Abstract
Albuminuria and reduced glomerular filtration rate are manifestations of chronic kidney disease (CKD) that predict end-stage renal disease, acute kidney injury, cardiovascular disease and death. We hypothesized that SNPs identified in association with the estimated glomerular filtration rate (eGFR) would also be associated with albuminuria. Within the CKDGen Consortium cohort (n= 31 580, European ancestry), we tested 16 eGFR-associated SNPs for association with the urinary albumin-to-creatinine ratio (UACR) and albuminuria [UACR >25 mg/g (women); 17 mg/g (men)]. In parallel, within the CARe Renal Consortium (n= 5569, African ancestry), we tested seven eGFR-associated SNPs for association with the UACR. We used a Bonferroni-corrected P-value of 0.003 (0.05/16) in CKDGen and 0.007 (0.05/7) in CARe. We also assessed whether the 16 eGFR SNPs were associated with the UACR in aggregate using a beta-weighted genotype score. In the CKDGen Consortium, the minor A allele of rs17319721 in the SHROOM3 gene, known to be associated with a lower eGFR, was associated with lower ln(UACR) levels (beta = −0.034, P-value = 0.0002). No additional eGFR-associated SNPs met the Bonferroni-corrected P-value threshold of 0.003 for either UACR or albuminuria. In the CARe Renal Consortium, there were no associations between SNPs and UACR with a P< 0.007. Although we found the genotype score to be associated with albuminuria (P= 0.0006), this result was driven almost entirely by the known SHROOM3 variant, rs17319721. Removal of rs17319721 resulted in a P-value 0.03, indicating a weak residual aggregate signal. No alleles, previously demonstrated to be associated with a lower eGFR, were associated with the UACR or albuminuria, suggesting that there may be distinct genetic components for these traits.
INTRODUCTION
Chronic kidney disease (CKD) affects ∼26 million adults in the USA (1). It is an illness of major public health importance as affected individuals experience a substantially increased risk of end-stage renal disease and need for dialysis, as well as a dramatic excess in cardiovascular morbidity and mortality (2). CKD is defined as persistent kidney damage marked either by the presence of albuminuria or a reduced estimated glomerular filtration rate (eGFR) (3). For any given level of eGFR, albuminuria is associated with increased CVD and all-cause mortality outcomes (4). Furthermore, the prevalence of albuminuria increases dramatically as the GFR falls, from <10% in those with a preserved GFR to almost 60% in those with advanced CKD (1). Furthermore, albuminuria is the strongest known risk factor for the progression of CKD, such that the risk of end-stage kidney disease is 10 times higher in people with a preserved GFR and albuminuria compared with those with the same GFR but no albuminuria (5).
We have previously shown that a reduced eGFR and albuminuria are associated with differential risk factor profiles (6). Further, only one-quarter of individuals with CKD have albuminuria (6). This is corroborated by data from the National Health and Nutrition Examination Surveys (NHANES), which showed that increased trends in the prevalence of albuminuria, but not CKD defined as an eGFR <60 ml/min/1.73 m2, can be explained by hypertension and diabetes (1). Taken together, these data suggest potential differential etiologies for albuminuria and a reduced GFR.
Genome-wide association studies (GWAS) have identified 16 loci for the eGFR in European populations from the CKDGen Consortium (7,8). We have additionally identified seven SNPs for the eGFR in African-American populations from the CARe Renal Consortium (9,10). Notably, only one locus at SHROOM3 has been identified previously as being associated with both the urinary albumin-to-creatinine ratio (UACR) and the eGFR, leading us to speculate that the genetic architecture of the eGFR and the UACR as identified through GWAS in population-based cohorts may be different (9). Indeed, other studies have shown that there is a weak genetic concordance between both traits (11). To investigate this more comprehensively, we performed a targeted SNP analysis using 16 eGFR-associated SNPs previously identified in Europeans (7) and 7 eGFR-associated SNPs previously identified in African-Americans (10) to determine whether these SNPs were also associated with the UACR. Given the known inverse associations between the eGFR and the UACR, we hypothesized that alleles associated with a lower eGFR would also be associated with an increased UACR. We tested this in the existing CKDGen albuminuria and CARe Renal Consortium data sets (9,10).
RESULTS
All study participants were of European (CKDGen Consortium) or African-American (CARe Renal Consortium) descent (9,10). Study sample characteristics can be found in Supplementary Material, Table S1 and S2.
