Abstract
Summary: Many important data in current biological science comprise hundreds, thousands or more individual results. These massive data require computational tools to navigate results and effectively interact with the content. Mobile device apps are an increasingly important tool in the everyday lives of scientists and non-scientists alike. These software present individuals with compact and efficient tools to interact with complex data at meetings or other locations remote from their main computing environment. We believe that apps will be important tools for biologists, geneticists and physicians to review content while participating in biomedical research or practicing medicine. We have developed a prototype app for displaying gene expression data using the iOS platform. To present the software engineering requirements, we review the model-view-controller schema for Apple's iOS. We apply this schema to a simple app for querying locally developed microarray gene expression data. The challenge of this application is to balance between storing content locally within the app versus obtaining it dynamically via a network connection.
Availability: The Hematopoietic Expression Viewer is available at http://www.shawlab.org/he_viewer. The source code for this project and any future information on how to obtain the app can be accessed at http://www.shawlab.org/he_viewer.
Contact: cashaw@bcm.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
Microarray and more recently developed gene expression technologies allow biologists to simultaneously examine all of the changes that occur in the transcriptome under different conditions. The resulting data are massive and difficult to interact with using spreadsheets and traditional manual curation. This wealth of data has required new methods for examining content and considering the differences in expression of individual genes across experimental conditions. One method of visualizing microarray results is using the ‘heat map’ display, such as those made available in GeneXplorer (Perou et al., 2000). Tools like these are instrumental, because they allow investigators to visualize the ensemble of expression data and to identify patterns that can be used to explain biological processes. Unfortunately, these methods do not provide drill-down visualization of individual gene and probe measurements. In cases where such analysis is required, either complex commercial end-user software or massive spreadsheets are the only currently viable methods of interacting with data.
Our team has previously generated a large database of gene expression patterns across 10 cell types in the hematopoietic system (Chambers et al., 2007). Hematopoietic stem cells are the adult stem cells that continually regenerate all constituents of the blood and represent the best studied adult stem cells. However, the genes and processes regulating hematopoiesis have not been completely defined.
Based on feedback from our experimental collaborators, we wished to provide a means of querying the data in a variety of non-traditional situations, such as the auditorium seating of a seminar, unplanned discussion of gene expression patterns with colleagues or other ‘on the go’ situations remote from the main computing environment. We developed a novel application, the Hematopoietic Expression Viewer, for use on the Apple iPhone. Although common in the general community, biomedical technical ‘apps’ are relatively rare. A literature search reveals two published examples, the TimeTree2 app (Kumar and Hedges, 2011) and the Immunological Genome Project app (Heng, 2008), which includes microarray results but is centered on the immunological mouse community.
The iOS (Apple's mobile operating system) Hematopoietic Expression Viewer allows ready access to the hematopoietic expression data, enhancing our ability to analyze and to identify genes that may be important in hematopoiesis, stem cell biology or blood malignancies among other uses.
The iOS ‘model-view-controller’ (MVC) software architecture was used to construct this app using the Apple development environment (Fig. 1). MVC is designed to increase modularity by separating tasks into separate constructs that interact in a highly stereotyped manner. The model helps ensure that modifications of one section will not break functionality in another area. MVC distributes responsibilities into the three distinct but related components for which it is named. The model holds the application state, the view handles presentation of the application state to the user, and the controller mediates between the two and alters the application's state in response to input that flows through the view. Communication between these elements requires a separate paradigm, the ‘event-listener’ (Fig. 1). An event is a predefined condition often resulting from user action, such as ‘searchBarSearchButtonClicked’, that is broadcast to any object subscribed as a listener. The controller's duties are primarily comprised of listening for events and taking appropriate actions in response. Most of these events are generated by the view, but they may also be produced by outside entities, such as the operating system, as in the case of an ‘applicationDidFinishLaunching’ event.
Fig. 1.
