Abstract
Background
Pelargonium is one of the most popular garden plants in the world. Moreover, it has a considerable economic importance in the ornamental plant market. Conventional cross-breeding strategies have generated a range of cultivars with excellent traits. However, gene transfer via Agrobacterium tumefaciens could be a helpful tool to further improve Pelargonium by enabling the introduction of new genes/traits. We report a simple and reliable protocol for the genetic transformation of Pelargonium spp. and the production of engineered long-life and male sterile Pelargonium zonale plants, using the pSAG12::ipt and PsEND1::barnase chimaeric genes respectively.
Results
The pSAG12::ipt transgenic plants showed delayed leaf senescence, increased branching and reduced internodal length, as compared to control plants. Leaves and flowers of the pSAG12::ipt plants were reduced in size and displayed a more intense coloration. In the transgenic lines carrying the PsEND1::barnase construct no pollen grains were observed in the modified anther structures, which developed instead of normal anthers. The locules of sterile anthers collapsed 3–4 days prior to floral anthesis and, in most cases, the undeveloped anther tissues underwent necrosis.
Conclusion
The chimaeric construct pSAG12::ipt can be useful in Pelargonium spp. to delay the senescence process and to modify plant architecture. In addition, the use of engineered male sterile plants would be especially useful to produce environmentally friendly transgenic plants carrying new traits by preventing gene flow between the genetically modified ornamentals and related plant species. These characteristics could be of interest, from a commercial point of view, both for pelargonium producers and consumers.
Keywords: Pelargonium zonale, Pelargonium peltatum, pSAG12 promoter, ipt gene, Engineered long-lived plants, Delayed senescence, Engineered male sterility, PsEND1 promoter, Barnase, Anther ablation, Biosafe ornamentals
Background
The genus Pelargonium (family Geraniaceae), with more than 200 species, represents one of the most popular garden plants around the world, having a considerable economic importance in the market of ornamental plants. The vegetative-propagated bedding plants from the two groups Pelargonium zonale (syn. Pelargonium x hortorum) and Pelargonium peltatum hybrids are the most cultivated potted plants. In addition, species of Pelargonium are important in the perfume industry and are cultivated and distilled for their scent.
Pelargonium spp. grow as annuals in temperate climates. Leaves are usually alternate and palmately lobed or pinnate, often on long stalks and sometimes with light or dark patterns. The erect stems bear five-petaled zygomorphic flowers in umbel-like clusters (pseudo umbels). These plants have been bred to produce a variety of flower shapes, ranging from star-shaped to funnel-shaped, and colors, such as white, pink, red, orange-red, fuchsia or deep purple. Pelargonium breeding programmers have yielded new flower colors and shapes, early and continuous flowering, good postharvest and market quality and pest and disease resistance.
Gene transfer by means of Agrobacterium tumefaciens enables the introduction of new genes from unrelated species and would be a helpful tool for further progress in pelargonium breeding. The first step for applying genetic transformation via Agrobacterium is to develop an efficient in vitro regeneration system for the target plant. It is also beneficial to develop methods to propagate explants from adult plants of genotypes tested for agronomic performance.
The vegetative-propagated pelargoniums are highly heterozygous resulting in variable offspring [1]. For P. zonale, as well as P. peltatum hybrids, several reports have been published dealing with in vitro regeneration from explants taken from adult plants, using either organogenesis [2-7] or somatic embryogenesis [5,7-11]. Agrobacterium-mediated transformation of different pelargonium genotypes was reported by several authors using different marker and reporter genes [1,4,9,12-14], however no reports on the use of the green fluorescent protein (gfp) gene as an in vivo selectable marker have been published. Gfp expression in transformed cells should be useful to identify transformation events at early stages, such that selectable marker genes (antibiotic or herbicide resistance) may not be required.
Cytokinins have been implicated in several aspects of plant development, including plant senescence [15-20], and are thought to be synthesized mainly in the roots and transported to the shoots via the xylem. Concentrations of endogenous cytokinins decline in plant tissues as senescence progresses. The cytokinin content of the xylem sap of sunflower (Helianthus annuus) and soybean (Glycine max) also decreases rapidly with the onset of senescence, which suggests that reduction in cytokinin transport from roots to shoots allows senescence to progress [21,22]. Moreover, exogenous application of cytokinins has been exploited commercially to extend the shelf life of freshly harvested vegetables and cut flowers [23].
Transgene-encoded cytokinin biosynthesis was initially studied in Nicotiana tabacum using constitutive or inducible overexpression of the isopentenyl phosphotransferase (ipt) gene of Agrobacterium tumefaciens. This enzyme catalyzes the rate-limiting step for de novo cytokinin biosynthesis in plants [24]. Isopentenyl AMP is the precursor of all other cytokinins, of which the three most commonly detected and physiologically active forms are isopentenyl adenine (IPA), zeatin (Z), and dihydrozeatin (DHZ) [25]. Overexpression of the ipt gene in transgenic plants led to elevated foliar cytokinin concentrations and delayed leaf senescence, but high cytokinin levels have been reported to be detrimental to growth and fertility [26-30]. To circumvent these effects, Gan and Amasino [31] devised a strategy, based on auto-regulated cytokinin production, which delayed leaf senescence in transgenic tobacco without altering other plant phenotypes. This strategy exploited a senescence-specific gene promoter (pSAG12) from an Arabidopsis thaliana gene [32], fused to the ipt gene (tmr gene from the Ti plasmid of A. tumefaciens) [33]. The pSAG12::ipt chimaeric gene was reported to be activated only at the onset of senescence in the lower mature leaves of tobacco. This approach resulted in cytokinin biosynthesis restricted to the leaves, which inhibited leaf senescence, preventing cytokinin overproduction. The ability to delay leaf senescence has potential for crop improvement. The effect of pSAG12::ipt expression in transgenic plants has been assessed in several Solanaceous crops [17], and in rice (Oryza sativa) [34], cauliflower (Brassica oleracea) [35] and lettuce (Lactuca sativa L. cv. Evola) [36,37]. In potted ornamental plants, like pelargoniums, a delay in the senescence process would be of interest both for consumers and producers.