UACR results
Association results for the 16 eGFR-associated SNPs with the UACR in the CKDGen Consortium can be found in Table 1. As previously reported, there was a significant association with rs17319721 in SHROOM3 on chromosome 4. The UACR levels were lower per copy of the A allele at rs17319721 (P-value = 0.0002), whereas the eGFR levels were also lower in our previously published analysis. No additional SNPs met the Bonferroni P-value threshold of 0.003 (0.05/16).
Table 1.
Association results for the urinary-to-albumin ratio (UACR) in European Americans from the CKDGen Consortium
| SNP ID | Nearest gene | Coded allele | Minor allele frequency | Direction of the coded allele effect on the GFR | The UACR β-coefficient per coded allele copy | Standard error | P-value |
|---|---|---|---|---|---|---|---|
| rs17319721 | SHROOM3 | A | 0.43 | – | −0.034 | 0.009 | 0.0002 |
| rs11959928 | DAB2 | A | 0.44 | – | −0.020 | 0.009 | 0.03 |
| rs1394125 | UBE2Q2 | A | 0.35 | – | −0.024 | 0.011 | 0.03 |
| rs10109414 | STC1 | T | 0.42 | – | −0.019 | 0.009 | 0.04 |
| rs1260326 | GCKR | T | 0.41 | + | 0.018 | 0.009 | 0.05 |
| rs267734 | ANXA9 | C | 0.2 | + | −0.018 | 0.011 | 0.10 |
| rs7805747 | PRKAG2 | A | 0.24 | – | 0.025 | 0.016 | 0.11 |
| rs4744712 | PIP5K1B | A | 0.39 | – | −0.014 | 0.009 | 0.12 |
| rs6420094 | SLC34A1 | G | 0.34 | – | −0.014 | 0.012 | 0.24 |
| rs626277 | DACH1 | C | 0.4 | + | 0.0113 | 0.010 | 0.25 |
| rs12460876 | SLC7A9 | C | 0.39 | + | −0.010 | 0.009 | 0.30 |
| rs653178 | ATXN2 | C | 0.5 | – | 0.009 | 0.009 | 0.34 |
| rs881858 | VEGFA | G | 0.28 | + | −0.010 | 0.011 | 0.37 |
| rs13538 | ALMS1 | G | 0.23 | + | 0.005 | 0.011 | 0.66 |
| rs12917707 | UMOD | T | 0.18 | + | 0.004 | 0.012 | 0.76 |
| rs347685 | TFDP2 | C | 0.28 | + | 0.003 | 0.010 | 0.77 |
The statistical significance defined using a Bonferroni correction for the 16 SNPs (0.05/16 = 0.003).
Albuminuria results
Association results for albuminuria from the CKDGen Consortium can be found in Table 2. Similar to what we observed for the UACR, only one SNP reached significance. The A allele at rs17319721 in SHROOM3 was associated with a lower odds ratio for albuminuria (P-value = 1.87E-06; OR = 0.88). No additional SNPs met the Bonferroni P-value threshold of 0.003.
Table 2.
Association results for albuminuria in European Americans from the CKDGen Consortium
| SNP ID | Nearest gene | Coded allele | Minor allele frequency | Directionality of the GFR related to the coded allele | Odds ratio related to the coded allele (95% confidence interval) | P-value* |
|---|---|---|---|---|---|---|
| rs17319721 | SHROOM3 | A | 0.43 | – | 0.88 (0.83–0.93) | 1.87E-06 |
| rs11959928 | DAB2 | A | 0.44 | – | 0.97 (0.92–1.02) | 0.26 |
| rs1394125 | UBE2Q2 | A | 0.35 | – | 0.95 (0.90–1.01) | 0.13 |
| rs10109414 | STC1 | T | 0.42 | – | 0.95 (0.91–1.00) | 0.07 |
| rs1260326 | GCKR | T | 0.41 | + | 1.07 (1.01–1.12) | 0.02 |
| rs267734 | ANXA9 | C | 0.2 | + | 1.03 (0.97–1.10) | 0.36 |
| rs7805747 | PRKAG2 | A | 0.24 | – | 1.07 (0.98–1.16) | 0.15 |
| rs4744712 | PIP5K1B | A | 0.39 | – | 0.96 (0.91–1.01) | 0.11 |
| rs6420094 | SLC34A1 | G | 0.34 | – | 1.01 (0.94–1.08) | 0.84 |
| rs626277 | DACH1 | C | 0.4 | + | 1.00 (0.95–1.06) | 0.93 |
| rs12460876 | SLC7A9 | C | 0.39 | + | 0.97 (0.92–1.03) | 0.30 |
| rs653178 | ATXN2 | C | 0.5 | – | 1.00 (0.94–1.05) | 0.88 |
| rs881858 | VEGFA | G | 0.28 | + | 1.00 (0.94–1.06) | 0.95 |
| rs13538 | ALMS1 | G | 0.23 | + | 1.08 (1.02–1.16) | 0.01 |
| rs12917707 | UMOD | T | 0.18 | + | 1.03 (0.96–1.10) | 0.46 |
| rs347685 | TFDP2 | C | 0.28 | + | 1.01 (0.95–1.07) | 0.74 |
Statistical significance defined using a Bonferroni correction for the 16 SNPs (0.05/16 = 0.003).
