Abstract
Decorin binding protein A (DBPA) is an important lipoprotein from the bacterium Borrelia burgdorferi, the causative agent of Lyme disease. Absence of DBPA drastically reduces the pathogenic potential of the bacterium and biochemical evidence indicates DBPA’s interactions with the glycosaminoglycan (GAG) portion of decorin are crucial to its function. We have solved the solution structure of DBPA and studied DBPA’s interactions with various forms of GAGs. DBPA is determined to be a helical bundle protein consisting of five helices held together by a strong hydrophobic core. The structure also possesses a basic patch formed by portions of two helices and two flexible linkers. Low molecular weight heparin-induced chemical shift perturbations for residues in the region as well as increases in signal intensities of select residues in their presence confirm residues in the pocket are perturbed by heparin binding. Dermatan sulfate (DS) fragments, the dominant GAG type found on decorin, were shown to have lower affinity than heparin but are still capable of binding DBPA.
Keywords: glycosaminoglycan, bacterial adhesin, GAG-protein interactions, solution NMR
Lyme disease is a vector-borne illness spread by ticks infected with the bacterium Borrelia burgdorferi. Although never entering the cell, the existence of these bacteria in the extracellular space can lead to local inflammations that produce symptoms from minor skin lesions to serious arthritis as well as heart and neural diseases if left untreated (1). One important aspect of Borrelia’s existence as an extra cellular bacterium is its extensive interactions with the extracelluar matrix. These interactions allow it to be extricated from blood and into surrounding tissues. In fact, the spread of Borrelia from blood to tissues is an pre-requisite to chronic infection, which is resistant to antibiotic treatments that have proven effective in the treatment of early stage Lyme disease (1–3). Development of a universal vaccine for the disease has proven difficult owing to the large genetic variability among different strains of Borrelia (1, 4). However, a few proteins have been identified as crucial for Borrelia infectivity in human and hold the potential for being useful therapeutic targets. One of the proteins is the lipoprotein decorin binding protein (DBP). DBPs are membrane anchored proteins expressed exclusively during the human host infection stage, and were identified for being the primary adhesin for the small proteoglycan decorin (5, 6), an essential component in all collagen based connective tissues and whose absence greatly impairs the dissemination of Borrelia burgdorferi (7). Both variants of DBP (DBPA and DBPB) have been shown to be important to the Borrelia infection process with the deletion of the genes coding the proteins leading to defects in dissemination and early survival of the bacterium, presumably due to their inability to bind decorin and evade the immune system (8–11).
Interactions of DBPs with decorin have already been characterized biochemically and the protein is found to interact strongly with the glycosaminoglycan (GAG) part of decorin (5, 12, 13). Leong and coworkers also investigated interactions of various strains of DBPA with GAGs using both in vitro and in vivo assays. Their studies demonstrated DBPA has affinity for both heparin and dermatan sulfate (DS), but not chondroitin sulfate (CS) (12–16). Interestingly, two studies also implicate the core protein of decorin in its interactions with DBPs (5, 14). To investigate the decorin-binding site on DBPA, sequence alignments and peptide based mutagenesis study have also been conducted. Several Lysine residues on the protein have been shown to be crucial (17, 18), consistent with the conclusion that the glycosaminoglycan (GAG) portion of decorin is responsible for most of the interactions with DBPs. Similarly, an inadvertent mutation produced in the study done by Benoit et al. also showed the Lysine-rich C-terminus of DBPA is also involved in binding GAGs (14). Although the protein sequence of DBPB is relatively uniform among different strains of Borrelia, the DBPA sequence differs considerably from strain to strain. Biochemical studies conducted on transgenic strains of Borrelia showed different DBPA sequences have type-specific interactions with GAGs, raising the possibility that different strains of Borrelia may target different tissue types (12).
Despite extensive biochemical and functional characterization, structural information on DBPs is scarce. It has been shown, by both secondary structure prediction and circular dichroism, that the protein is mostly α-helical. The secondary structure prediction also points to the existence of four helices in the protein, making the canonical four-helical bundle a likely conformation for DBPs. However, no high resolution structure of the protein is available. This makes the interpretation of the available biochemical data difficult and impedes our understanding of the mechanism by which DBPs achieve their GAG and decorin core protein recognition. In this paper, we present the solution structure of DBPA from Borrelia burgdorferi strain B31 and NMR characterization of its interactions with GAG fragments. DBPA is shown to be an all helical protein. However, its conformation is an unusual helical bundle consisting of five helices. The helical bundle is held together by a strong hydrophobic core while the surface of the protein is populated by both basic and acidic amino acids. Despite the presence of basic amino acids throughout DBPA, electrostatic potential map of the protein reveals a well defined basic pocket close to the C-terminus. In agreement with other biochemical studies, residues previously identified as crucial to GAG-DBPA interactions in other strains of DBPA are found in the pocket. Low molecular weight heparin-induced changes in chemical shifts and signal intensities of DBPA residues near the pocket provided proof that the basic patch most likely is involved in heparin binding. Interestingly, DS fragments did not induce chemical shift perturbations similar to heparin. Instead the presence of DS hexasaccharide (dp6) produced severe signal broadening in DBPA. Besides NMR titrations, gel mobility shift assay using fluorescently-tagged GAG fragments was also carried out to characterize the GAG length dependence of DBPA-GAG interactions. The assay showed conclusively that DBPA binds heparin with a higher affinity than DS and heparin fragments as short as a tetrasaccharide (dp4) can interact with the protein. Finally, after mutating the N-terminal cysteine to serine in order to prevent to dimerization, cystine chemical shift values confirmed that the remaining two cysteines at the C-terminal tail form an intra-molecular disulfide bond.
Experimental Procedure
Expression and purification of DBPA
An open reading frame coding the mature form of the B31 version of DBPA (residues 24 to 191) with the N-terminal cysteine mutated to serine was synthesized by Genscript Inc. (Piscataway, NJ) and cloned into the pHUE vector (19) as a chimera with His-tagged ubiquitin at the N-terminus. The plasmid was then transformed into BL21(DE3) and the transformed bacteria were grown to an OD600 of 0.8 at 37 C before induction with 0.5 mM IPTG, followed by 3 more hours of incubation at 30 C. After harvesting the culture, the resuspended cell pellet was lysed using sonication and fusion protein in the cleared supernatant was extracted through Ni-affinity chromatography using a 1 ml HisTrap column (GE Life Sciences). Specifically, cleared supernatant in 20 mM sodium phosphate buffer, pH 8.0, 10 mM imidazole, 500 mM NaCl was loaded onto the column at a flow rate of 1 ml/min. The column was washed with 35 mM imidazole for five minutes before an imidazole gradient of 35 to 500 mM was applied to elute the protein. After exchanging the buffer to 25 mM Tris, pH 8.0 and 100 mM NaCl, the purified fusion protein was digested with the ubiquitinase USP2 overnight at room temperature (19). The cleaved DBPA was separated from His-ubiquitin and His-tagged ubiquitinase by passing the mixture through a Ni-affinity column and collecting the flow through. Unlabeled protein was obtained by growing the culture in normal LB and isotopically labeled DBPAs were obtained by expressing the protein in M9 media supplemented with 15NH4Cl or 13C-glucose.