Engineered male sterility in ornamental plants has many applications such as hybrid seed production, elimination of pollen allergens, reduction of the need for deadheading to extend the flowering period, redirection of resources from seeds to vegetative growth and increase of flower longevity. The use of this technology could be especially useful to produce environmentally friendly transgenic ornamentals carrying new traits, as this modification would prevent gene flow between the genetically modified plants and related species [38-40]. Expression of the barnase gene under control of the anther-specific PsEND1 promoter [38,39,41,42] may be used to efficiently create male sterile versions of existing pelargonium cultivars without adversely affecting the respective phenotypes. The PsEND1 promoter shows specific expression in those tissues involved in anther architecture of many plant species. PsEND1 is a pea anther-specific gene that displays very early expression in the anther primordium cells. Later on, PsEND1 expression becomes restricted to the epidermis, connective, endothecium and middle layer, but it is never observed in the anther filament, tapetal cells or microsporocytes. The expression pattern of this gene continues until floral anthesis. In addition, the expression of barstar, an inhibitor of the ribonuclease barnase, has been used to restore fertility to plants with barnase-induced sterility [43,44] and to prevent the possible effects of ectopic barnase expression in engineered male and female sterile plants [40].
The main objectives of this work were to develop a simple and reliable Agrobacterium-mediated protocol for the genetic transformation of Pelargonium zonale and P. peltatum using the gfp gene as an in vivo marker, to test the effects of pSAG12::ipt expression on leaf senescence and plant morphology, and to engineer male sterility in this ornamental crop by ablating tissues essential for the anther development and subsequently for pollen growth using the PsEND1::barnase chimaeric gene. The potential commercial applications (increased shelf life, reduced plant architecture) of transgene-encoded auto-regulated cytokinin biosynthesis in this ornamental plant and the production of male sterile lines to prevent undesirable gene flow between the genetically modified plants and related species are also discussed.
Methods
Plant material and tissue culture
Pelargonium peltatum cv. Aranjuez (Figure 1a-b) and Pelargonium zonale (syn. Pelargonium x hortorum) cv. 370 (Figure 1c-d) in vitro propagated plants were used as source of explants. Young leaf explants (first to the fifth leaf from the apex) were harvested from 30–40 days old axenically grown plantlets propagated from shoot segments. Axenic plants were established by surface sterilization of 2 cm shoot segments processing an axillary bud excised from greenhouse grown plants. These nodal cuttings were first washed thoroughly with water, and then surface-sterilized by immersion in a 2.5% solution of sodium hypochlorite with 0.1% of 7X-O-matic detergent (Flow Laboratories) for 20 min and rinsed three times with sterile distilled water. Once sterilized, the nodal cuttings were cultured on Rooting Medium (RM) composed of Murashige and Skoog (MS) basal medium [45], 20 mg l-1 sucrose, 1 mg l-1 thiamine-HCl, 100 mg l-1 myo-inositol, 8 g l-1 agar and 0.1 mg l-1 IAA. Axenic plants obtained from shoots segments were propagated every two months in 580 ml culture vessels with 60 ml of RM and maintained as in vitro stock plants. All cultures were incubated at 25°C under a 16 h photoperiod with fluorescent light (60 μmol m-2 s-1 intensity).
Figure 1.
In vitro regeneration of transgenic plants via somatic embryogenesis in Pelargonium peltatum and via organogenesis in P. zonale. (a) P. peltatum WT plants. (b) P. peltatum inflorescence. (c) P. zonale WT plants. (d) P. zonale inflorescence. (e-f). Production of somatic embryos in a callus of P. peltatum cultivated in selective Morphogenesis Induction Medium (MIM). (g). P. peltatum developing embryos in selective Elongation Medium (EM). (h). P. peltatum transgenic plantlets in Rooting Medium (RM). (i-j). Adventitious buds in a callus of P. zonale in selective MIM. (k). P. zonale shoot elongation in selective EM. (l). P. zonale rooted plantlet in RM. (m-n). Detection of transformation events in both Pelargonium spp., GFP green fluorescence is clearly visible in the initial whitish callus with a disorganized growth. (o-p). Chlorophyll shows strong red autofluorescence that could mask the green fluorescence of transformed cells, it becomes increasingly difficult to identify in the subsequent organogenic callus and in the adventitious buds. (q-r). Green fluorescence was observed in regenerated shoots but is masked by the chlorophyll in the young leaves. (s-t). Green fluorescence can be observed in the periphery of young leaves where chlorophyll does not accumulate. (u-v). Green fluorescence is especially evident in the roots, where the chlorophyll is absent. In general, within the same organ, GFP detection varied in different tissues or cell types depending on their chlorophyll content.
In vitro plant regeneration was carried out using a Morphogenesis Induction Medium (MIM) composed of MS basal medium and Shahin [46] vitamins supplemented with 30 g l-1 sucrose, 100 mg l-1 myo-inositol, 8 g l-1 agar, IAA (0.01 mg l-1), TDZ (0,5 mg l-1) and 1 mg l-1 of Cu-sulphate. The pH was adjusted to 5.7 before autoclaving. The petiole proximal area of axenic leaves was cut into 1 cm2 pieces and cultured on MIM with the abaxial surface in contact with the medium. Regeneration in Pelargonium zonale was carried out via direct organogenesis and in Pelargonium peltatum via somatic embryogenesis.
A. tumefaciens strain and chimaeric gene constructs
Agrobacterium strain LBA4404 was used in all transformation experiments. LBA4404 cells were electroporated to carry different plasmids: i) a pBIN19 binary vector harboring, from the right to the left border, the nptII marker gene under the control of the nos promoter and the nos terminator, and the gfp-S65T[47] reporter gene under the control of a 2X 35SCaMV constitutive promoter and the nos terminator; ii) a plasmid derived from pBI101 harboring, from the right to the left border, the nptII marker gene under the control of the nos promoter and the nos terminator, and the barnase gene under the control of the PsEND1 promoter and the nos terminator (pBI101-PsEND1::barnase construct) [38], and iii) a plasmid (pVDH393) carrying the ipt gene under the control of the senescence inducible pSAG12 promoter and with the nos terminator, the reporter gene uidA (GUS-intron) under the control of the 35SCaMV promoter and with the 35SCaMV terminator and the nptII marker gene under the control of the 35SCaMV promoter and with the 35SCaMV terminator (pVDH393-pSAG12::ipt construct; supplied by company Van der Have, ND). Bacteria were grown at 28°C on solid LB plates supplemented with 40 mg l-1 rifampicin and 100 mg l-1 kanamycin. A single colony was used to inoculate 25 ml of LB liquid medium with the same antibiotics. Flasks were maintained at 28°C and 200 rpm for 24 h and later on were used to inoculate a liquid MS medium supplemented with 20 g l-1 sucrose, 100 mg l-1 myo-inositol, 1 mg l-1 thiamine-HCl, 100 mg l-1 2-(N-morpholino)ethane sulfonic acid (MES) and 0.2 mM acetosyringone dissolved in 70% ethanol (sterilized by filtration), which was cultured at 28°C for 12 h. Inoculation of explants was conducted when the bacterial culture reached an OD (600 nm) of 0.06.