African-American ancestry UACR results
Association results for the UACR in the CARe Renal Consortium can be found in Table 3. Of the seven variants tested, none was associated at P< 0.007.
Table 3.
Association results for the urinary-to-albumin ratio (UACR) in African-Americans based on GFR-associated SNPs in the CARe Renal Consortium9
| Lead SNP | Gene | Chromosome | UACR coded allele | Coded allele frequency | β-coefficient for the eGFR related to the coded allele | UACR beta | UACR P-value |
|---|---|---|---|---|---|---|---|
| rs6781340 | TFDP2 | 3 | T | 0.41 | −0.014 | 0.025 | 0.36 |
| rs1750571 | VEGFA | 6 | A | 0.07 | 0.023 | 0.037 | 0.44 |
| rs3738479 | ANXA9 | 1 | A | 0.39 | 0.013 | 0.024 | 0.38 |
| rs4293393 | UMOD | 16 | A | 0.81 | −0.013 | 0.019 | 0.57 |
| rs3822460 | DAB2 | 5 | T | 0.83 | −0.013 | −0.048 | 0.17 |
| rs13022873 | GCKR | 2 | A | 0.81 | 0.013 | −0.016 | 0.62 |
| rs12302645 | ATXN2 | 12 | A | 0.94 | −0.018 | −0.040 | 0.47 |
The statistical significance defined as P< 0.007.
Concordance between UACR and eGFR
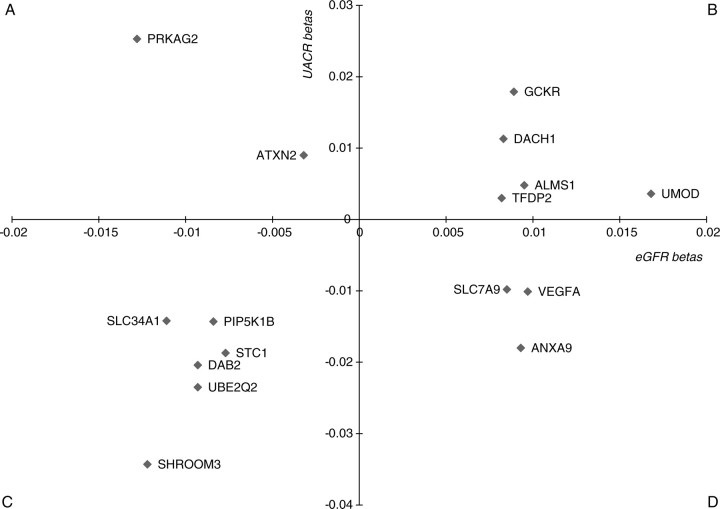
In general, higher eGFR levels are associated with lower UACR levels (for example, Framingham Heart Study correlation = −0.14; P-value <0.001), leading us to hypothesize that alleles associated with a lower eGFR would be associated with a higher UACR, most notably for rs17319721 at the SHROOM3 locus. However, in the CKDGen Consortium, of the 16 variants tested, only 5 were in this expected direction, whereas 11 were in the opposite direction (Fig. 1). We observed similar patterns of discordance in our African-American participants from the CARe Renal Consortium. Of the seven variants tested, only two showed associations in the hypothesized direction.
Figure 1.