Analytical size exclusion chromatography of DBPA
To characterize the oligomerization state of DBPA, DBPA was subjected to analytical size exclusion chromatography using a 160-mL Superdex 75 gel filtration column (GE Lifesciences). The column was first equilibrated in 50 mM sodium phosphate, pH 6.5, 300 mM NaCl. 100 µl of 0.6 mM DBPA was then injected onto the column and the flow rate was held at 0.7 mL/min for a total elution volume of 130 mL. The elution time of DBPA was compared to standards of known molecular weight (Bio-Rad, cat #. 151-1901)
Glutaraldehyde cross-linking assay
100 µL of solution containing either 5, 12.5, 25 or 50 µM of DBPA in 20 mM HEPES buffer, pH 7.5, was cross-linked by the addition of 5 µL of 2.5% glutaraldehyde and incubation at 37 C for 5 minutes. The reaction was quenched with 10 µL of 1 M Tris, pH 8.0 and 5 µg of protein from each reaction was analyzed by SDS-PAGE.
NMR data acquisition and analysis for DBPA structure
NMR data on DBPA were collected on Agilent Inova 800 MHz spectrometer equipped with cryogenically cooled triple resonance probe and Varian Inova 500 MHz spectrometer with room temperature probe. Most pulse sequences used were based on experiments included in the pulse sequence package BioPack (Agilent Inc.). For backbone assignment, HNCACB, HNCO, HNCOCA, HNCOCACB and HNCOCA spectra of 2H,13C,15N-labeled DBPA were collected. 15N and 13C-edited NOESYHSQC spectra of 13C, 15N-labeled DBPA were collected for structural determination. Assignments of methyl groups were done using the methyl HCCH-TOCSY experiments and assignments of other side chain protons were done using 13C-edited NOESYHSQC. HN and NC residual dipolar couplings (RDCs) of DBPA aligned in 7% neutral polyacrylamide gel were collected on 2H,13C,15N-labeled DBPA using J-modulated pulse sequences from Liu et al (20). Residue specific rotational correlation times were calculated using procedure outlined in Liu et al (21). Samples used in data collection contain 0.6 to 1 mM of DBPA and 50 mM sodium phosphate, pH 6.5. To confirm the absence of intermolecular NOEs, two dimensional 13C/15N filtered, 13C-edited NOESY spectrum of DBPA was collected on a 1 mM DBPA sample containing 0.5 mM 13C/15N labeled DBPA and 0.5 mM unlabeled DBPA. The pulse sequence used was the gCNfilnoesyChsqcA sequence from Biopack and 256 transients were accumulated for each FID. All data processing were done with NMRPipe (22) and analyzed in NMRView (23).
Structure determination
Backbone dihedral angles of well-ordered residues were calculated with TALOS (24). Isotopically-edited NOESYHSQC experiments of DBPA were first analyzed and assigned manually with the focus on finding unambiguous long range contacts. The partially-assigned NOESYHSQC peak lists were then used in CYANA’s default automatic structure determination protocol (25). The resulting structure and final constraint table were used in XPLOR-NIH for final refinement with RDCs (26). The ten structures with the least amount of NOE violations were used in the ensemble presented in the paper.
Production of low molecular weight GAG fragments
Heparin and DS were purchased from Sigma Aldrich. All GAGs were dialyzed and lyophilized to remove excess salt. Short GAG fragments were obtained through enzymatic partial depolymerization. Porcine mucosa heparin was depolymerized with heparinase I (IBEX Inc, Canada) until the digestion was 30% complete (27). Porcine mucosa DS fragments were also obtained through 30% partial digestion by Chondroitinase ABC (Sigma Aldrich) (28). In both cases, the fragments were separated based on size using a 2.5 cm-by-175 cm size exclusion chromatography column (Bio-Rad Biogel P10) and a flow rate 0.2 ml/min. Fractions containing same size fragments were pooled, desalted and lyophilized. The sizes of the DS fragments were verified with ESI-MS, and the size of heparin fragments were inferred using DS as a standard. The amount of GAG fragments was determined gravimetrically after lyophilization. The longest DS fragment isolated is dp14 and the longest heparin fragment isolated is dp10. Disaccharide composition of the GAGs were analyzed by exhaustively digesting the GAGs using the appropriate lyases and separating the resulting disaccharides by SAX-HPLC. The identities of the disaccharides were determined by comparing their elution times to standards with known structure.
Gel mobility shift assays for DBPA
Gel mobility shift assays were done according to Seo et al.(29) Briefly, DS and heparin fragments of lengths dp4, 6, 8 and 10 were fluorescently labeled with 2-aminoacridone according to Lyon et al.(30) Two µg of the fluorescently labeled GAG were then mixed with either one molar equivalence of DBPA in 50 mM sodium phosphate buffer, pH 6.5, 150 mM NaCl buffer or equal volume of the same buffer without the protein, and incubated for 30 mins. The mixtures were subjected to electrophoresis at 120 V for 20 minutes in 1% agarose gel with 40 mM Tris, 20 mM acetate, 1 mM EDTA, pH 8.0 buffer. The positions of the fragments were visualized using a UV panel.
Titrations of DBPA with heparin & DS
Titrations of DBPA with intact GAG polymer were done by adding 200 µg of intact polymer to a 400 µL sample containing 200 µM DBPA in 50 µg aliquots. For titrations of DBPA with heparin dp6, heparin dp10 and DS dp14, aliquots of 40 mM polysaccharide stock solution were added to 400 µL of 100 µM DBPA in 50 mM sodium phosphate, pH 6.5, 150 mM NaCl at concentrations of 0.3, 0.6, 0.9 1.2, 1.6 and 2.0 mM. For the titration of DBPA with DS dp6, aliquots of 40 mM DS dp6 stock solution was added to 400 µL of 200 µM DBPA at concentrations of 0.4, 1.0, and 1.6 mM. A 1H,15N-HSQC spectrum was acquired at each point to monitor the titrations. Chemical shift changes in both 1H and 15N dimensions were combined into a single normalized chemical shift value according to (31). Specifically, normalized chemical shift δn is defined by δn=(ΔδH 2+(1.7×ΔδN)2)^(1/2), where ΔδH and ΔδN represent chemical shift changes in Hz units for the 1H and 15N dimensions, respectively. Kd of binding was extracted by fitting plots of normalized chemical shift changes relative to ligand concentration using the 1-to-1 binding chemical shift fitting feature in the software xcrvfit (http://www.bionmr.ualberta.ca/bds/software/xcrvfit/). The DS dp6 concentration dependent signal decay rate was extracted by measuring HSQC peak intensities at different DS dp6 concentrations and fitting the plot of intensity vs. DS dp6 concentrations to the equation I([DS])=I(0)e−[DS]R, where [DS] is the concentration of DS dp6, I is the intensity of the signal and R is the rate constant governing the decay. Data for the titrations were acquired on a Varian 800 MHz Inova spectrometer.
Results
Oligomerization state of DBPA
Native DBPA containing the N-terminal cysteine was known to form dimers (5, 18). To prevent the dimerization of DBPA in this study, the N-terminal cysteine was mutated to serine. However, the broadness of DBPA’s NMR signals is typical of a larger protein. To test whether DBPA oligomers exist, DBPA was subjected to glutaraldehyde cross linking at pH 7.5. Significant dimer formation was detected at concentrations higher than 10 µM (figure S1A). Species corresponding to trimers were also seen at higher concentrations, indicating the protein can aggregate non-specifically at high concentrations. Since the crosslinking reaction is non-reversible, the results are only a qualitative indication that oligomers exist. More accurate characterization of oligomer fractions was carried out using size exclusion chromatography and NMR rotational correlation times. When DBPA was analyzed using size-exclusion chromatography, the protein eluted slightly earlier than the 17 kDa molecular weight standard and modest aggregation was seen (figure S1B). Thus size exclusion chromatography indicates the oligomerization equilibrium is heavily biased towards the formation of monomers. The rotational correlation times of the protein were determined to be around 15 ns for structured regions of DBPA (figure S2). Comparing with correlation times of proteins with known apparent molecular weight, this corresponds to a weight of 25 kDa. Since the monomer molecular weight is only 19 kDa, the larger apparent weight indicates roughly 30% of protein exist in dimer form under NMR conditions. These data corroborate the theory that although oligomers are present, they are the minor fraction. Since intermolecular NOEs mis-interpreted as intra-molecular NOEs are known to produce incorrect NMR structures (32), a 13C/15N filtered-13C edited NOESYHSQC experiment was performed on a 1 mM DBPA sample containing half 13C/15N-labeled DBPA and half unlabeled DBPA. No credible intermolecular NOE cross peaks were detected (figure S3). Some weak peaks corresponding to NOE contacts between 1H with chemical shifts of 4.1 ppm and 1.9 ppm were seen. However, these are most likely residual signals from strong intraresidue contacts between Hα and Hβ atoms. No cross peaks were seen in crucial regions where hydrophobic core protons are found, eliminating the possibility that the hydrophobic core structure, which largely determines the three-dimensional fold of the protein, is distorted due to intermolecular NOE contamination. The signal broadening observed in the NMR data is most likely caused by exchanges between oligomer and monomer. Because the density of DBPA on bacterial membrane is not known, it is unclear whether the aggregation occurs in vivo.