The promoter region of the PsEND1 gene (GenBank accession n.: AY324651) was previously cloned into the binary vector pBI101 where the −2736/-6 promoter fragment was fused to the coding sequence of the β-glucuronidase (uidA) reporter gene [41] PsEND1::barnase-barstar chimaeric gene: primers Ribo1 (5’-TAGGATCCCGACCATGGCACAGGTTATC-3’) and Inhi2 (5´-GCGAGCTCTTAAGAAAGTTGATGGTGATG-3´) were designed based on the published sequence of barnase and barstar genes [48] to amplify the barnase-barstar fragment and to introduce BamHI and SacI restriction sites. Barnase is a very active ribonuclease. Even a low level of expression from aberrant promoter sequences or run-off expression from neighboring genes during manipulations in E. coli or Agrobacterium would have prevented the survival of the bacteria. Therefore, the barstar gene which encodes an inhibitor of barnase is included in the construct. The PCR resulting fragment was cloned into the pGEM-T Easy (Promega) and later released with the BamHI and SacI enzymes. The BamHI-SacI fragment was cloned by replacement of the uidA coding sequence into the binary vector pBI101 generating the pBI101-PsEND1::barnase-barstar construct. The nos::ntpII plant selectable marker gene, which confers resistance to kanamycin in transgenic plants, was also introduced in the T-DNA.
Transformation and regeneration of transgenic plants
Transformation experiments were carried out using both Pelargonium zonale and Pelargonium peltatum leaf explants as starting material. Leaf explants were prepared from two months old axenic plants as described above. Explants were inoculated in groups of 25 explants with 50 ml of the bacterial suspension for 5 min and thereafter transferred to co-culture medium consisting of MIM supplemented with 0.2 mM acetosyringone. Explants were co-cultured for 2–3 days in the dark at 25°C, afterwards they were transferred into sterile glass jars containing a liquid washing medium (MS basal medium, 20 mg l-1 sucrose, 1 mg l-1 thiamine-HCl, 100 mg l-1 myo-inositol, 100 mg l-1 MES and 600 mg l-1 cefotaxime) and soaked for 5 min. The explants were subsequently blotted dry with sterile filter paper and subculture onto selective MIM supplemented with 300 mg l-1 timentin and 50 mg l-1 kanamycin for Agrobacterium eradication and selection of transgenic events, respectively. All antibiotics were filter sterilized and added to cooled media (45°C) before pouring into 9 cm diameter Petri dishes as 25 ml medium per plate. Control explants were treated in the same manner, except for the inoculation with Agrobacterium. Control groups were established and cultured on medium with and without kanamycin. All explants were subculture every 2 weeks onto the same fresh medium until shoots were long enough to be separated from the callus. After 2.5-3 months in culture, calli showing well developed morphogenetic structures (shoots in the case of P. zonale and somatic embryos in P. peltatum) were transferred to a selective Elongation Medium (EM: MS basal medium and Shahin vitamins, supplemented with 30 g l-1 sucrose, 100 mg l-1 myo-inositol, 8 g l-1 agar, 0.01 mg l-1 NAA, 0.1 mg l-1 6 BA, 1 mg l-1 of Cu-sulphate, 300 mg l-1 timentin and 50 mg l-1 kanamycin). After 1–1.5 months in EM, the shoots were cut and cultivated in Rooting Medium (RM). Regenerated plantlets with well-developed roots were transferred to plastic pots containing peat moss and perlite (3:1) as substrate and acclimatized in growth chambers initially covered with a transparent plastic to maintain humidity. Plants were cultivated under long day conditions (16-h light/8-h dark photoperiod) and then transferred to a greenhouse until they flowered. Transformation efficiency was estimated as the number of independent transformation events (one transgenic plant per explant) in relation to the total number of inoculated explants.
Ploidy level analysis
Pelargonium zonale and P. peltatum have 2n = 18 chromosomes. Modern cultivars have been obtained by intra- and inter-specific hybridization and usually they show high ploidy levels when compared with wild species. The ploidy level was evaluated by flow cytometry as described previously [49]. Leaf tissue from in vitro plants was used for nuclei isolation. Pieces of tissue (1 cm2) were chopped individually on a glass plate with a sharp razor blade in 200 μl of nuclei isolation buffer (Partec). The sample was then passed through a 50 μm nylon filter and 800 μl of staining solution (Partec), containing 1 mg l-1 DAPI (4,6-diamino-2-phenyl-indole), were added for DNA fluorescence. The DNA content of the isolated nuclei was measured using a Partec PAS-II flow cytometer equipped with a mercury lamp. Fluorophore excitation peak is below 420 nm and fluorescence emission peak for DAPI is between 435 and 500 nm. The data were plotted on a histogram where the horizontal axis shows DNA content (proportional to fluorescence) and the vertical axis shows nuclei number. About 5000 to 10000 nuclei were measured per sample. Analyses were carried with young leaves from adult plants of the original cultivar and young leaves from transgenic plants.