Scatter plot of SNP effects on the eGFR and the UACR. Quadrants labeled (A) (lower eGFR effect size, higher UACR effect size) and (D) (higher eGFR effect size, lower UACR effect size) represent associations consistent with the observed correlation of the eGFR and the UACR; quadrants labeled (B) (higher eGFR effect size, higher UACR effect size) and (C) (lower eGFR effect size, lower UACR effect size) represent associations inconsistent with the observed correlation of the traits.
Genotype score analysis
In the CKDGen Consortium, we found that the 16-SNP genotype score weighted by the beta-coefficient was associated with albuminuria (P= 0.0006). However, this result was driven almost entirely by the SHROOM3 variant, rs17319721, known a priori to be associated with albuminuria. The removal of rs17319721 resulted in a P-value of 0.03, indicating a weak residual aggregate signal.
DISCUSSION
We observed no robust associations beyond SHROOM3, which we have previously reported, between eGFR-associated SNPs and the UACR. A similar lack of association was observed between eGFR-associated SNPs and the UACR in the CARe Renal Consortium. Although the genotype score analysis suggested a weak aggregate association signal, taken together, these results suggest differential genetic underpinnings, as identified from population-based GWAS, for these traits.
Prior research supports our primary findings, which suggest a differential etiology between the eGFR and the UACR. First, both traits show unique associations with some clinical risk factors. For example, in a cross-classification analysis of the eGFR and albuminuria, we found that only a quarter of the participants had both a reduced eGFR and albuminuria (6). Participants with albuminuria in the absence of a reduced eGFR had a higher prevalence of smoking, diabetes and elevated triglycerides, whereas participants without albuminuria in the presence of a reduced eGFR had a lower prevalence of smoking and diabetes despite being older (6). Similar findings of differential risk factor associations with the eGFR when compared with the UACR have been observed in other studies including the Zuni Kidney Project (11). These data suggest that these traits may have disparate underlying biological mechanisms.
Furthermore, weak genetic correlations between the eGFR and the UACR also support a differential underpinning for both traits. Prior work has documented a low genetic correlation (r = −0.002) between the eGFR and the UACR (12). Differential genetic correlations between each trait and the clinical risk factors have previously been shown; for example, the Zuni Kidney Project observed significant genetic correlations of the UACR with blood pressure, whereas none was observed with regard to the eGFR (11). Other studies have also documented unique genetic correlations for the UACR and the eGFR with other clinical risk factors, such as diabetes, hypertension, HDL and LDL (13,14). These results additionally corroborate our findings that there may be a distinct genetic architecture between the UACR and the eGFR.
As albuminuria and a reduced GFR commonly coexist in advanced kidney disease, our finding of minimal overlap in the genetic underpinnings to these traits may appear counterintuitive from a clinical standpoint. However, it must be remembered that it is atypical for these traits to occur simultaneously in early kidney disease, such as is seen in the general population. For example, in NHANES, only 8% with a GFR >60 ml/min/1.73 m2 have albuminuria, whereas the rate rises to 58% for those with a GFR 15–29 ml/min/1.73 m2 (15). Furthermore, in kidney diseases characterized by albuminuria, such as diabetic nephropathy or focal segmental glomerulosclerosis, a normal or increased GFR is characteristic in early disease and a reduced GFR often only manifests as a late phenomenon (16). In comparison, diseases primarily characterized by a low GFR due to reduced functioning nephron mass, such as polycystic kidney disease, often do not manifest albuminuria until the disease is quite advanced (17). This time-varying relationship in the onset of these two traits is often due to a maladaptive response of one to the other, and distinct genetic influences would thus be expected to underlie them at these differing time-points. Our observations are consistent with these data, and support the existence of a complex, distinct and time-varying interplay of small to moderate genetic influences underlying these phenotypes in the general population. Importantly, the new understanding gained from such insights may ultimately lead to different approaches to disease prevention and treatment.
It is therefore surprising that rs17319721 in SHROOM3 is associated with both a lower UACR and lower eGFR in the general population. Thus, reasons for the joint associations between this variant may be due to pleiotropic associations of this gene on both traits. SNPs in SHROOM3 have previously been shown to be associated with serum magnesium concentrations (18), and these SNPs are in linkage disequilibrium with our lead SNP (r2= 0.85). Downstream effector proteins of SHROOM3 such as GTPase, Rho Kinases, Rap1 and myosin II may also act interdependently to contribute to the gene's functionality (19). To the best of our knowledge, SNPs in SHROOM3 have primarily been associated with renal traits, suggesting a possible renal pleiotropic specificity to its actions. Further functional work should focus on a better understanding of the mechanisms involved in the functional underpinnings of SHROOM3 in association with the UACR and the eGFR. Apart from pleiotropic actions, mechanisms may exist that are jointly associated with both a higher eGFR and higher UACR. For example, a higher GFR is associated with albuminuria in a variety of hyperfiltration states, including diabetes (20,21), sickle cell disease (22), hyperuricemia (23), hypertension (24) and primary aldosteronism (23).