Structure of DBPA
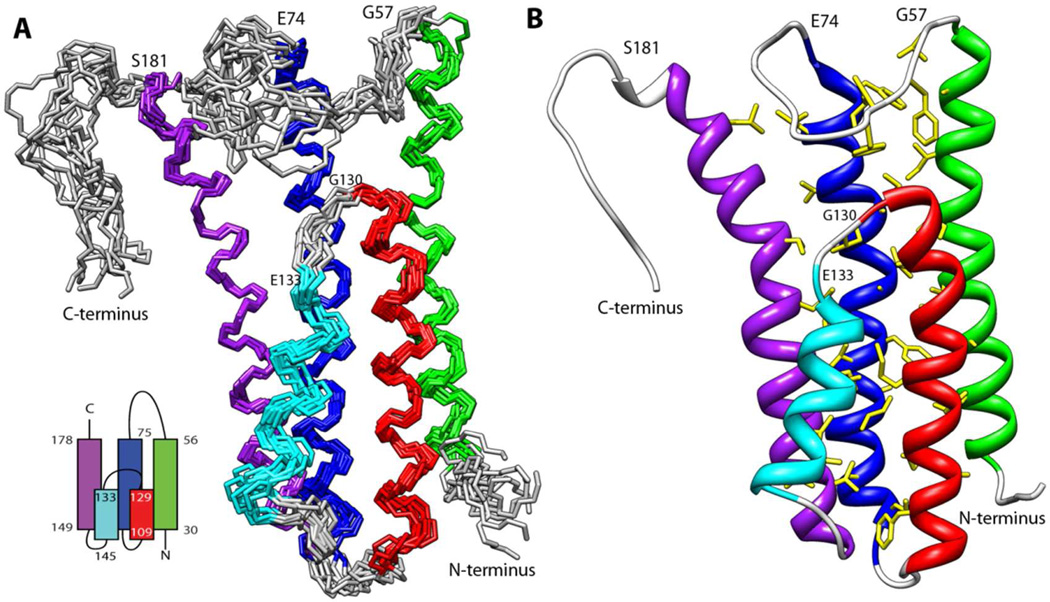
The mature form of DBPA (residues 24 to 191) was predicted to consist of four α-helices. Although the experimental data was consistent with DBPA being mostly helical, an unstructured region was found in one of the predicted helices (residue G130 to residue E133), leading to five helices in the protein instead of four. Further examination of the NOE contact pattern between the helices showed the two halves of the predicted third helix are anti-parallel to one another. Figure 1A shows the ensemble of structures most consistent with the experimental data. Table 1 contains the structural statistics for the ensemble. Overall, DBPA adopts a five helical bundle conformation. The partition of the predicted helix three into two anti-parallel helices altered the topology of the protein such that the long linker between helices one and two is proximate to the C-terminal tail. The helical bundle is held in place by a tightly packed hydrophobic core involving 31 residues (figure 1B). Helices one to three are perfectly parallel to each other while helices four and five are at an angle with the other helices. Helix two is central to the formation of the hydrophobic core with eleven residues in the helix making hydrophobic contacts with residues in helices one, three and five. Helices three and five also form parts of the core by making contacts with each other as well as helix two. The short helix four sits at the periphery of the core, making minor contacts with helices three and five only. The angled orientation of helix four and part of helix five are reflected in the values of their backbone HN residual dipolar couplings: RDCs for consecutive residues in both helices four and five experience large cyclic oscillations in their values, an indication that parts of helices are not perfectly parallel to a single dominant alignment axis (figure S4). Despite its small size and weak contacts to the core, rotational correlation time measurement shows helix four is as rigid as the rest of the protein (figure S2), confirming that its position relative to the rest of the protein is stable. However, helix four in the structure ensemble shows a backbone RMSD of 0.85 Å, larger than the 0.60 Å RMSD for residues in other helices. This coupled with the observation of smaller RDCs for residues in helix four and the fact that RDC values are very sensitive to the presence of motional averaging shows helix four may undergo more internal motion than other helices. A structural homology search using the Dali server (33) found no significant homology between the DBPA structure and those already in the PDB.
Figure 1.
A) Ensemble of 10 lowest energy structures of DBPA in solution. Helix one (residue 30 to 56) is colored green; helix two (residue 75 to 104) is colored blue; helix three (residue 109 to 129) is colored red; helix four (residue 133 to 145) is colored cyan; helix five (residue 149 to 178) is colored purple. Schematic topology of the protein is shown in the lower left corner. B) Ribbon depiction of DBPA with the side chains of amino acids in the hydrophobic core shown in yellow.
Table 1.
Structural statistics for the ensemble of DBPA structures
| NOE-based distance constraints | ||
| Total | 1171 | |
| intra-residue [i = j] | 81 | |
| sequential [| i - j | = 1] | 242 | |
| medium range [1 < | i - j | < 5] | 396 | |
| long range [| i - j | ≥ 5] | 452 | |
| NOE constraints per restrained residue a | 7.3 | |
| Residual dipolar couplings | ||
| H-N | 113 | |
| N-C | 103 | |
| Dihedral-angle constraints: | 210 | |
| Total structures computed | 100 | |
| Number of structures used | 10 | |
| Residual constraint violations b | ||
| Distance violations / structure | ||
| 0.1 – 0.2 Å | 15 | |
| 0.2 – 0.5 Å | 8 | |
| > 0.5 Å | 0 | |
| Dihedral angle violations / structure | ||
| 1 – 10 ° | 11 | |
| > 10 ° | 0 | |
| RDC violations / structure | ||
| 0–1 Hz | 3 | |
| > 1 Hz | 0 | |
| RMSD Values | ||
| all | orderedc | |
| All backbone atoms | 1.4 Å | 0.6 Å |
| All heavy atoms | 1.9 Å | 1.1 Å |
| Ramachandran Plot Summary from Procheck d | ||
| Most favoured regions | 98.0% | |
| Additionally allowed regions | 1.8% | |
| Generously allowed regions | 0.2% | |
| Disallowed regions | 0.0% |
There are 160 residues with conformationally restricting constraints
Calculated for all constraints for the given residues, using sum over r^-6
Residues with sum of phi and psi order parameters > 1.8 Ordered residue ranges: 31–55,76–103,110–128,134–142,145–179
Residues selected based on: Dihedral angle order parameter, with S(phi)+S(psi)>=1.8 Selected residue ranges: 31–55,76–103,110–128,134–142,145–179
Besides the helical regions, the protein also contains two large disordered segments: the flexible linker between helices one and two (residues G57 to E74) as well as a long C-terminal tail. Residues in both regions possessed lower rotational correlation times typical of flexible, dynamic segments (figure S2). The C-terminal tail contains the only two cysteines in the protein. Cβ chemical shifts of the cysteines in the absence of reducing agents indicate they are involved in the formation of disulphide bonds. To determine whether the disulfide bond is inter- or intra-molecular, SDS-PAGE was performed under non-reducing conditions and no dimers were seen. HSQC of the protein under reducing conditions (10 mM DTT) was also collected: although many C-terminal residues did experience a decrease in rotational correlation time, corresponding to an increase in dynamics, the overall correlation time for the protein did no change. These observations confirm that the disulfide bond formed is intra-molecular. Since the C-terminal tail is a potential GAG-binding segment in the protein, GAG-binding affinities of reduced and oxidized DBPA were tested: no significant difference was detected. However, the reduced C-terminal tail of the protein did show more signs of degradation than the oxidized version.