PCR analysis
Plant DNA used for PCR analysis was extracted from young leaves using the protocol of Rogers and Bendich [50]. PCR analysis was carried out for all transgene using the following primer pairs: for the nptII gene, forward primer KAN-1: (5’-AAG ATG GAT TGC ACG CAG GTT C-3’) and reverse primer KAN-2: (5’-GAA GAA CTC GTC AAG AAG GCG A-3’); for the uidA gene, forward primer GUS-1: (5’-ATC AGG AAG TGA TGG AGC ATC A-3’) and reverse primer GUS-2: (5’GGT GAT CGG ACG CGT CGG GTC G-3’); for the gfp gene, forward primer GFP-D: (5’-ATG GTG AGC CAA GGG CGA GGA-3’) and reverse primer GFP-R: (5’-GGA CCA TGT GAT CGC GCT TC-3)’; for the barnase-barstar genes, forward primer Ribo3 (5´-ACG GAC CAT TAT CAG ACC TTT AC-3´) and reverse primer Inhi3 (5´-CGC AGC CTT CCG CTT TCG C-3´); for ipt gene, forward primer IPTDIR (5’-GGT CCA ACT TGC ACA GGA AAG-3’ and reverse primer IPTREV: (5’-CCC TCC AAA GTT GAA CCA ACT C-3’). The PCR reactions were carried out in a total volume of 20 μl comprising 0.1-0.2 μg genomic DNA, 0.2 mM of each dNTPs, 1.5 mM MgCl2, 0.6 μM 5’ and 3’ primers and 0.5 U Taq DNA polymerase. For nptII analysis, DNA was denatured at 94°C for 5 min followed by 30 cycles (94°C for 30 s, 56°C for 45 s, 72°C for 1 min) and finally 10 min at 72°C. For uidA analysis, DNA was denatured at 94°C for 5 min followed by 30 cycles of (94°C for 30 s, 60°C for 45 s, 72°C for 1 min) and finally 10 min at 72°C. For gfp analysis, DNA was denatured at 94°C for 5 min followed by 35 cycles of (94°C for 30 s, 60°C for 30s, 72°C for 1 min) and finally 10 min at 72°C. For barnase-barstar and ipt analyses, DNA was denatured at 94°C for 5 min followed by 35 cycles of (94°C for 30 s, 55°C for 30s, 72°C for 1 min) and finally 10 min at 72°C. The expected products sizes were 781 bp for nptII, 1021 bp for uidA, 661 bp for gpf, 544 bp for barnase-barstar and 460 bp for ipt gene. PCR products were detected by UV light after electrophoresis on 1% w/v agarose ethidium bromide gels.
Real-time RT-PCR analysis
Total RNA was isolated from detached leaves of the P. zonale transgenic lines 3.4, 3.9, 4.3, 4.12 and from leaves of WT control plants using Plant RNA Purification Reagent protocol (Invitrogen Corporation, Carlsbad, CA). DNase treatment of the RNA preparations for real-time reverse transcription-polymerase chain reaction (Real-Time RT-PCR) was performed using the Turbo DNA-Free Kit (Ambion, Austin, TX) according to the manufacturer’s specifications. RNA concentration for each sample was measured spectrophotometrically (NanoDropTechnologies, Inc., Wilmington, DE) and the quality was visually assessed by formaldehyde-agarose gel electrophoresis. For first-strand synthesis, total RNA (1 μg) was reverse-transcribed in a 20 μl reaction mixture using the PrimerScript 1st strand cDNA Synthesis Kit (TAKARA; http.//http://www.takara-bio.co.jp). Two microliters of RT reaction were used for a Real-Time RT-PCR analysis with 300 nM of each primer mixed with the Power SYBR ® Green PCR Master Mix (Applied Biosystems) as indicated manufacturer’s instructions. The reactions were carried out into 96 weel-optical reaction plates using ABI PRISM 7500 Sequence Detection System and appropriated software (Applied Biosystems). The data were normalized using Pelargonium x hortorum PhACTIN7[51]. Relative expression values were calculated after normalizing against the maximum expression value. Primers were designed for all genes using PRIMER EXPRESS version 2.0 (Applied Biosystems) with default parameters. Primers used were as follow: ipt (isopentenyl transferase): ipt-qRT-DIR: 5’-GCTACCCAGAACCAGATCACG-3’ and Ipt-qRT-REV: 5’-ATCTGCGTCGAGCTGCAATA-3’ and PhACTIN7: PhACTIN7-qRT-DIR: 5’-TCCAGCAGATGTGGATTTCAAA-3’ and PhACT7-qRT-REV: 5’-TTGATGGGCCAGACTCATCAT-3’.
Analysis of gene expression for each sample was performed on three experimental repeats with real-time RT-PCR for both ipt and PhACTIN7. The actual value of each pair (ipt- PhACTIN7) is equal to 2-ΔΔCt. Each sample’s expression level relative to PhACTIN7 is the mean of three biological repeats. The relative expression level of the sample with the highest expression level was set to one and all other samples values were normalized to that value to generate the fold.
Light microscopy
Transformed explants were examined periodically for gfp expression under a fluorescence stereomicroscope (Leica MZ FLIII) equipped with a Leica Fluorescence Module GFP3 comprising a 470–440 nm Excitation Filter and a 525–550 nm Barrier Filter. A mercury lamp provided the light source. The red autofluorescence from chlorophyll was not blocked with any filter. Floral buds and stamens from both transgenic and WT plants were freshly harvested and dissected using forceps and scalpel. Light photographs of dissected flowers and stamens were obtained using a stereomicroscope (MZ8, Leica).
β-glucuronidase activity was determined histochemically according to Jefferson et al. [52]. Root, shoot and leaf segments from the putative transgenic plants were stained for 24 h at 37°C, cleared with 70% ethanol and observed under a stereomicroscope (MZ8 Leica).
Morphological measurements
Morphological measurements of vegetative growth were made to determine whether the barnase and ipt expression could affect different plant growth parameters. Measurements were taken in the greenhouse on T1 hemizygous transgenic plants and WT control plants. Plant height in flowering plants (distance from soil line to top of the tallest growing point), leaf length and width (average measurements from five fully expanded leaves), leaf petiole length, internodal length and number of inflorescences per plant were evaluated. Morphological measurements were taken over the course of several days on each plant as its first five flowers reached anthesis. Means differing significantly were compared using the Student test at a 5% probability level. Data variability was expressed as the mean ± SE.
Quantification of chlorophyll
Analysis of leaf senescence was conducted by extraction of chlorophyll in detached leaves incubated in darkness from WT control and pSAG12::ipt plants respectively. Using a porcelain mortar cooled with liquid nitrogen, samples were crushed to a fine powder. In 10 ml centrifuge tubes the samples were mixed with 100 mg of MgCO3 and 5 ml of 100% (v/v) acetone. Bleached leaf material was removed by centrifugation (5 min; 2,000 g) and 1 ml aliquots of supernatants transferred to new tubes. Chlorophyll (a + b) content of extracts was determined spectrophotometrically [53].
Results
In vitro regeneration from leaf explants of axenic P. peltatum and P. zonale plants and selection with kanamycin
To induce in vitro plant regeneration, leaf explants were taken from axenically-propagated plants and cultured in Morphogenesis Induction Medium (MIM). By using axenic Pelargonium peltatum and Pelargonium zonale plants as the source for explants, the percentage of regenerating explants reached 90% in P. peltatum and 80% in P. zonale. Plant regeneration was carried out via somatic embryogenesis in P. peltatum (Figure 1e-h) and via organogenesis in P. zonale (Figure 1i-l).
