomer. The rate of inactivation is dependent upon the oxidation state of trypanothione reductase, with the NADPH-reduced form being inactivated significantly faster than the oxidised form. Inactivation is slowed by clomipramine and a melarsen oxide-trypanothione adduct (both are competitive inhibitors) but accelerated by quinacrine. The structure of the trypanothione reductase-quinacrine mustard adduct was determined to 2.7 Å, revealing two molecules of inhibitor bound in the trypanothione-binding site. The acridine moieties interact with each other through π-stacking effects, and one acridine interacts in a similar fashion with a tryptophan residue. These interactions provide a molecular explanation for the differing effects of clomipramine and quinacrine on inactivation by quinacrine mustard. Synergism with quinacrine occurs as a result of these planar acridines being able to stack together in the active site cleft, thereby gaining an increased number of binding interactions, whereas antagonism occurs with non-planar molecules, such as clomipramine, where stacking is not possible.
Keywords: enzyme-inhibitor complex, Trypanosoma cruzi, trypanothione reductase, quinacrine mustard, X-ray diffraction
INTRODUCTION
Parasitic protozoa of the order Kinetoplastida are the causative agents of several serious tropical diseases. These comprise Chagas disease (caused by Trypanosoma cruzi), sleeping sickness (by T. brucei spp) and the leishmaniases, caused by parasites of the genus Leishmania (1). Despite these infections causing in excess of 120,000 deaths per annum (2) comparatively little research effort is directed towards these diseases. As a result of this disregard, current chemotherapy is poor and treatment failure common, due to widespread drug resistance and severe side effects.
The development of new drugs to treat these diseases would be aided by the discovery of compounds that interfere with processes that are essential to the parasites, but absent from their human hosts. A promising target for the design of such inhibitors is the thiol metabolism of these protozoa. This is dependent upon trypanothione (T[SH]2 or N1,N8-bis(glutathionyl)spermidine), a low molecular mass dithiol that is absent from humans but used by the kinetoplastids to maintain their intracellular redox balance (3). This thiol acts to counteract environmental stress through a variety of enzymatic and non-enzymatic reactions, and has been implicated in acquired resistance to chemotherapeutic agents (4, 5). Many of these protective reactions oxidise T[SH]2 to trypanothione disulphide (T[S]2), which is then recycled back to T[SH]2 by trypanothione reductase (TryR), an NADPH-dependent disulphide oxidoreductase. TryR is thought to be the key enzyme in these organisms’ redox metabolism, acting as the sole route of reducing equivalents from the NADP+/NADPH couple to thiol-containing species (6). The fundamental importance of TryR activity to parasite survival has been demonstrated by both genetic manipulations and the discovery that the targets of several of the current antiprotozoal drugs are enzymes involved in trypanothione metabolism (7-9).
In contrast, in the mammalian host, the major low-molecular mass thiol is glutathione (GSH), with the enzyme glutathione reductase acting to reduce glutathione disulphide (GSSG) (10). Although TryR and glutathione reductase are closely related and show approximately 40% sequence identity (11), each enzyme is highly specific for its respective substrate (12). The three dimensional structure of a T. cruzi TryR-T[S]2 complex has been solved, demonstrating that this specific recognition of T[S]2 is mainly determined by the negative charge and hydrophobic surfaces within the large TryR active site (13). The marked differences between the TryR and glutathione reductase active sites therefore make selective inhibition possible and a large number of TryR-specific inhibitors have already been identified (14). Many of these TryR inhibitors are tricyclic compounds that are thought to interact with the hydrophobic polyamine binding site within the active site (15). One of the best characterised of these inhibitors is quinacrine, an acridine derivative that is an effective and selective inhibitor of TryR (16)(17).
The development of more potent TryR inhibitors would be facilitated by high-resolution structures of TryR-inhibitor complexes. Although one structure of the T. cruzi TryR-quinacrine complex has been solved, the resulting model was unfortunately of low resolution and only showed a single quinacrine molecule bound in one of the four active sites that are present in the asymmetric unit (18). This model is inconsistent with subsequent studies that showed that enzyme inhibition is produced by multiple quinacrine molecules binding to the enzyme (19). These conflicting data encouraged us to extend these studies using quinacrine mustard (QM) as a high-affinity probe to characterise the quinacrine binding sites in TryR. This is an alkylating derivative of quinacrine in which the quinacrine diethylamino group has been replaced by the reactive bis-(2-chloroethyl)amino mustard group (Figure 1). Here we show that QM is an active-site directed irreversible inhibitor of TryR. A structure of the resulting covalent TryR-QM adduct was obtained that demonstrated that two QM molecules stack in the TryR active site. The kinetics of the inactivation reaction were studied in detail and these data were found to be entirely consistent with the crystallographic model of the interactions between QM ligands. These results therefore provide a structural explanation for the multi-site inhibition of TryR by quinacrine (19) with implications for the future design of tight-binding and irreversible inhibitors.
Figure 1.
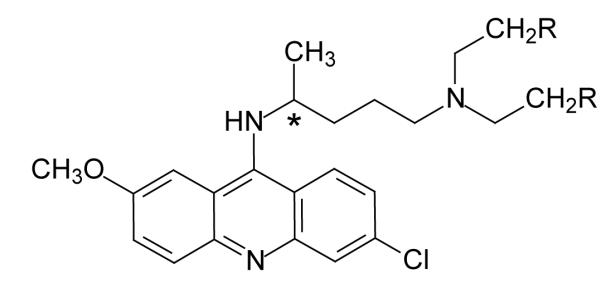
Structure of quinacrine (R=H) and quinacrine mustard (R=Cl). Asterisk indicates the chiral center C-19.
EXPERIMENTAL PROCEDURES
Materials
Recombinant T. cruzi TryR was purified as previously described (20). Glutathione reductase was purified to homogeneity from outdated human blood donations (21). Trypanothione was purchased from Bachem, Germany. Quinacrine and QM were obtained from Sigma. Co-substrate NADPH was purchased from Melford Laboratories. All other reagents were standard commercial products and of the highest available purity.
Trypanothione reductase assay
The concentrations of stock solutions of T[S]2 were determined by titration with TryR and determining the change in NADPH absorption at 340 nm, following total substrate depletion. TryR was quantified spectrophotometrically using the absorption coefficient of 11.3 mM−1 cm−1 at 464 nm (22). TryR activity was assayed at 27°C using either a Shimadzu UV-2401 or a UV-1601 temperature-controlled spectrophotometer in 1 ml assays containing 40 mM (Na+)HEPES pH 7.5, 1 mM EDTA, 150 μM NADPH and 100 μM T[S]2 unless stated otherwise.