The strengths of this study include large albuminuria data sets from the CKDGen Consortium and the CARe Renal Consortium. The large sample sizes and the targeted gene approach increased the power to detect associations with the UACR and albuminuria, and the complementary samples allowed evaluation within both European and African ancestry populations. We also used a panel of well-established SNPs for the eGFR. Some limitations warrant mention. First, our African ancestry data set for the UACR was underpowered. Nonetheless, it is the largest data set of its kind in participants of African ancestry and makes an important point about the multi-ethnic nature of our results. Secondly, whereas our study indicates that there is little overlap in the common genetic determinants of the eGFR and the UACR in the general population, similar analyses in cohorts enriched for more advanced kidney disease or containing more cases of combined CKD and albuminuria may yet identify novel genes. Finally, as both the eGFR and the UACR may vary over time, testing for their association with longitudinal traits may yield different results. However, we were underpowered to test this hypothesis.
Apart from the SHROOM3 locus, we observed no robust associations between the eGFR-associated SNPs and the UACR in both Europeans and African-Americans, suggesting that there may be distinct genetic components to these traits as identified by population-based GWAS.
MATERIALS AND METHODS
Overall design study
Genetic association testing for the UACR and albuminuria was performed in the CKDGen cohorts of European ancestry using existing GWAS meta-analysis data sets for the UACR and albuminuria (9). Additional association testing for the UACR was performed in the CARe cohorts of African-American ancestry (10).
Study exposure
Sixteen variants previously identified in a two-stage GWAS in association with the eGFR were specifically queried for association with the UACR and albuminuria in the CKDGen Consortium (n= 31 580) (7). Seven eGFR SNPs previously validated in a GWAS using participants of African ancestry were queried for association with the UACR in the CARe Renal Consortium (n= 5569) (10). Imputation scores for the 16 and 7 SNPs, respectively, are shown in Supplementary Material, Table S3.
Outcomes
For the CKDGen Consortium and CARe studies, the quantitative trait UACR was calculated in each participating study. The UACR was log-transformed for analysis; sex-specific residuals that were age-adjusted were calculated as previously described (9). For the CKDGen studies, the dichotomous trait albuminuria was defined by a UACR >17 mg/g for men and >25 mg/g for women (25,26). For the CARe studies, albuminuria was not secondarily analyzed due to the relatively small sample size.
Statistical methods
We used previously published meta-analysis data to look- up the results of the 16 eGFR SNPs in European Americans and the 7 eGFR SNPs in African -Americans for association with the UACR. To correct for multiple testing, we used a Bonferroni corrected P-value of 0.05/16 (0.003) in the CKDGen Consortium and 0.05/7 (0.007) in the CARe Consortium. In the CKDGen Consortium, we had 66% power to detect a beta-coefficient of 0.034 in a sample of 31 580 for an alpha of 0.003 (0.05/16). The power was lower in CARe due to the smaller sample size. To improve the power, we assessed whether the 16 GFR SNPs in aggregate were associated with the UACR using a beta-weighted genotype score, as has been used in comparable analyses of blood pressure and lung function variants (27,28).
SUPPLEMENTARY MATERIAL
FUNDING
This work was partially supported by the National Heart, Lung and Blood Institute's Framingham Heart Study (contract no. N01-HC-25195).