Since the plasmids used for transformation experiments carried the nptII gene as the selectable marker, it was necessary to determine the suitable concentration of kanamycin for the selection of transgenic events. Leaf explants from axenic plants were cultured in selective MIM with different concentrations of kanamycin (0, 25, 50 and 100 mg l-1). A concentration of 50 mg l-1 was sufficient to inhibit the growth of non-transformed cells in leaf explants of both cultivars. Additionally, regenerated shoots were cultured in selective Elongation Medium (EM) with different kanamycin concentrations (0, 25, 50 and 100 mg l-1). According to our results (data not shown), a kanamycin concentration of 50 mg l-1 was used in EM to select transgenic plantlets for both cultivars.
Genetic transformation experiments
Genetic transformation experiments of P. zonale and P. peltatum were conducted using leaf explants from axenic plants. In the initial transformation experiments, explants from both Pelargonium spp. were inoculated with Agrobacterium tumefaciens strain LBA4404 carrying the pBIN19 binary vector harbouring, between the left and right border, the nptII marker gene under the control of the nos promoter and the nos terminator and the gfp-S65T reporter gene under the control of a 2x35SCaMV constitutive promoter and the nos terminator. The explants were cultured in selective MIM with 50 mg l-1 kanamycin. After one month of culture, explants began to develop morphogenetic calli on the cut surface of the leaf explants. Non-transformed calli became necrotic after 30–50 days in selective medium. After 2.5-3 months in selective MIM, the calli differentiated adventitious shoots in the case of P. zonale and somatic embryos in P. peltatum, which were excised and cultured in selective EM. In some cases, several independent transformation events were identified within the same explant (adventitious shoots on opposite sides of the same explant), but only one plant per explant was recovered to be sure that all the selected plants were derived from independent transformation events. After 1–1.5 moths of culture in selective EM, the shoots were excised and cultured in Rooting Medium (RM). Finally, 6 independent rooted plants of P. zonale and 6 of P. peltatum were selected and transferred to pots. Transgenic plants were acclimatized in a growth chamber and transferred to the greenhouse, where they subsequently flowered normally under long day conditions. Transformation efficiency, estimated as the number of independent transformation events (one transgenic plant per explant) in relation to the total number of inoculated explants, ranged between 2-3% in both cultivars.
To produce long-life and male sterile pelargonium plants, we used the same transformation protocol described above and axenic leaves of P. zonale as the starting material. The leaf explants were co-cultivated with A. tumefaciens strain LBA4404 carrying the chimaeric constructs pVDH393-pSAG12::ipt and pBI101-PsEND1::barnase respectively.
GFP as an in vivo selectable marker
While the nptII gene was employed for the selection of transgenic plants, the gfp gene expression was also examined during the transformation process in order to evaluate the ability of gfp as in vivo selectable marker. The expression of the gfp gene in the leaf explants was observed 40–50 days after inoculation (when adventitious buds or somatic embryos begin to appear) and also in the transgenic plants obtained. The gfp expression in transformed cells should be useful to select transformation events at early stages, so that antibiotic selection marker genes may not be required. However, in the case of Pelargonium spp., if the selection based only on gfp expression, a significant number of transgenic plants could be undetected due to the presence of chlorophyll in the green tissues. This problem is related to the fluorescence visualization in tissues with high chlorophyll content, as in the case of leaves. Chlorophyll shows strong red autofluorescence that could mask the green fluorescence of a few cells. In both Pelargonium spp., green fluorescence is clearly visible in the initial whitish callus with disorganized growth (Figure 1m-n), while it becomes increasingly difficult to identify in the subsequent organogenic callus and in the adventitious buds (Figure 1o-p). All kanamycin resistant plants showed green fluorescence in different organs and tissues. Green fluorescence was observed in regenerated shoots and plantlets (Figure 1q-r), on the border of leaves (Figure 1s-t) and especially in the roots, where the chlorophyll is absent (Figure 1u-v). In general, within the same organ, gfp detection varied in different tissues or cell types depending on their chlorophyll content.
Ploidy level analysis
The ploidy level of plantlets regenerated in selective medium was analyzed and the data were compared with those of the original materials. Analyses were carried out with control leaves from adult plants of the original cultivar and young leaves from transgenic plants (Figure 2a-b). Only diploid plants showing identical ploidy level as the original ones were selected.
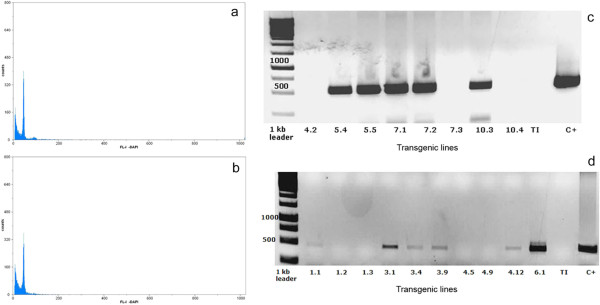
Figure 2.
Ploidy level and PCR analyses of transgenic pelargonium plants. Histograms of number of nuclei per channel as a function of relative fluorescence intensity resulting from flow cytometry analysis of nuclei stained with DAPI. Nuclei were isolated from leaf samples of diploid WT control (a) and transgenic (b) plants. (c). Identification of the barnase-barstar transgene (544 bp fragment) by PCR in different P. zonale male sterile plants. C + (positive control: pBI101-PsEND1::barnase-barstar) and TI (negative control). (d). Identification of the ipt transgene (460 bp fragment) by PCR in different P. zonale pSAG12::ipt transgenic plants. C + (positive control: pVDH393-pSAG12::ipt) and TI (negative control).
PCR analysis of the transgenes and selection of transgenic lines
The presence of the nptII, uidA, gfp, barnase-barstar and ipt transgenes in the selected transgenic lines of both P. zonale and P. peltatum was confirmed by PCR. Figure 2 shows the detection by PCR analysis of the barnase-barstar (Figure 2c) and ipt transgenes (Figure 2d) in different transgenic plants of P. zonale.
Based on the PCR analysis, we selected 12 independent lines of P. zonale, 6 carrying the pVDH393-pSAG12::ipt construct and 6 with the pBI101-PsEND1::barnase chimaeric gene for further phenotypic and molecular analyses.