Quinacrine mustard inactivation kinetics
Inactivation reactions were carried out in assay buffer containing 40 mM (Na+) HEPES pH 7.5, 1 mM EDTA at room temperature, using 1 μM TryR or NADPH-reduced TryR and varying amounts of QM. Residual activity of inactivation mixtures was determined at various time points by initiating TryR assays with 10-μl samples (100-fold dilution) of this inactivation mixture. The resulting data were then fitted using non-linear regression to a single exponential decay function using the program GraFit (Erithacus Software Limited) written by R.J. Leatherbarrow. All activities are expressed as a percentage of the activity of a parallel control lacking QM.
Measurement of QM-TryR stoichiometry
NADPH-reduced TryR (1 μM) was inactivated with 10 μM QM and aliquots removed at intervals. Inactivation was quenched by adding 2-mercaptoethanol to a final concentration of 25 mM and residual activities determined as described above. Trichloroacetic acid was added to each sample (20% (w/v) final concentration) and incubated for 2 h at 4°C. Denatured protein was recovered by centrifugation (10,000 g, 10 min, 4°C) and washed twice with acetone (−20°C) to remove residual QM and FAD. The protein pellet was then digested with proteinase K (0.1 mg ml−1) in 40 mM HEPES, pH 7.5, 1 mM EDTA for 4 h at 56°C and covalently bound QM was quantified spectrophotometrically using a millimolar absorption coefficient of 7.9 mM−1 cm−1 at 424 nm.
Synthesis of the adduct of melarsen oxide with dihydrotrypanothione (Mel T)
T[SH]2 was produced from T[S]2 by reduction with a 1.5-fold excess of tris-(2-carboxyethyl)phosphine dissolved in Buffer A (10 mM ammonium acetate pH 7.8). The melarsen oxide-trypanothione adduct (Mel T) was prepared by mixing 10 μmole T[SH]2 with 10 μmole melarsen oxide in 0.85 ml of buffer A. The resulting mixture was then applied at 1 ml min−1 to a 1 ml (0.5 × 5 cm) Pharmacia Mono S HR 5/5 cation exchange column that had been pre-equilibrated with 5 column volumes of buffer A. After washing the column with 5 column volumes of buffer A, MelT was eluted by a linear gradient of 0-100% buffer B (500 mM ammonium acetate pH 7.8). Under these conditions, Mel T eluted at approximately 100 mM ammonium acetate. The product was then lyophilised, redissolved in 1 mM HCl and stored at −20°C
Mass spectrometric analysis of QM-TryR adduct
A sample of TryR (20 μM) was reduced with NADPH (150 μM) mixed with 100 μM QM and inactivation monitored by enzymatic assay as described above. When 95% inactivation had been achieved, the reaction was stopped by addition of 2-mercaptoethanol to a final concentration 25 mM. Another control sample was run in parallel without QM. Both samples were dialysed extensively against water, diluted two-fold with 40 mM NH4HCO3 and digested with trypsin (6.25 μg ml−1) overnight at 30°C. Aliquots (1 μl) were spotted onto a MALDI sample target and allowed to dry before addition of 1 μl of matrix (10 mg ml−1 α-cyano-4-hydroxycinnamic acid in 50:50 water acetonitrile / 0.1% (w/v) trifluoracetic acid). The dried samples were then analyzed by MALDI-TOF MS in reflectron mode. The peptide masses identified were then compared to a theoretical digest of the protein.
Crystallographic methods
A sample of 1 mg TryR (20 nmoles active site) in a volume of 1.2 ml was reduced with 500 nmol NADPH and inactivated with 40 nmoles QM. Upon complete inactivation, the reaction was quenched by the addition of 2-mercaptoethanol to 10 mM, the protein exchanged into 100 mM (Na+) malate pH 6 and concentrated to 15 mg ml−1. Crystallisation experiments concentrated on previously reported conditions (23), namely 13 mg ml−1 TryR-QM adduct in (Na+) malate buffer at pH 6.0 and ca. 20% (w/v) PEG 8000 as precipitant. Only one suitable crystal was obtained, the rest were twinned or too small for use. Since we were unable to identify cryo-protection conditions for these TryR crystals, the sample was placed in a capillary with a drop of mother liquor for use in diffraction experiments. Data were collected on BM14 at the European Synchrotron Radiation Facility at room temperature from a single crystal (approximate dimensions 0.05 × 0.05 × 0.3 mm) with a MAR image plate as detector and X-ray wavelength set at 1.00 Å. The space group is P43 with unit cell dimensions of a = 93.1 Å, c = 156.6 Å and the crystal was found to be isomorphous with those previously obtained of the enzyme by itself or in complex with substrate (13, 23). All data were processed, reduced and scaled using the HKL suite of programs (24) then analysed using the CCP4 software package(25). Four percent of the diffraction data, 1228 reflections, were reserved for cross-validation using the R-free parameter (26). Data measurements ceased when radiation damage became evident, which limited the quantity of data that could be obtained, the relevant statistics being provided in Table 1.
Table 1.
Diffraction data, refinement and model statistics for the TryR inhibitor complex.
| Resolution range (Å) | 21 - 2.7 | |
|---|---|---|
| No. of measurements/unique | 86089 / 29073 | |
| Coverage overall (%) | 79 (82) | |
| I/σ(I) all data | 9.4 (2.6) | |
| Rsym overall (%) | 0.08 (0.34) | |
| R-work /R-free (%) | 0.19 / 0.25 | |
| Wilson B (Å2) | 46 | |
| Average isotropic thermal parameters (Å2) |
overall main chain/side chain |
46 44 / 48 |
| solvents/ FAD groups | 38 / 34 | |
| inhibitors | 57 | |
| rms bond lengths and angle associated distances (Å) |
0.006 0.025 |
Numbers in parentheses correspond to the highest resolution shell, a bin of about 0.1Å.
The asymmetric unit contains a functional homodimer, the monomers of which were placed by rigid-body refinement using REFMAC (27). Subsequent positional and temperature factor refinement employing tight non-crystallographic symmetry (NCS) restraints, a bulk solvent correction and anisotropic scaling produced a model suitable for rebuilding in the graphics program O (28). Several rounds of rebuilding, the conservative addition of ordered solvent molecules and refinement produced a model with R and R-free values less than 0.30. At this point interpretation of omit electron density in each active site was made using the coordinates for quinacrine (Reference code QUIANC10, (29) obtained from the Cambridge Structural Database (30). Geometric restraints for refinement were generated taking into account the likely protonation state at pH 6.0. Although the electron density clearly indicates the orientation of the tricyclic ring systems, it is not of sufficient clarity to distinguish between S- and R-isomers at C19, both of which are present in the CSD entry QUIANC10. Accordingly, the S-isomer was arbitrarily chosen for model building. Minimal residual difference density and temperature factors consistent with those of the rest of the structure were obtained by refining the quinacrine moieties at half-occupancy. The final model contains a TryR dimer, four quinacrine moieties, two FAD molecules, one maleic acid and 53 ordered water molecules. The average temperature factor for all atoms is consistent with the Wilson thermal parameter derived from the diffraction data processing. The Ramachandran analysis (31, 32) indicated that 99.7% of the residues are in a favourable conformation. Additional experimental details are presented in Table 1.