Supplementary Material
ACKNOWLEDGEMENT
This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Coresh J., Selvin E., Stevens L.A., Manzi J., Kusek J.W., Eggers P., Van L.F., Levey A.S. Prevalence of chronic kidney disease in the United States. JAMA. 2007;298:2038–2047. doi: 10.1001/jama.298.17.2038. [DOI] [PubMed] [Google Scholar]
- 2.Go A.S., Chertow G.M., Fan D., McCulloch C.E., Hsu C.Y. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N. Engl. J. Med. 2004;351:1296–1305. doi: 10.1056/NEJMoa041031. [DOI] [PubMed] [Google Scholar]
- 3.National Kidney Foundation. K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am. J. Kidney Dis. 2002;39(2 Suppl. 1):S1–266. [PubMed] [Google Scholar]
- 4.Matsushita K., van der Velde M., Astor B.C., Woodward M., Levey A.S., de Jong P.E., Coresh J., Gansevoort R.T. Association of estimated glomerular filtration rate and albuminuria with all-cause and cardiovascular mortality in general population cohorts: a collaborative meta-analysis. Lancet. 2010;375:2073–2081. doi: 10.1016/S0140-6736(10)60674-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keith D.S., Nichols G.A., Gullion C.M., Brown J.B., Smith D.H. Longitudinal follow-up and outcomes among a population with chronic kidney disease in a large managed care organization. Arch. Intern. Med. 2004;164:659–663. doi: 10.1001/archinte.164.6.659. [DOI] [PubMed] [Google Scholar]
- 6.Foster M.C., Hwang S.J., Larson M.G., Parikh N.I., Meigs J.B., Vasan R.S., Wang T.J., Levy D., Fox C.S. Cross-classification of microalbuminuria and reduced glomerular filtration rate: associations between cardiovascular disease risk factors and clinical outcomes. Arch. Intern. Med. 2007;167:1386–1392. doi: 10.1001/archinte.167.13.1386. [DOI] [PubMed] [Google Scholar]
- 7.Kottgen A., Pattaro C., Boger C.A., Fuchsberger C., Olden M., Glazer N.L., Parsa A., Gao X., Yang Q., Smith A.V., et al. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kottgen A., Glazer N.L., Dehghan A., Hwang S.J., Katz R., Li M., Yang Q., Gudnason V., Launer L.J., Harris T.B., et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat. Genet. 2009;41:712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boger C.A., Chen M.H., Tin A., Olden M., Kottgen A., de Boer I.H., Fuchsberger C., O'Seaghdha C.M., Pattaro C., Teumer A., et al. CUBN is a gene locus for albuminuria. J. Am. Soc. Nephrol. 2011;22:555–570. doi: 10.1681/ASN.2010060598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C.T., Garnaas M.K., Tin A., Kottgen A., Franceschini N., Peralta C.A., de Boer I.H., Lu X., Atkinson E., Ding J., et al. Genetic association for renal traits among participants of African ancestry reveals new loci for renal function. PLoS. Genet. 2011;7:e1002264. doi: 10.1371/journal.pgen.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacCluer J.W., Scavini M., Shah V.O., Cole S.A., Laston S.L., Voruganti V.S., Paine S.S., Eaton A.J., Comuzzie A.G., Tentori F., et al. Heritability of measures of kidney disease among Zuni Indians: the Zuni Kidney Project. Am. J. Kidney Dis. 2010;56:289–302. doi: 10.1053/j.ajkd.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leon J.M., Freedman B.I., Miller M.B., North K.E., Hunt S.C., Eckfeldt J.H., Lewis C.E., Kraja A.T., Djousse L., Arnett D.K. Genome scan of glomerular filtration rate and albuminuria: the HyperGEN study. Nephrol. Dial. Transplant. 2007;22:763–771. doi: 10.1093/ndt/gfl674. [DOI] [PubMed] [Google Scholar]
- 13.Fogarty D.G., Rich S.S., Hanna L., Warram J.H., Krolewski A.S. Urinary albumin excretion in families with type 2 diabetes is heritable and genetically correlated to blood pressure. Kidney Int. 2000;57:250–257. doi: 10.1046/j.1523-1755.2000.00833.x. [DOI] [PubMed] [Google Scholar]
- 14.Placha G., Poznik G.D., Dunn J., Smiles A., Krolewski B., Glew T., Puppala S., Schneider J., Rogus J.J., Rich S.S., et al. A genome-wide linkage scan for genes controlling variation in renal function estimated by serum cystatin C levels in extended families with type 2 diabetes. Diabetes. 2006;55:3358–3365. doi: 10.2337/db06-0781. [DOI] [PubMed] [Google Scholar]