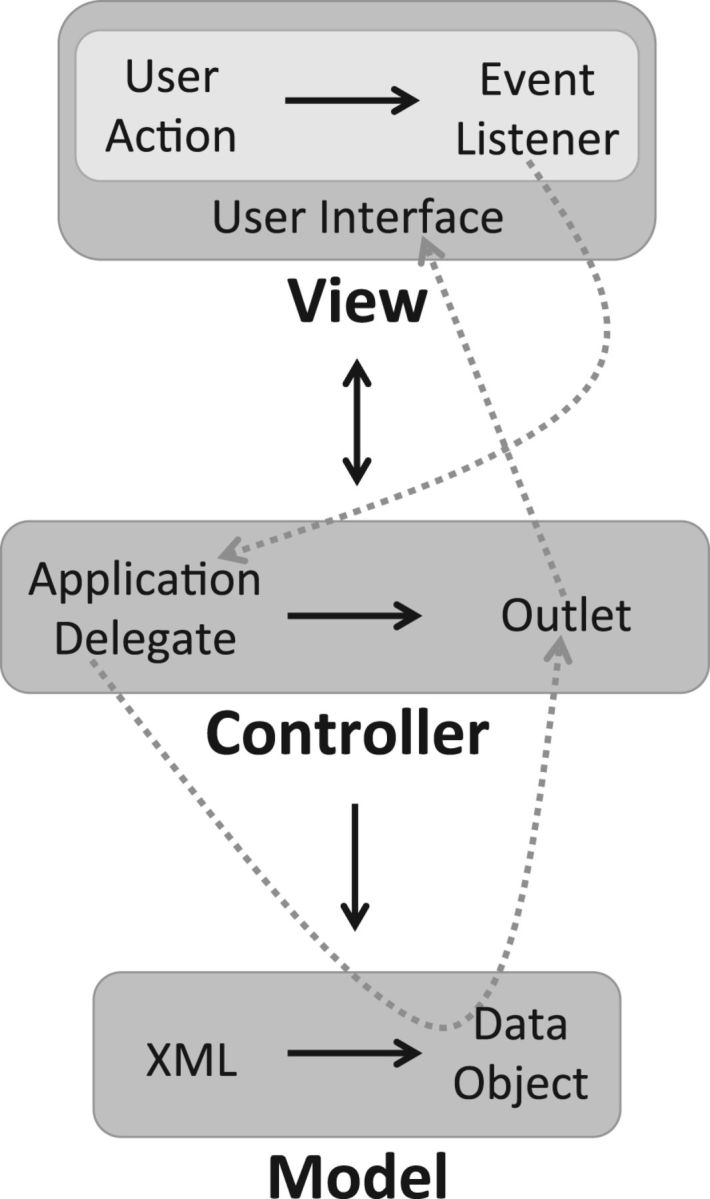
Model-view-controller
A challenge we encountered at several stages of the development process was the resource limitation imposed by the hardware. In particular, load times were a crucial challenge. The load time for our first prototype, for example, lasted nearly an entire minute. We responded to this challenge by changing the storage architecture for our data from a local SQLite database to a much more specific XML-based data structure called a ‘property list dictionary’. The modularity of the MVC architecture allowed us to accomplish this with relatively few changes to the rest of the code and decreased the load time by 75%.
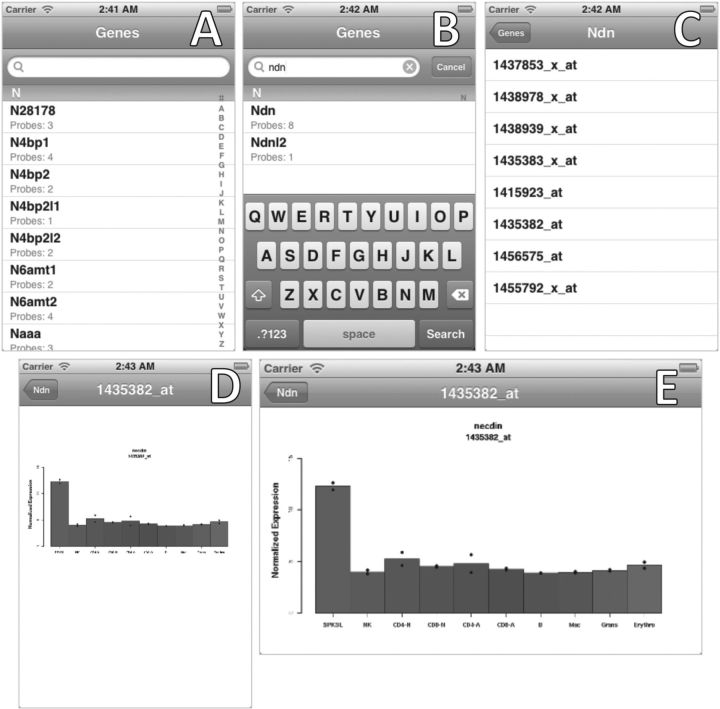
To use the Hematopoietic Expression Viewer, a user opens the app and is presented with a list of genes, each with an indicator of the quantity of probes used to measure corresponding expression levels. The user then either scrolls to the gene of interest, by sliding a finger along the gene list directly or along the alphabetized section list on the right of the screen, or searches for the any part of the gene name using the fuzzy search box at the top of the screen. Once the gene has been located, the user taps the gene and is presented with another screen. This screen displays a list of the probes that target that gene. The user then taps the probe of interest and is taken to a screen that displays a bar chart of the probed gene expression in a variety of key hematopoietic cell types. The display can be rotated to enable an enlarged view of the chart. The user can navigate to previous screens to review alternate data by tapping customized ‘back’ buttons at the top left corners of screens (Fig. 2).
Fig. 2.
Hematopoietic Expression Viewer app querying example gene Ndn. (A) Section N clicked. (B) Gene Ndn searched. (C) Ndn clicked. (D) Ndn probe clicked. (E) Expression graph roated and enlarged
The ‘Post-PC’ era requires biologists to consider new methods of sharing and distributing data. This challenge is made more difficult by the massive data that are now the standard experimental paradigm for current investigations. This challenge is particularly salient when drill-down probe-level information must remain in some context, such as the change in expression levels over time or for specific cell types. We developed a prototype iOS application to share gene expression data with our colleagues and the broader community. This style of app is likely to be repeated by ourselves and others.
ACKNOWLEDGEMENTS
We thank all Shaw laboratory members for helpful comments. The splash page for the app was devised by Stuart Chambers and the Goodell laboratory at BCM.
Funding: R.A.J. was supported by R25GM56929 and NEIT32EY07102. C.A.S. and E.S.C. were supported by RO1HL102482. M.A.G. was supported by DK092883.
Conflict of Interest: none declared.
REFERENCES
- Chambers S.M., et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng T.S. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- Kumar S., Hedges S.B. TimeTree2: species divergence times on the iPhone. Bioinformatics. 2011;27:2023–2024. doi: 10.1093/bioinformatics/btr315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou C.M., et al. Molecular portraits of human breast tumors. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]