RESULTS
Reaction of quinacrine mustard with trypanothione reductase
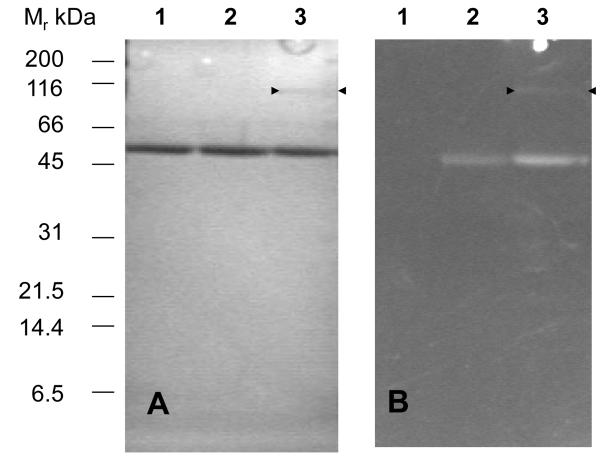
Both oxidized (E) and NADPH-reduced (EH2) forms of TryR show time-dependent inactivation by 20 μM QM (Figure 2). This effect was specific for TryR since exposure of reduced human glutathione reductase to 100 μM QM for 30 min did not cause any inhibition. Inactivation can be rapidly quenched, but not reversed, by the addition of a large excess of 2-mercaptoethanol to remove any un-reacted inhibitor (Figure 2A, arrow). Moreover, inactivation could not be reversed by dilution of the enzyme-inhibitor complex or by extensive dialysis (not shown). However, QM-inactivated TryR could be readily visualised under ultraviolet light in SDS-PAGE gels due to the intense fluorescence of the acridine moiety (Figure 3). In addition to a fluorescent band at 55 kDa corresponding to the TryR monomer, a faint band at 110 kDa is visible in NADPH-reduced samples, suggesting that a minor amount of cross-linking between subunits of the native homodimer also occurs. Together these findings indicate that inactivation involves covalent modification of the enzyme.
Figure 2. Effect of trypanothione reductase redox state on inactivation by quinacrine mustard and protection by the competitive inhibitor Mel T.
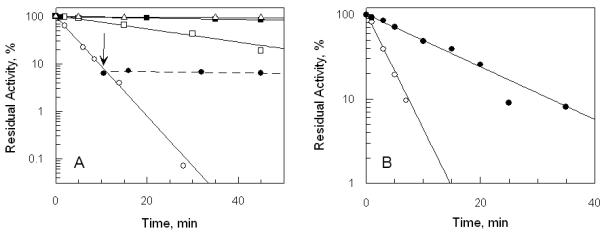
Panel A, effect of redox state. Samples of 1 μM TryR were incubated at room temperature with or without 150 μM NADPH or 20 μM QM and residual activity determined by 100-fold dilution into assay buffer as described in Experimental Procedures. Residual activity is expressed as a percentage of the activity at zero time without addition of QM. No additions:  ; NADPH alone: ■; QM alone: □. NADPH plus QM: ○. Inactivation of TryR in the presence of NADPH and QM was quenched after 10 minutes (arrow) by adding excess 2-mercaptoethanol (25 mM): ●. Glutathione disulphide (2 mM) was included in these latter assays in order to oxidize any residual 2-mercaptoethanol which would otherwise directly reduce the substrate T[S]2. Panel B, protection of trypanothione reductase by Mel T. TryR (1 μM) was incubated with 20 μM QM and 150 μM NADPH, plus or minus 40 μM Mel T. Residual enzyme activity was determined as above. QM alone: ○; QM plus Mel T: ●.
; NADPH alone: ■; QM alone: □. NADPH plus QM: ○. Inactivation of TryR in the presence of NADPH and QM was quenched after 10 minutes (arrow) by adding excess 2-mercaptoethanol (25 mM): ●. Glutathione disulphide (2 mM) was included in these latter assays in order to oxidize any residual 2-mercaptoethanol which would otherwise directly reduce the substrate T[S]2. Panel B, protection of trypanothione reductase by Mel T. TryR (1 μM) was incubated with 20 μM QM and 150 μM NADPH, plus or minus 40 μM Mel T. Residual enzyme activity was determined as above. QM alone: ○; QM plus Mel T: ●.
Figure 3. SDS-PAGE of trypanothione reductase inactivated by quinacrine mustard.
TryR (0.1 mg ml−1) was incubated with or without NADPH (150 μM) and QM (20 μM) for 10 min and the reaction quenched by the addition of excess 2-mercaptoethanol (25 mM). Samples were separated by 10% SDS PAGE and examined under UV-illumination followed by staining with Coomassie blue. Panel A: Coomassie stained gel: lane 1, untreated control; lane 2, plus QM without NADPH; lane 3; plus QM with NADPH. The arrow heads point to an additional faint band at 110 kDa. Panel B: UV-illumination, lanes as in panel A.
Loss of TryR enzyme activity follows pseudo-first-order kinetics for both E and EH2 forms of the enzyme. Inactivation of the EH2 form is about 8-fold faster than that of the E form (Figure 2A), suggesting that the sulphydryl groups of the redox-active Cys53 and/or Cys58 in the active site could be a preferential target for alkylation. To test this possibility we examined the effects of competitive inhibitors of the T[S]2-binding site on the kinetics of inactivation (Figure 2B). Mel T, a conjugate of T[SH]2 and melarsen oxide, is a competitive inhibitor of TryR from T. brucei with a Ki value of 9 μM (33). Under our experimental conditions, Mel T was found to be a linear competitive inhibitor of T. cruzi TryR (Ki = 15.5 ± 0.8 μM). As shown in Figure 2B, Mel T protects the EH2 form of the enzyme from inactivation by QM. The pseudo-first-order rate constants for inactivation of EH2 enzyme in the presence (k’obs = 0.072 ± 0.004 min−1) and absence (kobs= 0.32 ± 0.03 min−1) of Mel T (i = 40 μM) were fitted to the following equation, which describes protection by a simple competitive inhibitor (34):
| Equation 1 |
This yielded a Ki for Mel T of 12 μM, in good agreement with the directly determined value of 15.5 μM. This result suggests that QM alkylates TryR within the T[S]2-binding site.