- 15.Coresh J., Astor B.C., Greene T., Eknoyan G., Levey A.S. Prevalence of chronic kidney disease and decreased kidney function in the adult US population: Third National Health and Nutrition Examination Survey. Am. J. Kidney Dis. 2003;41:1–12. doi: 10.1053/ajkd.2003.50007. [DOI] [PubMed] [Google Scholar]
- 16.Ritz E., Zeng X.X., Rychlik I. Clinical manifestation and natural history of diabetic nephropathy. Contrib. Nephrol. 2011;170:19–27. doi: 10.1159/000324939. [DOI] [PubMed] [Google Scholar]
- 17.Wilkie P. Adult polycystic kidney disease: diagnosis presentation and genetic implications. Scott. Med. J. 1992;37:71–73. doi: 10.1177/003693309203700303. [DOI] [PubMed] [Google Scholar]
- 18.Meyer T.E., Verwoert G.C., Hwang S.J., Glazer N.L., Smith A.V., van Rooij F.J., Ehret G.B., Boerwinkle E., Felix J.F., Leak T.S., et al. Genome-wide association studies of serum magnesium, potassium, and sodium concentrations identify six Loci influencing serum magnesium levels. PLoS. Genet. 2010;6:1–8. doi: 10.1371/journal.pgen.1001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishimura T., Takeichi M. Shroom3-mediated recruitment of Rho kinases to the apical cell junctions regulates epithelial and neuroepithelial planar remodeling. Development. 2008;135:1493–1502. doi: 10.1242/dev.019646. [DOI] [PubMed] [Google Scholar]
- 20.Chiarelli F., Verrotti A., Morgese G. Glomerular hyperfiltration increases the risk of developing microalbuminuria in diabetic children. Pediatr. Nephrol. 1995;9:154–158. doi: 10.1007/BF00860729. [DOI] [PubMed] [Google Scholar]
- 21.Amin R., Turner C., van Aken S., Bahu T.K., Watts A., Lindsell D.R., Dalton R.N., Dunger D.B. The relationship between microalbuminuria and glomerular filtration rate in young type 1 diabetic subjects: the Oxford Regional Prospective Study. Kidney Int. 2005;68:1740–1749. doi: 10.1111/j.1523-1755.2005.00590.x. [DOI] [PubMed] [Google Scholar]
- 22.Thompson J., Reid M., Hambleton I., Serjeant G.R. Albuminuria and renal function in homozygous sickle cell disease: observations from a cohort study. Arch. Intern. Med. 2007;167:701–708. doi: 10.1001/archinte.167.7.701. [DOI] [PubMed] [Google Scholar]
- 23.Lee J.E., Kim Y.G., Choi Y.H., Huh W., Kim D.J., Oh H.Y. Serum uric acid is associated with microalbuminuria in prehypertension. Hypertension. 2006;47:962–967. doi: 10.1161/01.HYP.0000210550.97398.c2. [DOI] [PubMed] [Google Scholar]
- 24.Palatini P., Benetti E., Zanier A., Santonastaso M., Mazzer A., Cozzio S., Zanata G., De T.R., Zaninotto M. Cystatin C as predictor of microalbuminuria in the early stage of hypertension. Nephron Clin. Pract. 2009;113:c309–c314. doi: 10.1159/000235949. [DOI] [PubMed] [Google Scholar]
- 25.Mattix H.J., Hsu C.Y., Shaykevich S., Curhan G. Use of the albumin/creatinine ratio to detect microalbuminuria: implications of sex and race. J. Am. Soc. Nephrol. 2002;13:1034–1039. doi: 10.1681/ASN.V1341034. [DOI] [PubMed] [Google Scholar]
- 26.Warram J.H., Gearin G., Laffel L., Krolewski A.S. Effect of duration of type I diabetes on the prevalence of stages of diabetic nephropathy defined by urinary albumin/creatinine ratio. J. Am. Soc. Nephrol. 1996;7:930–937. doi: 10.1681/ASN.V76930. [DOI] [PubMed] [Google Scholar]
- 27.Soler A.M., Wain L.V., Repapi E., Obeidat M., Sayers I., Burton P.R., Johnson T., Zhao J.H., Albrecht E., Dominiczak A.F., et al. Effect of 5 genetic variants associated with lung function on the risk of COPD, and their joint effects on lung function. Am. J. Respir. Crit. Care Med. 2011;184:786–795. doi: 10.1164/rccm.201102-0192OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D., Verwoert G.C., Hwang S.J., et al. International Consortium for Blood Pressure Genome-Wide Association Studies. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature Genet. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.