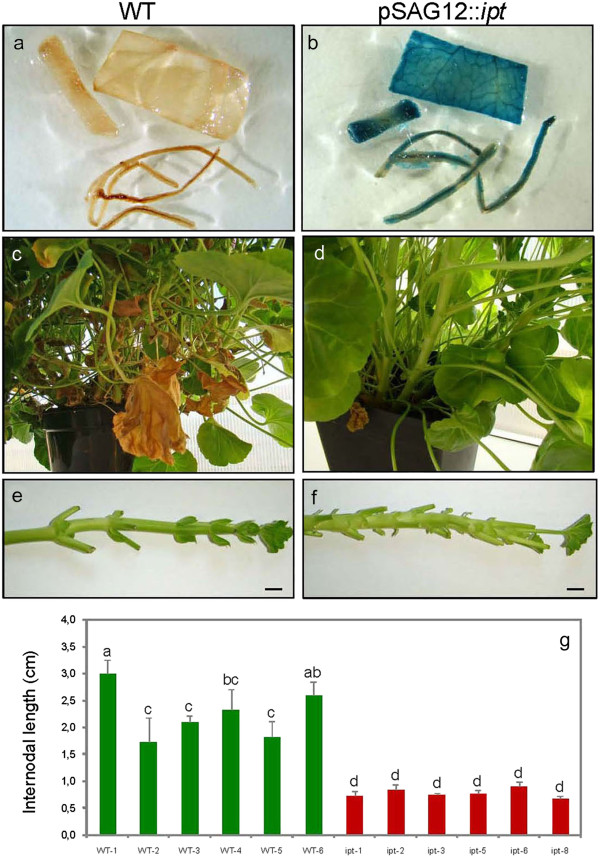
The selected transgenic lines of P. zonale carrying the pSAG12::ipt construct were also evaluated for GUS production, because they all carry the uidA reporter gene under the control of the 35SCaMV promoter. In Figure 3a-b, constitutive GUS expression can be observed in stems, leaves and roots of the pSAG12::ipt plants in comparison with control plants (WT).
Figure 3.
Reduced internode length and increased branching in Pelargonium zonale pSAG12::ipt transgenic plants. (a-b). Constitutive GUS expression in leaf, stem and root fragments in the pSAG12::ipt transgenic plants as additional proof of their genetic transformation. (c-d). Non-transformed WT potted plant (left) compared with a pSAG12::ipt transgenic plant (right). The transgenic plant showed delayed leaf senescence, more evident in the basal leaves and increased branching. (e-f). The pSAG12::ipt stems showed reduced internodal length (right) when compared with control WT plants. (g). Statistical analysis of the important differences in internodal length among 6 WT plants and 6 transgenic lines. The transgenic pSAG12::ipt plants showed a more compact architecture that the WT phenotype. Bars: 1 cm.
Morphological measurements in selected transgenic lines
Morphological measurements of vegetative growth were taken over the course of several days for each plant as its first five flowers reached anthesis in order to determine whether transgene expression could affect different growth parameters. The measured parameters included plant height at flowering, leaf width, leaf length, leaf petiole length, node number, internodal length and number of inflorescences per plant. Our results indicate that the vegetative growth and flowering of the transgenic plants were not modified in a significant manner by barnase expression in the anthers (not shown). Therefore, it seems that there is no ectopic effect of the barnase gene in vegetative or reproductive plant tissues other than anthers, corroborating the tissue-specificity of the PsEND1 promoter. In contrast, the transgenic pSAG12::ipt plants showed delayed leaf senescence, which is more evident in the basal leaves (Figure 3c-d), increased branching and shorter internodes than the WT, producing a phenotype that displays a more compact architecture of the whole plant (Figure 3e-f).
Delayed senescence in P. zonale pVDH393-pSAG12::ipt transgenic plants
After rooting, the in vitro transgenic plants were acclimatized and cultivated in the greenhouse to evaluate their possible delayed senescence. Transgenic plants showed delayed leaf senescence, increased branching and reduced internodal length when compared with control WT plants (not transformed). Figure 3g shows the important differences in internodal length among 6 WT plants and 6 transgenic lines. The transgenic pSAG12::ipt plants showed a phenotype with more compact architecture that the WT phenotype.
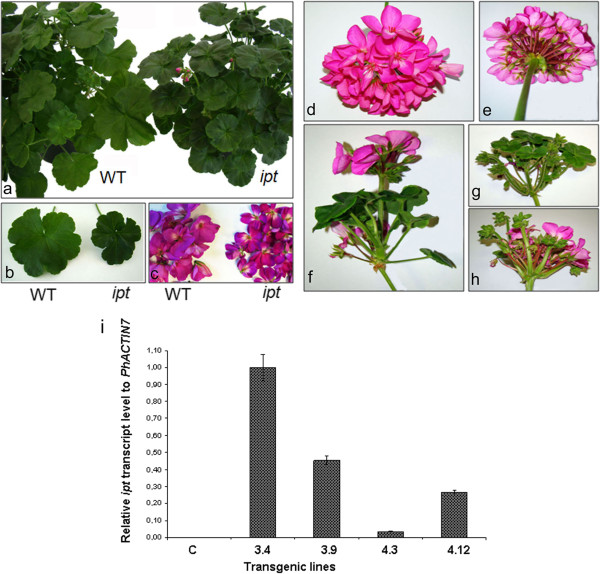
Another phenotypic difference between the WT and the transgenic plants was the smaller leaf size observed in the pSAG12::ipt plants (Figure 4a-b). In the inflorescences, the flower phenotypes were quite similar, although the pSAG12::ipt flowers were also reduced in size when compared with WT flowers (Figure 4c). Transgenic leaves and flowers showed a more intense color than the non-transformed ones, probably as a consequence of their reduced size. In some transgenic adult plants, we occasionally observed alterations in the inflorescence development when comparing transgenic with WT inflorescences (Figure 4d-e). In some pSAG12::ipt inflorescences, flowers coexist with new vegetative structures, which are produced at the same time as flowers or new inflorescences (Figure 4f-g-h). This occasional phenomenon could be due to a large number of copies or the insertion position of the pSAG12::ipt transgene in the plant genome. To elucidate if there was a correlation between the observed phenotype and the expression level of the exogenous gene in these plants, we carried out real-time RT-PCR experiments with four selected transgenic lines showing delayed senescence (3.4, 3.9, 4.3 and 4.12). The transgenic line 3.4 showed an inflorescence reversion phenotype. Our results indicated that the expression level of the transgene is higher in line 3.4 when compared with the other ones (Figure 4i). However, this phenotype could be considered as an undesirable collateral effect from a commercial point of view, for this reason, transgenic lines showing occasional inflorescence reversion were discarded.
Figure 4.