Stoichiometry and sites of alkylation by quinacrine mustard
In order to determine the stoichiometry of the inactivation process, samples of TryR were reduced with NADPH and exposed to QM for different time intervals, the reaction quenched by the addition of excess 2-mercaptoethanol and the resulting samples assayed for residual activity. Alkylated protein was precipitated with trichloroacetic acid, washed to remove unbound QM and re-dissolved by digestion with proteinase K for spectrophotometric determination of bound QM (Figure 4). The number of moles of QM bound per mole active site shows a limit of 1.7 ± 0.2, consistent with 2 mole QM being covalently bound per mole of inactive TryR monomer. Moreover, since 50% inactivation is associated with 1 QM bound per total monomer, it can be deduced that the majority of the inactive EH2 form of the enzyme has 2 QM bound per monomer.
Figure 4. Determination of stoichiometry of alkylation by quinacrine mustard.
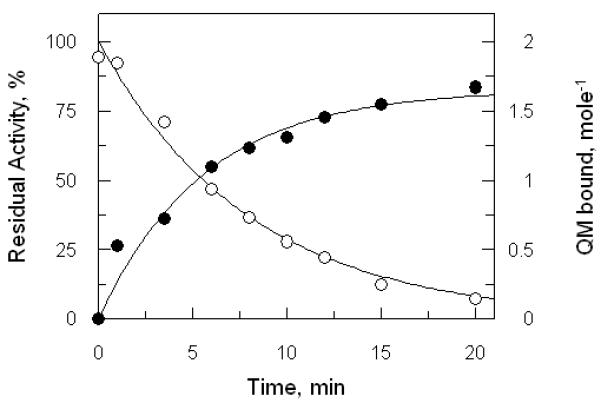
A sample of NADPH-reduced TryR (1 μM) was inactivated by 10 μM QM. Aliquots were removed at intervals, quenched with 2-mercaptoethanol and residual activities were determined as described in Figure 2. Residual activity is fitted to the equation for a single exponential decay (open circles). The remainder of each quenched sample was denatured with trichloroacetic acid to remove bound FAD, washed and re-dissolved by proteolytic digestion. Residual covalently bound QM was determined spectrophotometrically as described in Experimental Procedures (closed circles).
In order to determine the sites of alkylation, protein samples were inactivated as above, quenched with mercaptoethanol, dialysed and digested with trypsin. The resulting MALDI-TOF MS spectra clearly showed that the peptide fragment C (residues 32 to 61, 3041.6 Da) bearing Cys53 and Cys 58 disappeared on incubation with QM, indicating that this fragment was undergoing modification. However, although a number of new peptide fragments were observed, we were unable to unambiguously identify a predicted product (e.g. mono-alkylated peptideC + QM – HCl; mass 3774 Da; bis-alkylated peptide C + QM −2HCl, mass 3437 Da; or a mixed adduct between peptide C from TryR, QM and 2-mercaptoethanol, mass 3516 Da). Although modification of this peptide could have suppressed its ionisation, another possible explanation for this anomalous behaviour is discussed below.
Inhibition kinetics of quinacrine mustard
Inactivation of TryR by QM is also dose-dependent for the E and EH2 forms of the enzyme (Figure 5A and 5B, respectively). Formation of the QM-TryR covalent adduct could either proceed as a simple bimolecular reaction, or through a reversible binary complex. The difference between a general chemical modifier and an active-site-directed irreversible inhibitor is that the latter can form significant amounts of dissociable complex with enzyme (EI) prior to irreversible covalent modification of the enzyme (E*I) as shown in the following scheme(35):
The apparent rate of irreversible inhibition (kobs) is described by the following equation, where Ki is the dissociation constant for QM and k2 is the rate constant for conversion of EI to E*I. This equation takes the same form as the Michaelis-Menten equation giving a rectangular hyperbola, when kobs is plotted against inhibitor concentration (i) (36, 37).
| Equation 2 |
When i << Ki reduces to
| Equation 3 |
from which the ‘specificity constant for inhibition’ (k2/Ki) can be determined. In the oxidised (E form) of the enzyme, a plot of kobs versus [QM] shows a linear relationship over the range of inhibitor concentrations that we were able to examine (Figure 3C, closed circles). This result suggests that either no appreciable amount of reversible EI complex is formed, or that Ki is greater than 40 μM. The slope of this plot yields k2/Ki = 15.8 M−1 s−1 for the E-form. In contrast, the NADPH-reduced TryR (EH2 form) gives a hyperbolic plot (Figure 5C, open circles), indicating that a reversible TryR-QM complex is formed prior to irreversible chemical modification of the enzyme. Fitting the data to the above equation yields an apparent Ki of 41 ± 8 μM and k2 of 0.69 ± 0.09 min−1 for QM. (k2/Ki = 280 M−1 s−1). Assuming that the residue(s) alkylated in the E form is also available for alkylation in the EH2 form, then subtraction of the inactivation rate present in the E-form from the EH2-form, gives the kinetics of inactivation at the redox-active cysteine site (Figure C, diamond symbols). This yields an apparent Ki of 36 ± 8 μM and k2 of 0.58 ± 0.08 min−1 for QM. (k2/Ki = 269 M−1 s−1), which is 17-fold faster than for the E-form of the enzyme.
Figure 5. Kinetics of inactivation of trypanothione reductase by quinacrine mustard.