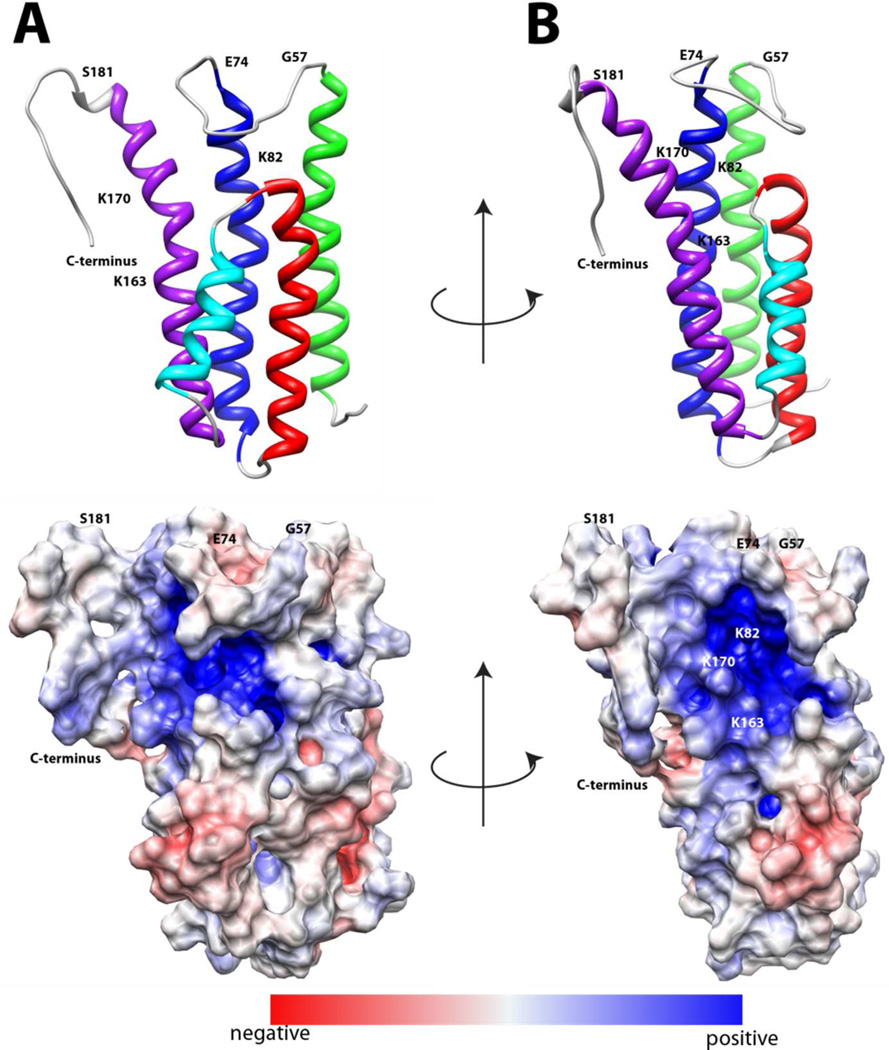
DBPA possesses a large number of basic residues typical of GAG-binding proteins. They are spread throughout the protein, but a particularly high concentration can be found on the last helix. The protein also contains a significant number of acidic residues, which are interspersed among the basic residues, leading to a neutralization of the charges in some areas. Figure 2 is the electrostatic potential map of the protein. A large basic patch composed of the second half of the helices two and five, the C-terminal tail, as well as the linker between helices one and two can be seen. Despite basic residues in other regions of the protein, they appear to have neutral to weakly acidic potential. In agreement with previous studies, residues known to be important in DBPA-GAG interactions, which include Lysines 82,163, 170 and the C-terminal tail (14, 17, 18), are found in the basic patch (figure 2).
Figure 2.
Electrostatic potential surface map of DBPA in two orientations. Red indicates acidic regions and blue indicates basic regions. A) the protein is in the same orientation as figure 1A. B) the protein is rotated 90 degrees about the vertical axis.
Interactions of DBPA with GAGs
To gain further insight into DBPA’s GAG-binding mechanism and to confirm the role of the basic pocket in binding GAGs, DBPA’s interactions with DS and heparin, the two types of GAGs known to bind DBPA were examined. Addition of heterogeneous polysaccharides of both species of GAGs reduced the NMR signals of DBPA greatly and without discrimination, but none were able to produce observable changes in DBPA chemical shifts. This phenomenon is often seen in GAG-binding proteins and the most plausible interpretation of the results is that interaction of DBPA with the long GAG chains led to extreme signal broadening of GAG-bound proteins due to the large size of the complex produced through a combination of GAG-induced oligomerization and multiple ligand binding, thus only free DBPA contributes to detectable NMR signals. This shows DBPA indeed interacts with heparin and DS, but gives no information on the GAG-binding epitope of DBPA. To study the GAG-binding epitope on DBPA, DBPA’s interactions with low molecular weight GAGs were examined. The use of short fragments not only reduces molecular weight of the polysaccharides, but also diminishes GAG-induced oligomerization of protein. However, since multivalency of GAGs is a crucial factor in protein-GAG interactions, short fragments of GAG are expected to have lower binding affinity compared to native polysaccharides. For this study, GAG-DBPA interaction’s dependence on GAG lengths is investigated using gel mobility shift assays, and DBPA was also titrated with heparin dp6 and dp10 as well as DS dp6 and dp14. Although the sulfation patterns of the fragments used are heterogeneous, disaccharide analyses of the GAGs can provide some insights into sulfation density of the fragments. Disaccharide analysis showed heparin contains 70% trisulfated disaccharide units with the rest being mostly disulfated disaccharides. The DS fragments analyzed with SAX-HPLC showed more than 90 % of disaccharides from DS are monosulfated, confirming previously published analysis on DS from the same source (28). In agreement with the law of probability, 30 % of dp6 fragments contain at least one disulfated unit and 50 % of dp14 fragments contain at least one disulfated unit. These distributions show the DS used is sufficiently uniform that chondroitinase ABC cleavage sites are not significantly biased by sulfation patterns.