Reduced leaf and flower size in Pelargonium zonale pSAG12::ipt transgenic plants. Occasional inflorescence reversion. (a). The pSAG12::ipt transgenic plants (right) showed a more compact architecture with reduced organs than the WT plants (left). (b). Reduced leaf size in the pSAG12::ipt plants (right) compared with a WT leaf (left). (c). The flowers of the pSAG12::ipt plants were also reduced in size (right) when compared with WT flowers (left). In adult plants cultivated at the greenhouse, we did not observe alterations in the inflorescence development when compared the WT inflorescences with the transgenic ones (d-e). Occasionally, in some pSAG12::ipt inflorescences, flowers coexist with new vegetative structures, which are produced at the same time as flowers or new inflorescences, indicating inflorescence reversion (f-g-h). (i). Real-time RT-PCR analysis of pSAG12::ipt transcript levels in detached leaves from the transgenic lines 3.4, 3.9, 4.3 and 4.12. Each sample’s expression level relative to Pelargonium x hortorum PhACTIN7 is the mean of three biological repeats. C: control WT leaves.
All of P. zonale pSAG12::ipt transgenic plants cultivated in the greenhouse exhibited delayed senescence when compared with WT control plants, especially at the basal leaves. In Figure 3, a comparison between adult basal leaves (more than 5 months) from pSAG12::ipt transgenic plants (Figure 3d) and basal leaves of non-transformed plants with a similar age (Figure 3c) was showed. A high number of adult leaves from control plants exhibited an evident senescence phenotype while similar leaves at the same positions in the transgenic plants remained green and fully expanded.
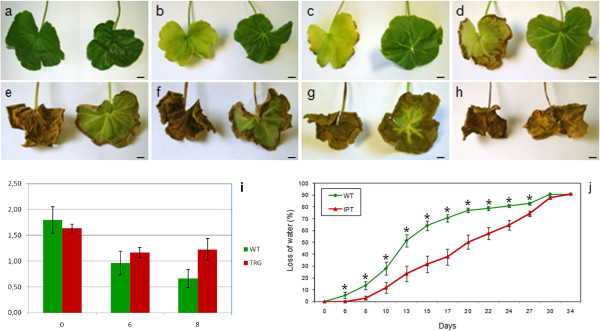
To better characterize and determine the delay of senescence in the transgenic plants, young and healthy leaves of similar age from both transgenic and control plants were detached and their petioles were placed in glass tubes with water at 28°C in darkness. The analysis of these leaves over time showed that leaves from the pSAG12::ipt transgenic plants remained green during more time than the controls. In Figure 5, we show a comparison of detached leaves from the control (left) and pSAG12::ipt (right) plants at 0 (a), 6 (b), 8 (c), 17 (d), 22 (e), 24 (f), 27 (g) and 34 (h) days of incubation in darkness. While the WT leaves exhibited evident symptoms of chlorophyll degradation after 6 days of incubation in darkness, the transgenic leaves exhibited similar symptoms after 22 days of incubation, indicating a delay in the senescence process. Likewise, necrotic symptoms appeared earlier in the WT leaves (~ 8 days) than in the transgenic ones (~ 20 days). Chlorophyll quantification assays in the detached leaves at 0, 6 and 8 days corroborated these observations (Figure 5i). For instance, the decline in chlorophyll (a + b) content after 8 days of incubation in darkness of WT and pSAG12::ipt leaves was 63.0% and 25.2% respectively, corroborating the delay in the senescence process in the transgenic plants. Moreover, water loss during the time course was lower in leaves from transgenic plants than from WT ones (Figure 5j). The differences begin to be significant after 6 days of incubation in darkness. After 15 days there was a 65% of loss of fresh weight in the WT leaves, while in the transgenic leaves the loss of weight was only a 32%. After 22 days the loss of fresh weight in the WT leaves reached the 80% of their initial weight, while the loss of water in the transgenic leaves was the 65% of their initial weight. After 30 days, the values of WT and transgenic leaves were similar. These data reinforce the idea that the chimaeric construct pSAG12::ipt could be useful in Pelargonium spp. to delay the senescence process and to produce long-lived plants.
Figure 5.
Senescence delay in detached leaves of WT Pelargonium zonale and pSAG12::ipt transgenic plants. Detached leaves of control (left) and pSAG12::ipt (right) plants at 0 (a), 6 (b), 8 (c), 17 (d), 22 (e), 24 (f), 27 (g) and 34 (h) days of incubation in darkness. (i) Mean concentration (±SE) of chlorophyll a + b (mg/g fresh weight) from detached leaves of control (WT) and pSAG12::ipt (TRG) plants at 0, 6 and 8 days of incubation in darkness. (j) Senescence delay of detached leaves from pSAG12::ipt plants. Fresh weight changes in detached leaves of WT P. zonale and a transgenic line carrying the pSAG12::ipt chimaeric gene over the time course analyzed. Data are the means of sixteen leaves ± SE. Bars: 1 cm.
Early anther ablation in P. zonale transgenic plants results in efficient male sterility
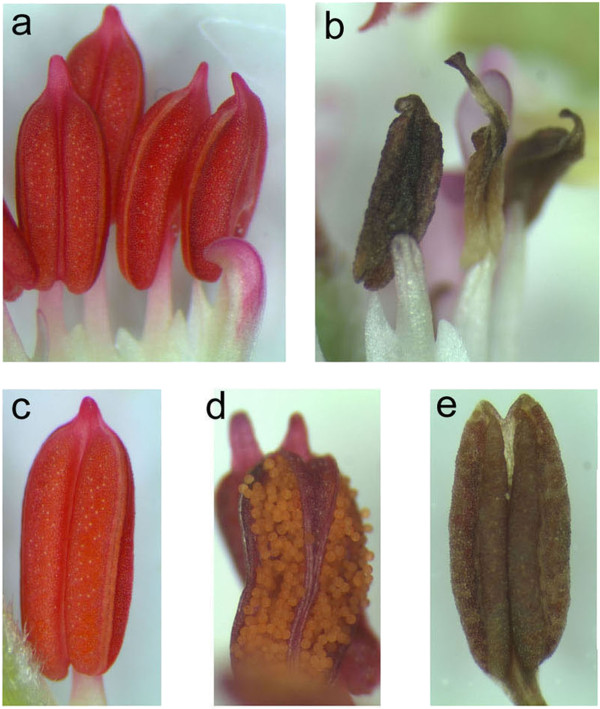
Transgenic P. zonale PsEND1::barnase plants showed normal vegetative development and flowering. However, anthers from transgenic lines carrying the chimaeric PsEND1::barnase gene construct showed dramatic differences in development when compared with non-transformed WT anthers. Anthers at different stages of development were examined by light microscopy. In WT anthers from flowers one day prior to anthesis (Figure 6a), the locules were fully developed, showing the normal shape (Figure 6c) and containing viable pollen grains which are visible during anthesis (Figure 6d), whereas the transgenic flowers showed collapsed structures at the end of a short filament in the place of a normal four-lobed anther (Figure 6b) with a fully expanded filament.