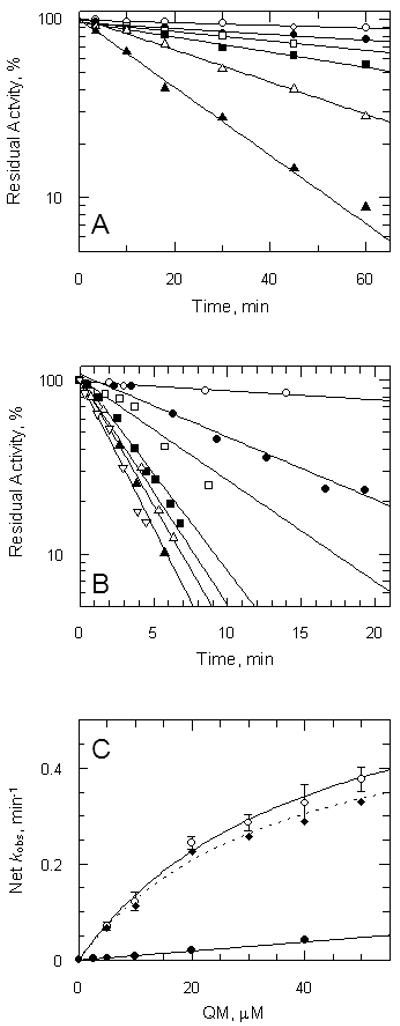
Panel A, oxidised enzyme. TryR (1 μM) was incubated at room temperature with addition of QM at time zero. Aliquots were removed at intervals and residual activity determined by 100-fold dilution into assay buffer containing 150 μM NADPH as described in the Experimental Procedures. Residual activity is expressed as a percentage of the activity at zero time without addition of QM. Lines are the non-linear fits for a single exponential decay. Concentrations of QM were: zero, ○; 2.5: ●; 5 □; 10: ■: 20: 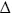 ; and 40 μM: ▲. Panel B, reduced enzyme. TryR (1 μM) was preincubated with 150 μM NADPH for 5 min prior to addition of QM. Concentrations of QM were: zero, ○; 5, ●; 10, □; 20, ■, 30,
; and 40 μM: ▲. Panel B, reduced enzyme. TryR (1 μM) was preincubated with 150 μM NADPH for 5 min prior to addition of QM. Concentrations of QM were: zero, ○; 5, ●; 10, □; 20, ■, 30, 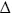 ; 40, ▲; and 50 μM,
; 40, ▲; and 50 μM, 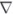 . Assays were performed and analyzed as described above. Panel C, rate of inactivation of TryR as a function of QM concentration. Pseudo-first-order rates of inactivation (kobs) were obtained from data in panels A and B and corrected for inactivation in the absence of inhibitor (Net kobs). Error bars are standard errors of the mean determined by non-linear regression. Data for oxidised enzyme (●) are fitted by linear regression; data for NADPH-reduced TryR (○) are fitted to equation 2 (see text). The diamond symbols represent the net rate for the reduced enzyme corrected for the linear rate observed for the oxidised enzyme.
. Assays were performed and analyzed as described above. Panel C, rate of inactivation of TryR as a function of QM concentration. Pseudo-first-order rates of inactivation (kobs) were obtained from data in panels A and B and corrected for inactivation in the absence of inhibitor (Net kobs). Error bars are standard errors of the mean determined by non-linear regression. Data for oxidised enzyme (●) are fitted by linear regression; data for NADPH-reduced TryR (○) are fitted to equation 2 (see text). The diamond symbols represent the net rate for the reduced enzyme corrected for the linear rate observed for the oxidised enzyme.
Protection and acceleration of QM inhibition by tricyclic inhibitors
The observation that an EI complex is formed as an intermediate in the reaction between the EH2 form of TryR and QM suggests that inactivation could be prevented by competition with quinacrine. However, when increasing concentrations of quinacrine were co-incubated with NADPH-reduced TryR and 10 μM QM, the rate of inactivation was initially increased over that produced by QM alone (Figure 6A, open circles). This effect was greatest at a quinacrine to QM ratio of 2.5:1, producing a 1.4-fold increase in the rate of TryR inactivation. Interestingly, this effect decreased when higher quinacrine concentrations were used. In contrast, no evidence of any rate enhancement was observed for the TryR incubated in the absence of NADPH (Figure 6A, closed circles). These data were fitted to equation 1 by non-linear regression using the explicit errors for each net kobs, yielding a Ki of 208 ± 45 μM. This result is consistent with the previous conclusion that the Ki for the E-form of TryR is > 40 μM (Figure 5C). To confirm the validity of this method, a similar experiment was done with the tricyclic competitive inhibitor clomipramine (Figure 6B). Clomipramine strongly protected both the E and EH2 forms from inactivation by QM, yielding Ki-values of 8.5 ± 1.7 μM and 7.5 ± 1.6 μM, respectively. Both values are consistant with the previously reported Ki of 6.5 ± 0.6 μM determined at pH 7.25 (38) and the value of 3.5 ± 0.2 μM determined via direct assay under our assay conditions (not shown). The unusual behaviour of quinacrine is consistent with the ability of this inhibitor to self-associate in solution (39), in contrast with clomipramine.
Figure 6. Effect of quinacrine and clomipramine on inactivation of TryR by QM.
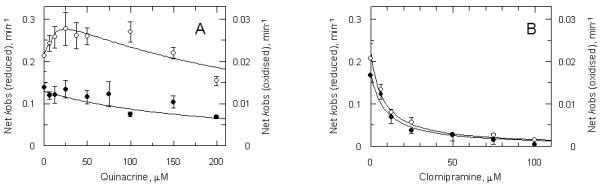
TryR (1 μM) was incubated with 10 μM QM, plus or minus NADPH and either quinacrine or clomipramine. Aliquots were removed at intervals and residual activity determined by 100-fold dilution into assay buffer as described in Materials and Methods. First order rate constants were determined by non-linear regression. Panel A: open circles, plus NADPH and quinacrine; closed circles, plus quinacrine alone. Panel B: open circles, plus NADPH and clomipramine; closed circles plus clomipramine alone.
Crystallographic studies on enzyme-quinacrine mustard interactions
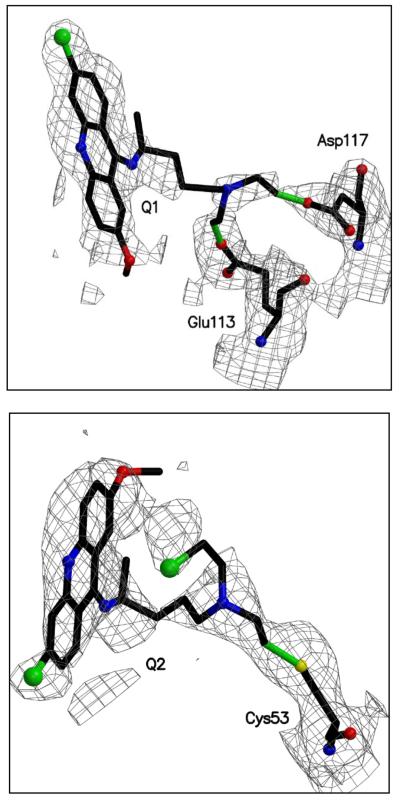
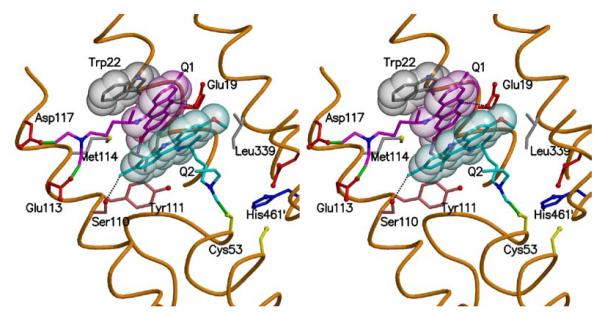
The crystal structure of QM-inactivated TryR was determined. The final model contains a TryR dimer, four quinacrine moieties, two FAD molecules, one maleic acid and 53 ordered water molecules. Figure 7 shows the fit of the model to the electron density and additional details are presented in Table 1. Since the conformations and interactions in the two active sites contained in the asymmetric unit were restrained on the basis of non-crystallographic symmetry, we only provide details for one. Two molecules of the quinacrine-based inhibitor, termed Q1 and Q2, are located in each active site where the substrate T[S]2 normally binds. Both interact with residues from a single subunit (Figure 8). Q1 binds under a hydrophobic lid created by Trp22. The acridine is side-on participating in van der Waals interactions with the side chains of Leu18, Met114, Tyr111 and π-stacking interactions with the tryptophan lid on one side and the ring system of Q2 on the other. The methoxy substituent of Q1 is placed between the side chains of Leu18 and Met114 (for clarity Leu18 is omitted from the figure). At the other end of the acridine, the chlorine substituent is directed out towards solvent and is some 5.5 Å distant from any protein atoms. The acridine nitrogen is likely to be protonated and forming a hydrogen bond with OE1 of Glu19 at a distance of 3.2 Å. The aliphatic tail of Q1, lines up alongside the side chain of Met114 and extends over towards a helix that lines one side of the active site. The two chlorine substituents on C-26 and C-28 have been lost by reaction with the enzyme and covalent linkages between Q1 and the acidic Glu113 and Asp117 side chains have been formed.