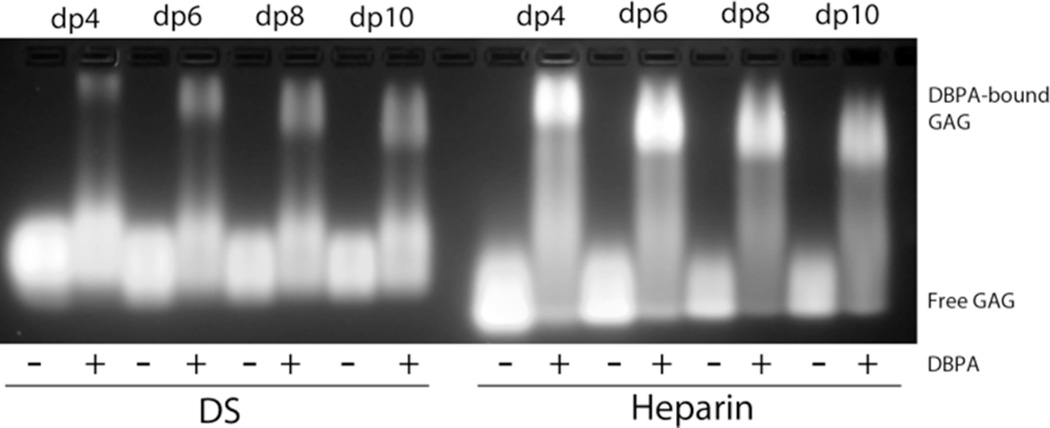
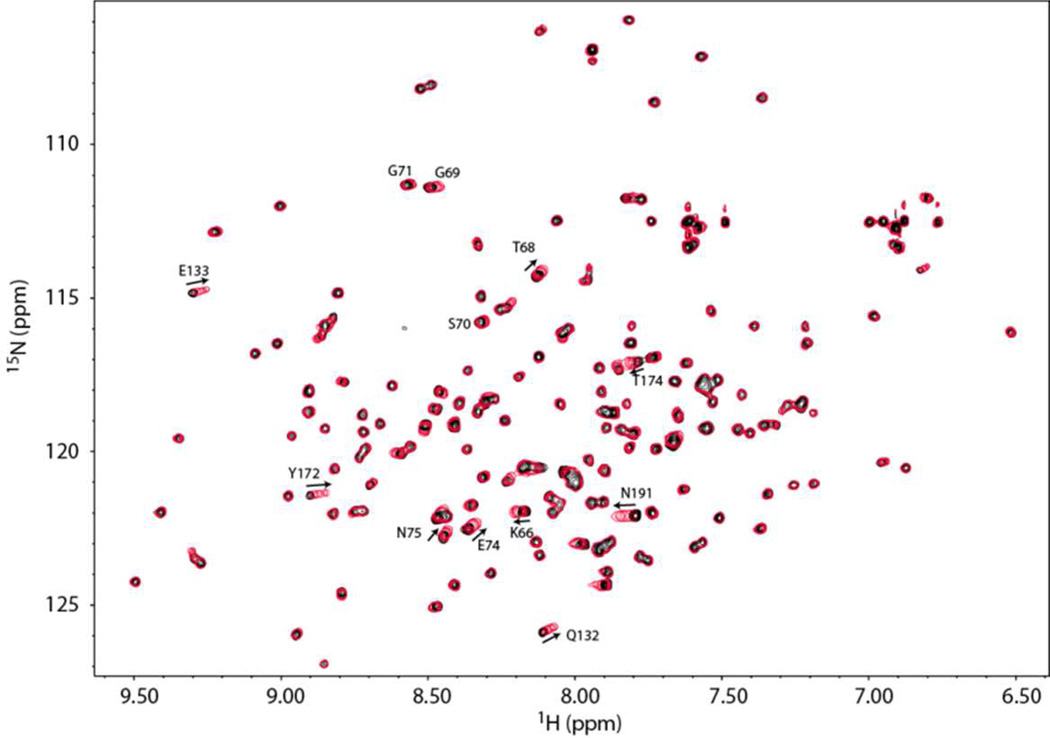
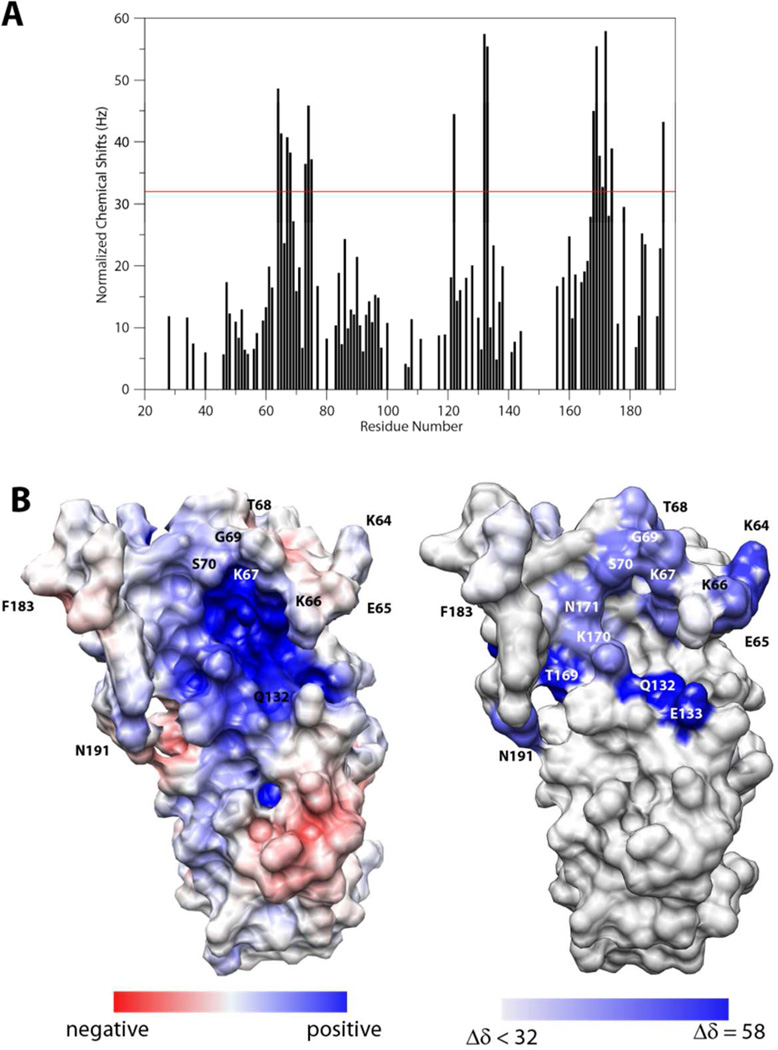
A qualitative characterization of DBPA-GAG interactions was carried out using gel mobility shift assay and fluorescently labeled GAG fragments according to Seo et al.(29) (figure 3) Migrations of all heparin fragments were affected significantly by the presence of DBPA. However, only a small percentage of DS fragments were shifted by DBPA, even at a length of dp10. These observations agreed with previous studies carried out using native GAG polymers, which showed heparin is a more potent inhibitor of bacterial adhesion than DS (15, 34). To quantitatively measure the interaction affinity and to map out the GAG binding epitope on DBPA, the protein was titrated with several GAG fragments of fixed length and monitored using NMR. Figure 4 is the overlay of 1H, 15N-HSQC spectra acquired at each titration point of DBPA-heparin dp10 titration. Interactions of DBPA with heparin dp10 fall in the fast exchange time scale and small, but significant chemical shift changes can be seen in several residues. Figure 5A shows residue-specific perturbations to 1H-15N chemical shifts by the heparin dp10 and figure 5B shows the same information mapped onto the surface of DBPA. The most perturbed residues include K64, D65, K67, T68, S73, E74, N75, V122, Q132, E133, H168 to K170, and N191. Many of the residues reside in the flexible region from residue G57 to E74. Most of the residues are located at or near the basic patch in the electrostatic potential surface map, identifying the basic patch as the most likely heparin binding epitope of DBPA. One especially notable change not illustrated by the chemical shift mapping was that residues G69 to G71 in the flexible linker showed significant increases in signal intensity. Signals corresponding to G71 and G69 increased 30 % while S70 increased 50 %. This is in contrast to signal intensity loss of 10 to 50 % experienced by other residues. The most plausible explanation for the observation is that heparin-binding in the vicinity of these residues has either prevented the labile amide protons from exchanging with solvents as exchanges between solvent protons and amide protons attenuate signal intensity, or the binding of heparin shifted the dynamic time scale of the flexible linker into a regime that no longer induce signal broadening. This occurrence shows the flexible linker may play a significant role in binding heparin. Examination of the flexible linker sequence showed it contains a BXBB motif (K64, D65, K66, K67), which is often associated with GAG-binding. The existence of such a motif therefore further enhances the role of the linker in GAG interactions. Titration of DBPA with heparin dp6 produced similar chemical shift migration changes (figure S5A) and using the chemical shift changes of N191 and Q132, which are among the most significant in the protein, the Kd of binding for dp10 was calculated to be in the 1–4 mM range (figure S5B). Surprisingly, the Kd of binding for the shorter dp6 fragment is comparable (figure S5B).
Figure 3.
Gel mobility shift assay of heparin and DS fragments in the presence and absence of DBPA. Mobility of fluorescently tagged GAG fragments is greatly retarded by their binding to DBPA.
Figure 4.
1H,15N-HSQC overlays of DBPA in the presence of different concentrations of heparin dp10. Signals experiencing large migration or large increase in signal intensity (G69, S70 & G71) are indicated with residue number and direction of migration. Solid black contour represent the starting HSQC of DBPA in the absence of heparin dp10. Each subsequent red single contour represents HSQC of DBPA at a different concentration of heparin dp10. The concentrations of heparin dp10 are 0.3, 0.6, 0.9, 1.2, 1.6 and 2.0 mM. The DBPA concentration is 0.1 mM.
Figure 5.
A) Normalized chemical shift perturbations to backbone amide nitrogen and proton by heparin dp10. The red line represents the value of perturbation 1.5 standard deviations higher than the average. B) Comparison of electrostatic potential map of DBPA (left) and heparin perturbation map of DBPA. The DBPA perturbation mapped is colored according to the chemical shift perturbation values shown in A. The most perturbed residues are shown in deep blue.
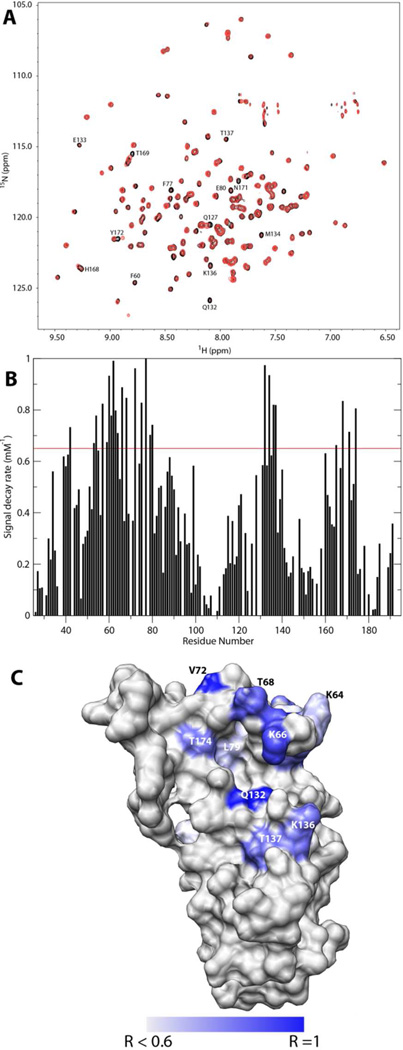
Limited amounts of DS dp6 and dp14 were also obtained by partial depolymerization of DS using chondroitinase ABC. DS dp6 induced very little chemical shift changes when added to DBPA (figure 6A) even at a protein-GAG ratio of 1:8, the increases in the peak intensity of residues G69 to G71 seen in the heparin titration were not observed either. Only residues Q127, T169 and Y172 demonstrated detectable chemical shift changes. However, large decreases in peak intensities were observed for a selective set of residues during the titration. Through a yet-not-well-understood mechanism, the decreases in peak intensities appear to form a single-exponential decay curve when plotted against DS dp6 concentrations (figure S6). This allowed the GAG-induced rate of signal decrease to be extracted for each residue in the protein by fitting the curve using the equation I([DS])=I(0)e−[DS]R, where [DS] is the concentration of DS dp6, I is the intensity of the signal and R is the rate constant governing the decay. Figure 6B shows the plot of R, rate of signal decrease, against residue number and figure 6C shows the same information visualized on the surface representation of DBPA. The profile is very similar to the plot of heparin-induced chemical shift perturbation. Residues that experienced large decreases in intensity include F60, A62, K66 V72, F77, Q132, M134, K136, T137, H168 and T174. Almost all of which can be located around the basic pocket, with a very high concentration found in the flexible linker and helix five. Of the three residues that experienced large GAG induced signal intensity boosts, G69 and G71 showed modest decline in signal intensity while S70’s signal intensity remained almost constant. The trend followed by these three residues is contrary to those surrounding them in the flexible linker, most of whom experienced large signal decreases. Owing to the lack of observed chemical shift changes in the titration, no dissociation constant of binding can be extracted. Titration of DBPA with DS dp14 also led to signal decreases similar to those seen in the DS dp6 titration. Not surprisingly, the rate and extent of signal intensity loss were slightly larger in the case of DS dp14 (figure S7).
Figure 6.
A) Superimpositions of HSQCs of 0.2 mM DBPA with no DS dp6 (black) and with 1.2 mM of DS dp6 (red). B) Plot of residue-specific signal decay rate. C) Locations of residues experiencing severe DS-induced signal decay. Residues having a decay rate of 0.6 mM−1 or higher are colored in increasingly deeper shades of blue. R stands for the decay rate. DBPA is in the same orientation as figure 2B.
Discussion
GAGs are a frequent target of pathogens during their infection processes. An ability to inhibit pathogen-GAG interaction is therefore a promising path for therapeutic intervention. However, the many important roles GAGs play in cellular processes force any intervening therapies to target microbe-GAG interactions specifically. Detailed structural characterizations of these pathogen-GAG interactions can therefore provide valuable information on the specificities of contacts. The Borrelia protein DBPA is well known for the crucial role it plays in the spread of Lyme disease and results outlined in this paper will shed light on possible exploitation of its interactions with GAGs as a path to combat Lyme disease.
Because DBPA was predicted to have four helices and an attempt at high resolution structure prediction using backbone chemical shifts and the program CS-ROSETTA (35) also showed the four helical-bundle as the most likely conformation, it is plausible that DBPA evolved from ancestral four-helical bundle proteins. Its present unconventional five-helix configuration maybe a consequence of its function as a GAG-binding protein since this allows the crucial GAG-binding regions to form a unified binding epitope. Titration of DBPA with heparin and DS also corroborate the five-helical arrangement because this arrangement places the most perturbed residues (C-terminus and the flexible linker) in the same area around the basic pocket, whereas the four-helical bundle conformation puts them on different ends of the protein. Sequences of DBPA differ greatly among different strains of Borrelia. The structure presented in this study represents DBPA from the strain B31. However, most biochemical data are obtained on strain N40 of Borrelia burgdorferi. Homology models of the N40 structure constructed with the B31 structure showed that N40 DBPA preserved most of the basic residues in B31, but the BXBB motif in the flexible linker is missing. Since the results from this study indicate the linker is important in DBPA-GAG interactions, this may offer a rational explanation to the observation that N40 DBPA has a lower affinity for DS and decorin than B31 DBPA (14).