Figure 7. The difference density maps of both inhibitor molecules binding to subunit A.
All atoms shown were omitted from the structure factor calculation. The density is contoured at the 1σ level of that observed in the unit cell and is depicted in chicken wire, green bonds indicate the covalent attachment between the inhibitor and the protein.
Figure 8. Sites of alkylation and orientation of quinacrine mustard in the trypanothione disulphide binding site.
A stereo view of the active site. The main chain around the active site cleft is a brown thread passing through the CA positions and selected side chains are shown. Trp22 is grey, Q1 pink and Q2 cyan and the van der Waals surface is depicted as translucent spheres. Green bonds represent the covalent links between the inhibitors and the protein and two potential hydrogen bonding interactions are represented by dashed lines. Figure produced with MOLSCRIPT (46) and RASTER3D (47).
Q2, as mentioned above, stacks under Q1 and is aligned such that the acridine nitrogen and methoxy substituent are directed out away from the protein and able to interact with water molecules. The chlorine substituent of Q2 is placed near the tail of Q1 and forms a hydrogen bond of length 3.0 Å with the hydroxyl group of Ser110. The tail of Q2 lies along a depression formed by the side chains of Ser15, Tyr111 and Ile339 at the base of the TryR active site. One of the chlorine substituents on the tail has been lost and a covalent link formed to SG of Cys53, part of the enzyme’s redox-active centre. The other chlorine has been retained and is tucked under the ring system of Q2.
DISCUSSION
The previous study of Jacoby et al. (18) on a weakly diffracting monoclinic crystal form of T. cruzi TryR soaked in a quinacrine solution was limited by the poor quality of the diffraction data. Of the four active sites present in the asymmetric unit, the resulting model had only one site occupied by the ligand and this at occupancy of 0.3. These data are not publicly available, but coordinates were kindly provided by the authors for comparative purposes. Their analysis only identified a single inhibitor molecule bound near the hydrophobic pocket mainly formed by the Trp22 and Met114 residues. The acridine was modelled in a position perpendicular to and overlapping with Q1 and Q2 in our structure, with the ring nitrogen and chlorine substituent pointing toward the side chain of Trp22. The considerable differences observed between the two structures may be a consequence of the different inhibitor molecules, with the QM capable of covalent modification of the protein.
However, we note that a previous kinetic analysis of several acridine derivatives indicated that more than one molecule of these inhibitors bind per TryR monomer (19). Unfortunately these data did not allow determination of the positions or number of interaction sites and it was assumed that one inhibitor bound in the active site in the fashion suggested in the previous model of a quinacrine complex. A second inhibitor was speculated to bind in a cavity at the dimer interface or to a hydrophobic patch at the edge of the active site. Moreover, multiple interaction sites of tricyclic molecules have been identified in a previous study that used photoaffinity labelling to examine the interaction of the tricyclic drug fluphenazine with TryR (40). These experiments indicated multiple binding sites for tricyclic molecules in the TryR active site, yielding fluphenazine / TryR monomer molar ratios in the range of 2-5. This result was interpreted in light of the known ability of phenothiazine drugs to stack in solution, a property that they share with quinacrine (39). Our data extend these findings by showing that the two molecules of QM which react to inactivate TryR actually bind in the active site, with no evidence for interaction elsewhere on the protein.
The observation that one of the chloroethyl groups on Q2 is unreacted in the native protein offers an explanation for two apparent discrepancies in our biochemical and mass spectrometry studies. First, the small amount of cross-linked dimer observed only in the EH2 form of TryR could arise during denaturation of the protein for SDS-PAGE. Second, digestion with trypsin would allow the mono-alkylated peptide fragment to cross-link with other peptides in the mixture, accounting for our failure to observe the predicted products by mass spectrometry.
The structure of this QM-TryR adduct represents an end-point of an alkylation reaction and might not therefore represent the actual sites for the non-covalent quinacrine interaction. However, several lines of kinetic evidence presented above indicate that the positions in which the QM are found in the crystal structure represent the actual sites at which non-reactive acridine molecules interact with TryR. Notably, the observation that a reversible EI complex (Ki 36 μM) is formed between QM and the EH2 form of TryR indicates that the inhibitor does bind to the active site region. Further support for this conclusion is provided by the protection afforded by the T[S]2 mimic, Mel T. Since it is probable that the binding mode of the arsenic-T[SH]2 conjugate Mel T is similar to that of the natural T[S]2 substrate, the ability of this inhibitor to protect against QM inactivation can be interpreted using the structure of the TryR-T[S]2 complex (13). In this structure both the Q1 and Q2 binding sites are occupied by the spermidine component of the T[S]2 substrate.
Unfortunately, the large size of the TryR substrate-binding cleft means that many different orientations of small ligands are possible. However, the synergistic effect of quinacrine on QM inactivation at low concentrations and the antagonistic effect at high concentrations clearly indicate that the two regions of the active site which bind these inhibitors can interact. This interaction appears to depend upon the planar tricyclic ring system since the competitive inhibitor clomipramine, which possesses a non-planar ring, obeys simple one-site competitive kinetics and protects TryR against QM inactivation. These data are therefore entirely consistent with the observed QM binding sites, which are adjacent to each other and allow stacking of the acridine ring systems.
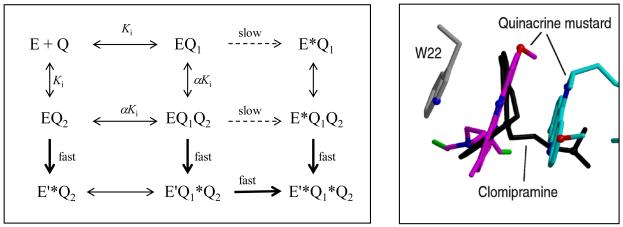
Based on the structure of the alkylated enzyme and our kinetic data we propose a model to explain the different effects of quinacrine and clomipramine on the QM inactivation reaction (Figure 9). Since inactivation of the EH2 enzyme proceeds rapidly through the reaction of a mustard group with the Cys53, occupation of the Q2 binding site by QM will be required for this reaction to occur. However, the majority of the contacts made by the QM moiety in this position are through van der Waals’ interactions with the acridine ring of the Q1 ligand. This implies that the affinity of site Q2 will be enhanced by an increase in the occupancy of site Q1. Based on the inactivation kinetics of QM with the oxidised enzyme, the affinity of this site is low (Ki > 40 μM). Hence, addition of quinacrine to a QM inactivation reaction will titrate site Q1 and may allow more quinacrine and QM to bind to site Q2, thus producing an increase in the overall rate of inactivation. Since Cys53 is only available for alkylation in the EH2 form, enhanced inactivation is not observed in the E form. At higher concentrations of quinacrine, QM will be increasingly excluded from both sites, resulting in decreased inactivation. In contrast, the occupation of site Q1 by clomipramine would be predicted to simultaneously reduce the occupancy of site Q2 and thus reduce the rate of inactivation, since the non-planar ring of this inhibitor would not allow π-stacking interactions.
Figure 9. Kinetic and structural models of the effects of clomipramine and quinacrine on inactivation of trypanothione reductase by quinacrine mustard.
The left hand panel shows a random sequential mechanism for the formation of the doubly alkylated end-product (E’*Q1*Q2). The two quinacrine-binding sites are distinguished as Q1 and Q2. Covalently modified sites are shown with an asterisk. E and E’ refer to oxidised (E) and reduced (EH2) forms of the enzyme. Binding to the first site affects binding to the second site by a factor α. Alkylation at the Q1 site is slow (dashed arrow) compared to the Q2 site (bold arrow). The right hand panel shows clomipramine overlaid on the quinacrine mustard site Q1, to illustrate how this simple competitive inhibitor could exclude quinacrine from both binding sites.
The description of two interacting binding sites in the active site of TryR has important implications for drug design and may allow the design of far more potent inhibitory molecules than can be produced from the simple tricyclic scaffold. This could be achieved through the design of molecules that could bind simultaneously to both of these sites. Indeed, previous studies may have unwittingly exploited this approach; for example, the finding that bis-substituted polyamine derivatives are significantly better TryR inhibitors than their mono-substituted equivalents (41). Importantly, these polyamine derivatives include some of the most potent TryR inhibitors which have yet been described, with N1,N12-bis-(5-bromo-3-indole acetyl)spermine having a Ki of just 76 nM against T. cruzi TryR (42). However, the flexibility of these spermine and spermidine derivatives could produce an entropic penalty upon binding. The identification of adjacent tricyclic binding surfaces may therefore allow the construction of conformationally constrained inhibitors with affinity for both sites, resulting in even tighter binding.
Although TryR provides an attractive target for selective inhibition, purely competitive inhibitors of this enzyme may not be ideal as drug candidates, unless of low nanomolar affinity. Since there are extremely high concentrations of T[SH]2 in the parasite cell (reaching 5 mM in L. donovani (43)), inhibition of TryR could result in the accumulation of sufficient T[S]2 to partially reverse the effects of a competitive inhibitor (44). Indeed, more than 90% inhibition of TryR is required to kill T. brucei (45) and L. donovani can survive an 85% reduction in TryR activity with no effect upon either their viability or their ability to metabolise H2O2 (8). Collectively, these data suggest that the ideal TryR inhibitor would be one that showed either tight-binding or irreversible inhibitory activities. Although the mustard groups render QM too reactive to be considered as a lead compound, the detailed mechanistic and structural data provided in this study may allow the design of less toxic compounds with similar activities.
Acknowledgments
AHF and WNH are funded by the Wellcome Trust and CSB is a BBSRC David Phillips Research Fellow. Tim Vickers was funded by a Wellcome Trust Vacation Scholarship. We would like to thank Mark Cunningham and Matt Berriman for assistance in the preliminary stages of this project and Dougie Lamont of the Post Genomics and Molecular Interactions Centre, University of Dundee for the MS studies. We acknowledge the use of the Engineering and Physical Sciences Research Council chemical database service hosted at Daresbury laboratory, access to the European Synchrotron Radiation Facility, discussions with colleagues, Luise Krauth-Siegel for the provision of coordinates for the TryR-mepacrine complex and excellent support from Gordon Leonard at the synchrotron.