Despite the presence of a multitude of basic amino acids throughout the protein, a well-defined, highly basic pocket does exist on DBPA. The most crucial GAG-binding residues identified by mutagenesis studies on N40 DBPA are fully preserved in the B31 version, and all three are found within the basic patch. Residues K163 and K170, together with residue R166, form a basic stretch on one face of helix five, reminiscent of motifs commonly found in other GAG-binding helices, and the positions of the basic amino acids in the sequence (BXXBXXTB) is similar but not identical to consensus GAG-binding sequences identified previously (36). Other residues forming the basic pocket include K66, K67 and K82. K136, a residue in the short linker between helices three & four is located at the periphery of the pocket and may also contributes to the basic electropotential in the region.
Ligand-induced atom chemical shift changes are often a good way to confirm the identity of the binding epitope. The heparin titration of DBPA showed the most perturbed residues are either in the C-terminus or the linker between helices one and two, both are close to the basic pocket and implicated in GAG-binding (14, 17). Perturbations to many residues in the linker demonstrate that linker plays an important role in binding GAGs. Furthermore, the increases in NMR signal intensities of linker residues G69 to G71 in the presence of heparin provides additional evidence for the interactions. Despite the changes heparin brought to residues in the linker, these residues were not identified as GAG binding in the peptide-based study of strain N40 DBPA’s interaction with GAGs (17). Because the N40 version of DBPA lacks the BXBB motif, this indicates linker residues’ interactions with GAGs is dependent on the BXBB motif. The Kd of DBPA’s interaction with heparin dp10 is in the low mM range. This is in complete agreement with previous studies that showed heparin dp10 and dp12 are unable to displace Borrelia burgdorferi strain N40 from Vero cells (15). However, assuming the free energies of binding are additive when two ligands are covalently linked, the current Kd for heparin dp10 implies a heparin fragments of approximately 20 monosaccharides will be sufficient to produce a low µM binding dissociation constant. This is also in agreement with the studies done by Leong et al. (15)
The gel mobility shift assay showed mostly monosulfated DS has a lower DBPA affinity than heparin. This offers an explanation to DS’s failure to induce chemical shift changes in the titrations. However, DS fragments do interact with DBPA since their presence produced severe attenuation of signal intensities. Remarkably, residues experiencing the most signal decay are also located around the basic pocket, similar to the chemical shift perturbation patterns seen in DBPA-heparin titration. Since it has been well established that protein-ligand interactions can produce apparent increases in transverse relaxation rates of those residues perturbed by ligands, these observations provide some confirmation that these parts of DBPA maybe involved in binding DS. Intriguingly, most signals appear to decay exponentially with respect to DS concentration. This cannot be easily explained by the two state exchange theory, which stipulates that exchanges between free and bound state of the protein lead to an apparent relaxation rate increase of (p1*p2)*Δω2/kex, where p1 and p2 represent the free and bound population fractions, Δω represents the change in chemical shift between free and bound states and kex is the exchange rate constant. However, it is worth noting that when the population of ligand-bound protein is low (< 10 %), the increase in transverse relaxation rate is proximately linear with respect to ligand concentration in a two state system, which in turn leads to the exponential decay of the signal with respect to GAG concentrations. If true, this argument will lead to the conclusion that short fragments of DS have very weak affinity for DBPA. This by no means imply DS is not the target for DBPA. Similar to heparin, affinity of DS fragments for a GAG-binding protein are related to their length. DS chains in vivo are often very long, with those on decorin containing ~ 150 monosaccharides, which would have much higher affinity for DBPA than fragments used in this study. It also should be noted that DS purchased for this study were extracted from porcine intestinal mucosa. These have been shown to consist almost entirely of monosulfated units (28) whereas decorin GAGs typically contain regions with higher sulfation density (37, 38), thus further boosting their affinity for DBPA. The fact that the decorin core protein is also known to play a role in its interaction with DBPA means the combined effect of DBPA-DS and DBPA-core protein interactions should make decorin DBPA’s dominant binding partner in vivo.
Results from this study constitute a first attempt to define the GAG-binding mechanism of DBPA. Effort is now underway to isolate longer fragments of heparin and DS, which may allow more well defined structural details of the interaction to be elucidated through either traditional techniques or newer integrated approaches that have been utilized effectively in other studies (39).
Supplementary Material
ACKNOWLEDGMENT
I want to thank Dr. James H. Prestegard & Dr. John Glushka of Complex Carbohydrate Research Center for the use of their 800 MHz spectrometer and Judit Losonczi & Evgeny Tishchenko of Agilent Corporation for the use of their 800 MHz spectrometer. This research is supported by grants from the National Institute of General Medical Sciences K99/R00 program (5R00GM088483). Grants from the National Center for Research Resources (5P41RR005351-23) and the National Institute of General Medical Sciences (8 P41 GM103390-23) from the National Institutes of Health provided support for the Resource for Integrated Glycotechnology.
ABBREVIATIONS
- DBP
Decorin binding protein
- DBPA
Decorin binding protein A
- DBPB
Decorin binding protein B
- GAG
Glycosaminoglycan
- DS
Dermatan sulfate
- CS
Chondroitin sulfate
- HSQC
Heteronuclear single quantum coherence
- RDC
Residual dipolar coupling
- SAX
Strong anion exchange
- SDS-PAGE
Sodium dodecylsulfate polyacrylamide gel electrophoresis
Footnotes
Accession Codes: Coordinates for the ensemble of ten DBPA structures has been deposited in PDB under the accession code 2lqu. Atom chemical shift assignment and RDC values have been deposited in BMRB under the accession code 18329.