Abbreviations
- QM
quinacrine mustard
- TryR
trypanothione reductase
- GR
glutathione reductase
- T[SH]2
dihydrotrypanothione (N1,N8-bis(glutathionyl)spermidine)
- T[S]2
trypanothione disulphide
- Mel T
adduct of melarsen oxide with dihydrotrypanothione
REFERENCES
- 1.Manson-Bahr PEC, Bell DR. Manson’s Tropical Diseases. 19 Ed Baillière Tindall, London, UK: 1987. [Google Scholar]
- 2.World Health Organization . The World health report 2002: Reducing risks, promoting healthy life. World Health Organization; Geneva: 2002. Statistical Annex. [Google Scholar]
- 3.Fairlamb AH, Blackburn P, Ulrich P, Chait BT, Cerami A. Science. 1985;227:1485–1487. doi: 10.1126/science.3883489. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt A, Krauth-Siegel RL. Curr. Top. Med. Chem. 2002;2:1239–1259. doi: 10.2174/1568026023393048. [DOI] [PubMed] [Google Scholar]
- 5.Ouellette M, Papadopoulou B. Parasitol. Today. 1993;9:150–153. doi: 10.1016/0169-4758(93)90135-3. [DOI] [PubMed] [Google Scholar]
- 6.Fairlamb AH, Cerami A. Annu. Rev. Microbiol. 1992;46:695–729. doi: 10.1146/annurev.mi.46.100192.003403. [DOI] [PubMed] [Google Scholar]
- 7.Dumas C, Ouellette M, Tovar J, Cunningham ML, Fairlamb AH, Tamar S, Olivier M, Papadopoulou B. EMBO J. 1997;16:2590–2598. doi: 10.1093/emboj/16.10.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tovar J, Cunningham ML, Smith AC, Croft SL, Fairlamb AH. Proc. Natl. Acad. Sci. USA. 1998;95:5311–5316. doi: 10.1073/pnas.95.9.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fries DS, Fairlamb AH. Antiprotozoal Agents. In: Abraham DJ, editor. Burger’s Medicinal Chemistry and Drug Discovery, volume 5: Chemotherapeutic Agents. Wiley and Sons; New York: 2003. [Google Scholar]
- 10.Meister A, Anderson ME. Annu. Rev. Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 11.Aboagye-Kwarteng T, Smith K, Fairlamb AH. Mol. Microbiol. 1992;6:3089–3099. doi: 10.1111/j.1365-2958.1992.tb01766.x. [DOI] [PubMed] [Google Scholar]
- 12.Henderson GB, Fairlamb AH, Ulrich P, Cerami A. Biochemistry. 1987;26:3023–3027. doi: 10.1021/bi00385a011. [DOI] [PubMed] [Google Scholar]
- 13.Bond CS, Zhang YH, Berriman M, Cunningham ML, Fairlamb AH, Hunter WN. Structure. 1999;7:81–89. doi: 10.1016/s0969-2126(99)80011-2. [DOI] [PubMed] [Google Scholar]
- 14.Augustyns K, Amssoms K, Yamani A, Rajan PK, Haemers A. Curr. Pharm. Des. 2001;7:1117–1141. doi: 10.2174/1381612013397564. [DOI] [PubMed] [Google Scholar]
- 15.Garforth J, Yin H, McKie JH, Douglas KT, Fairlamb AH. J. Enzyme Inhib. 1997;12:161–173. doi: 10.3109/14756369709029312. [DOI] [PubMed] [Google Scholar]
- 16.Krauth-Siegel RL, Lohrer H, Bucheler US, Schirmer RH. The antioxidant enzymes glutathione reductase and trypanothione reductase as drug targets. In: Coombs GH, North MJ, editors. Biochemical Protozoology. Taylor and Francis; London: 1991. [Google Scholar]
- 17.Chibale K, Haupt H, Kendrick H, Yardley V, Saravanamuthu A, Fairlamb AH, Croft SL. Bioorg. Med. Chem. Lett. 2001;11:2655–2657. doi: 10.1016/s0960-894x(01)00528-5. [DOI] [PubMed] [Google Scholar]
- 18.Jacoby EM, Schlichting I, Lantwin CB, Kabsch W, Krauth-Siegel RL. Proteins. 1996;24:73–80. doi: 10.1002/(SICI)1097-0134(199601)24:1<73::AID-PROT5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 19.Bonse S, SantelliRouvier C, Barbe J, Krauth-Siegel RL. J. Med. Chem. 1999;42:5448–5454. doi: 10.1021/jm990386s. [DOI] [PubMed] [Google Scholar]
- 20.Borges A, Cunningham ML, Tovar J, Fairlamb AH. Eur. J. Biochem. 1995;228:745–752. doi: 10.1111/j.1432-1033.1995.tb20319.x. [DOI] [PubMed] [Google Scholar]
- 21.Worthington DJ, Rosemeyer MA. Eur. J. Biochem. 1974;48:167–177. doi: 10.1111/j.1432-1033.1974.tb03754.x. [DOI] [PubMed] [Google Scholar]
- 22.Shames SL, Fairlamb AH, Cerami A, Walsh CT. Biochemistry. 1986;25:3519–3526. doi: 10.1021/bi00360a007. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Bond CS, Bailey S, Cunningham ML, Fairlamb AH, Hunter WN. Protein Science. 1996;5:52–61. doi: 10.1002/pro.5560050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Collaborative Computational Project, N. 4 Acta Crystallogr. D. 1994;50:760–763. [Google Scholar]
- 26.Brunger AT. Acta Crystallogr. D. 1993;49:24–36. doi: 10.1107/S0907444992007352. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Courseille C, Busetta B, Hospital M. Acta Crystallogr. B. 1973;29:2349–2355. [Google Scholar]
- 30.Fletcher DA, McMeeking RF, Parkin D. J. Chem. Inf. Comput. Sci. 1996;36:746–749. [Google Scholar]
- 31.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 32.Kleywegt GJ, Jones TA. Structure. 1996;4:1395–1400. doi: 10.1016/s0969-2126(96)00147-5. [DOI] [PubMed] [Google Scholar]
- 33.Fairlamb AH, Henderson GB, Cerami A. Proc. Natl. Acad. Sci. USA. 1989;86:2607–2611. doi: 10.1073/pnas.86.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cunningham ML, Fairlamb AH. Eur. J. Biochem. 1995;230:460–468. doi: 10.1111/j.1432-1033.1995.tb20583.x. [DOI] [PubMed] [Google Scholar]
- 35.Brocklehurst K. Biochem. J. 1979;181:775–778. doi: 10.1042/bj1810775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung MJ, Metcalf BW. Biochem. Biophys. Res. Commun. 1975;67:301–306. doi: 10.1016/0006-291x(75)90316-2. [DOI] [PubMed] [Google Scholar]
- 37.Carlson GM. Biochim. Biophys. Acta. 1984;789:347–350. doi: 10.1016/0167-4838(84)90191-2. [DOI] [PubMed] [Google Scholar]
- 38.Benson TJ, McKie JH, Garforth J, Borges A, Fairlamb AH, Douglas KT. Biochem. J. 1992;286:9–11. doi: 10.1042/bj2860009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Attwood D. Adv. Colloid Interfac. 1995;55:271–303. [Google Scholar]
- 40.Yin H, Chan C, Garforth J, Douglas KT, Bolgar MS, Gaskell SJ, Fairlamb AH. Chem. Commun. (Camb. ) 1996;1:973–974. [Google Scholar]
- 41.O’Sullivan MC, Zhou Q. Bioorg. Med. Chem. Lett. 1995;5:1957–1960. [Google Scholar]
- 42.Chitkul B, Bradley M. Bioorg. Med. Chem. Lett. 2000;10:2367–2369. doi: 10.1016/s0960-894x(00)00471-6. [DOI] [PubMed] [Google Scholar]
- 43.Ariyanayagam MR, Fairlamb AH. Mol. Biochem. Parasitol. 2001;115:189–198. doi: 10.1016/s0166-6851(01)00285-7. [DOI] [PubMed] [Google Scholar]
- 44.Zani CL, Fairlamb AH. Mem. Inst. Oswaldo Cruz. 2003;98:565–568. doi: 10.1590/s0074-02762003000400026. [DOI] [PubMed] [Google Scholar]
- 45.Krieger S, Schwarz W, Ariyanayagam MR, Fairlamb AH, Krauth-Siegel RL, Clayton C. Mol. Microbiol. 2000;35:542–552. doi: 10.1046/j.1365-2958.2000.01721.x. [DOI] [PubMed] [Google Scholar]
- 46.Kraulis PJ. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- 47.Merritt EA, Murphy MEP. Acta Crystallogr. D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]