SUPPORTING INFORMATION. Information on SDS-PAGE and SEC analysis of DBPA oligomerization, residue-specific 15N rotational correlation time of DBPA, HN RDC of aligned DBPA and 13C/15N-filtered, 13C-edited NOESY spectrum of DBPA, 15N-HSQCs of DBPA-heparin dp6 and DBPA-DS dp14 titrations, fittings of Kd for heparin-DBPA interactions and signal intensity decay rates seen in the DBPA-DS dp6 titrations is included in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Krupka M, Zachova K, Weigl E, Raska M. Prevention of Lyme Disease: Promising Research or Sisyphean Task? Archivum Immunologiae et Therapiae Experimentalis. 2011;59:261–275. doi: 10.1007/s00005-011-0128-z. [DOI] [PubMed] [Google Scholar]
- 2.Fallon BA, Levin ES, Schweitzer PJ, Hardesty D. Inflammation and central nervous system Lyme disease. Neurobiol. Dis. 2010;37:534–541. doi: 10.1016/j.nbd.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 3.Halperin JJ. Nervous system Lyme disease. J Neurol Sci. 1998;153:182–191. doi: 10.1016/s0022-510x(97)00290-6. [DOI] [PubMed] [Google Scholar]
- 4.Schuijt TJ, Hovius JW, van der Poll T, van Dam AP, Fikrig E. Lyme borreliosis vaccination: the facts, the challenge, the future. Trends Parasitol. 2011;27:40–47. doi: 10.1016/j.pt.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Guo BP, Brown EL, Dorward DW, Rosenberg LC, Hook M. Decorin-binding adhesins from Borrelia burgdorferi. Molecular microbiology. 1998;30:711–723. doi: 10.1046/j.1365-2958.1998.01103.x. [DOI] [PubMed] [Google Scholar]
- 6.Guo BP, Norris SJ, Rosenberg LC, Hook M. Adherence of Borrelia burgdorferi to the proteoglycan decorin. Infection and immunity. 1995;63:3467–3472. doi: 10.1128/iai.63.9.3467-3472.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown EL, Wooten RM, Johnson BJ, Iozzo RV, Smith A, Dolan MC, Guo BP, Weis JJ, Hook M. Resistance to Lyme disease in decorin-deficient mice. J Clin Invest. 2001;107:845–852. doi: 10.1172/JCI11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y, Xu Q, McShan K, Liang FT. Both decorin-binding proteins A and B are critical for the overall virulence of Borrelia burgdorferi. Infection and immunity. 2008;76:1239–1246. doi: 10.1128/IAI.00897-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi Y, Xu Q, Seemanaplli SV, McShan K, Liang FT. Common and unique contributions of decorin-binding proteins A and B to the overall virulence of Borrelia burgdorferi. PloS one. 2008;3:e3340. doi: 10.1371/journal.pone.0003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blevins JS, Hagman KE, Norgard MV. Assessment of decorin-binding protein A to the infectivity of Borrelia burgdorferi in the murine models of needle and tick infection. BMC Microbiol. 2008;8 doi: 10.1186/1471-2180-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weening EH, Parveen N, Trzeciakowski JP, Leong JM, Hoeoek M, Skare JT. Borrelia burgdorferi Lacking DbpBA Exhibits an Early Survival Defect during Experimental Infection. Infection and immunity. 2008;76:5694–5705. doi: 10.1128/IAI.00690-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer JR, Parveen N, Magoun L, Leong JM. Decorin-binding proteins A and B confer distinct mammalian cell type-specific attachment by Borrelia burgdorferi, the Lyme disease spirochete. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7307–7312. doi: 10.1073/pnas.1231043100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parveen N, Caimano M, Radolf JD, Leong JM. Adaptation of the Lyme disease spirochaete to the mammalian host environment results in enhanced glycosaminoglycan and host cell binding. Molecular microbiology. 2003;47:1433–1444. doi: 10.1046/j.1365-2958.2003.03388.x. [DOI] [PubMed] [Google Scholar]
- 14.Benoit VM, Fischer JR, Lin YP, Parveen N, Leong JM. Allelic variation of the Lyme disease spirochete adhesin DbpA influences spirochetal binding to decorin, dermatan sulfate, and mammalian cells. Infection and immunity. 2011;79:3501–3509. doi: 10.1128/IAI.00163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leong JM, Robbins D, Rosenfeld L, Lahiri B, Parveen N. Structural requirements for glycosaminoglycan recognition by the Lyme disease spirochete, Borrelia burgdorferi. Infection and immunity. 1998;66:6045–6048. doi: 10.1128/iai.66.12.6045-6048.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leong JM, Wang H, Magoun L, Field JA, Morrissey PE, Robbins D, Tatro JB, Coburn J, Parveen N. Different classes of proteoglycans contribute to the attachment of Borrelia burgdorferi to cultured endothelial and brain cells. Infection and immunity. 1998;66:994–999. doi: 10.1128/iai.66.3.994-999.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pikas DS, Brown EL, Gurusiddappa S, Lee LY, Xu Y, Hook M. Decorin-binding sites in the adhesin DbpA from Borrelia burgdorferi: a synthetic peptide approach. The Journal of biological chemistry. 2003;278:30920–30926. doi: 10.1074/jbc.M303979200. [DOI] [PubMed] [Google Scholar]
- 18.Brown EL, Guo BP, O'Neal P, Hook M. Adherence of Borrelia burgdorferi. Identification of critical lysine residues in DbpA required for decorin binding. The Journal of biological chemistry. 1999;274:26272–26278. doi: 10.1074/jbc.274.37.26272. [DOI] [PubMed] [Google Scholar]
- 19.Catanzariti AM, Soboleva TA, Jans DA, Board PG, Baker RT. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Science. 2004;13:1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Prestegard JH. Measurement of one and two bond N-C couplings in large proteins by TROSY-based J-modulation experiments. J Magn Reson. 2009;200:109–118. doi: 10.1016/j.jmr.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu YZ, Prestegard JH. Direct measurement of dipole-dipole/CSA cross-correlated relaxation by a constant-time experiment. Journal of Magnetic Resonance. 2008;193:23–31. doi: 10.1016/j.jmr.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. J Biomol Nmr. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 23.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 24.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol Nmr. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 25.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 26.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 27.Xiao Z, Zhao W, Yang B, Zhang Z, Guan H, Linhardt RJ. Heparinase 1 selectivity for the 3,6-di-O-sulfo-2-deoxy-2-sulfamido-alpha-D-glucopyranose (1,4) 2-O-sulfo-alpha-L-idopyranosyluronic acid (GlcNS3S6S-IdoA2S) linkages. Glycobiology. 2011;21:13–22. doi: 10.1093/glycob/cwq123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang HO, Gunay NS, Toida T, Kuberan B, Yu G, Kim YS, Linhardt RJ. Preparation and structural determination of dermatan sulfate-derived oligosaccharides. Glycobiology. 2000;10:1033–1039. doi: 10.1093/glycob/10.10.1033. [DOI] [PubMed] [Google Scholar]
- 29.Seo ES, Blaum BS, Vargues T, De Cecco M, Deakin JA, Lyon M, Barran PE, Campopiano DJ, Uhrin D. Interaction of human beta-defensin 2 (HBD2) with glycosaminoglycans. Biochemistry. 2010;49:10486–10495. doi: 10.1021/bi1011749. [DOI] [PubMed] [Google Scholar]
- 30.Lyon M, Deakin JA, Lietha D, Gherardi E, Gallagher JT. The Interactions of Hepatocyte Growth Factor/Scatter Factor and Its NK1 and NK2 Variants with Glycosaminoglycans Using a Modified Gel Mobility Shift Assay. Journal of Biological Chemistry. 2004;279:43560–43567. doi: 10.1074/jbc.M408510200. [DOI] [PubMed] [Google Scholar]
- 31.Farmer BT, 2nd, Constantine KL, Goldfarb V, Friedrichs MS, Wittekind M, Yanchunas J, Jr, Robertson JG, Mueller L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat Struct Biol. 1996;3:995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
- 32.Nabuurs SB, Spronk CAEM, Vuister GW, Vriend G. Traditional biomolecular structure determination by NMR spectroscopy allows for major errors. Plos Comput Biol. 2006;2:71–79. doi: 10.1371/journal.pcbi.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parveen N, Robbins D, Leong JM. Strain variation in glycosaminoglycan recognition influences cell-type-specific binding by lyme disease spirochetes. Infect Immun. 1999;67:1743–1749. doi: 10.1128/iai.67.4.1743-1749.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci U S A. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hileman RE, Fromm JR, Weiler JM, Linhardt RJ. Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. Bioessays. 1998;20:156–167. doi: 10.1002/(SICI)1521-1878(199802)20:2<156::AID-BIES8>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 37.Laremore TN, Ly M, Zhang Z, Solakyildirim K, McCallum SA, Owens RT, Linhardt RJ. Domain structure elucidation of human decorin glycosaminoglycans. Biochem J. 2010;431:199–205. doi: 10.1042/BJ20100788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zamfir AD, Flangea C, Sisu E, Serb AF, Dinca N, Bruckner P, Seidler DG. Analysis of novel over- and under-sulfated glycosaminoglycan sequences by enzyme cleavage and multiple stage MS. Proteomics. 2009;9:3435–3444. doi: 10.1002/pmic.200800440. [DOI] [PubMed] [Google Scholar]
- 39.Blaum BS, Deakin JA, Johansson CM, Herbert AP, Barlow PN, Lyon M, Uhrin D. Lysine and arginine side chains in glycosaminoglycan-protein complexes investigated by NMR, cross-linking, and mass spectrometry: a case study of the factor H-heparin interaction. J Am Chem Soc. 2010;132:6374–6381. doi: 10.1021/ja1000517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.