Figure 6.
Engineered anther ablation in the Pelargonium zonale PsEND1::barnase plants. (a). In WT anthers from flowers one day prior to anthesis, the locules were fully developed, showing the normal shape. (b). Transgenic PsEND1::barnase anthers showing collapsed structures at the end of a short filament in the place of a normal four-lobed anther with a fully expanded filament (c). (d) Viable pollen grains in a WT anther which are visible during anthesis. (e), No pollen grains were observed in the modified anther structures developed instead of normal anthers in the lines carrying the male-sterility construct. The locules of sterile anthers from flowers 3–4 days prior to anthesis were narrow and unexpanded. In most cases, the undeveloped anther, composed of the ablated anther tissues, becomes a necrotic structure that never dehisces.
No pollen grains were observed in the modified anther structures which developed instead of normal anthers in the lines carrying the male sterility construct. The locules of sterile anthers were narrow and unexpanded in flowers 3–4 days prior to anthesis. In most cases, the undeveloped anther becomes a necrotic structure, composed of ablated anther tissues, that never dehisces (Figure 6e). The cross-pollination of the male-sterile plants with pollen from wild-type plants usually produced normal fruits and seeds, indicating that female fertility was not affected. In some transgenic lines, we detected a small reduction in seed production (data not shown), these lines were discarded.
Discussion
Classical breeding programmes applied to produce new or improved varieties of floricultural species resulted in a range of cultivars with excellent traits, such as colour, shape, fragrance, vase life in cut-flower species, rooting potential or overall plant morphology. However, some of these aims have not yet been achieved in many ornamental species. Gene transfer by means of Agrobacterium tumefaciens enables the introduction of new genes/traits from unrelated species and would be a helpful tool in pelargonium breeding.
Efficient regeneration protocols that could be applicable to a broad spectrum of different genotypes are a prerequisite for developing a transformation system for a plant species or genotype [1]. We have developed a simple and reliable in vitro regeneration protocol for the genetic transformation of Pelargonium spp. The two Pelargonium genotypes used in the present study showed different regeneration ability. Using the same culture medium, regeneration was carried out via somatic embryogenesis in P. peltatum and via organogenesis in P. zonale. Interestingly, the percentage of regenerating explants obtained from both genotypes was similar (90% in P. peltatum and 80% in P. zonale) and also the transformation efficiency (2-3%).
We have also evaluated the use of the gfp gene as an in vivo selectable marker in Pelargonium. It has been reported that gfp expression in transformed cells is useful to select transformation events in early stages, so that antibiotic selection is not needed [47,54-61]. In the case of Pelargonium spp., the use of gfp as a selectable in vivo marker gene is restricted to identify transformation events, because at late developmental stages it becomes undetectable due to the presence of chlorophyll in the green tissues (leaves). In adult plants, gfp expression is only detectable in non green tissues like roots, petals or anther filaments.
Using this transformation protocol, we introduced two new traits in P. zonale cv. 370, one to produce long-life plants by inducing the ipt gene during plant senescence and the other to produce male sterile plants without pollen. During the initial transformation assays we used the gfp reporter gene to in vivo identify the transformations events and to evaluate transformation efficiency in the regenerating plantlets. The gfp gene has been used to successfully transform sugar cane, tobacco, maize, lettuce [54], walnut [55], citrus [56], peach [57], potato [58], pear [59], carrot [60] and other species [61]. In P. zonale, we have observed gfp expression along the regeneration process, but, at times longer than three weeks after inoculation, we could not correlate explants that regenerated transgenic plants with green fluorescence. Low levels of gfp fluorescence coincided in time with increased content of chlorophyll and the red autofluorescence of chlorophyll interferes with the gfp green fluorescence [62]. This interference could also be caused by pigments that are opaque to exciting UV or blue light [61]. Some authors state that transformation efficiencies based on resistance to a selective marker are probably underestimating the actual rate of regenerated transgenic plants [63]. Therefore PCR analysis has been proposed to identify transgenic plants in addition to the use of selectable or visual screening markers [63]. In the present work, the gfp gene was useful to confirm genetic transformation of P. zonale in vivo. All transgenic plants showed green fluorescence in nearly all tissues analyzed but there were large differences in green fluorescence between organs and tissues, depending on the chlorophyll content of each one. PCR analysis corroborated the presence of transgenes in the regenerated plantlets.
In genetic transformation experiments, the analyses generally focus on the molecular characterization of the transgenic plants but the ploidy level of the transgenic material is checked in relatively few cases. The confirmation of the ploidy level in transgenic material is particularly important during the selection of transgenic lines. Our results indicate that leaf tissues of the Pelargonium cultivars used here have a diploid number of chromosomes and regeneration from these explants leads to plants with the same ploidy level as adult material of the original cultivar.
Transgenic pSAG12::ipt plants showed delayed leaf senescence, which was more evident at the basal leaves. They also showed increased branching and reduced internodal length when compared with non transformed plants. In addition, the transgenic pSAG12::ipt plants displayed a more compact architecture than the WT plants. Other interesting phenotypic difference among the WT and the transgenic pSAG12::ipt plants was the reduction of transgenic leaf size. The plant architecture was compact in the transgenic plants, including tight inflorescences and flowers. In some pSAG12::ipt inflorescences, flowers coexisted with new vegetative structures which are produced at the same time as flowers or new inflorescences. This occasional phenomenon might be due to a change in the determination of the floral meristem leading to inflorescence reversion and new vegetative organs instead of flowers in the inflorescences.
Inflorescence or flower reversion may occur when the level of the floral signal is insufficient for the completion of flower development and the suppression of indeterminacy [64]. Flower and inflorescence reversion involve a switch from floral development back to vegetative development, thus rendering flowering a phase in an ongoing growth pattern rather than a terminal act of the meristem. Although it can be considered an unusual event, it is linked to environmental conditions and is most often a response to conditions opposite to those that induce flowering. A clear-cut reversion to leaf production has been described in Impatiens balsamina[65]. In I. balsamina, a leaf-derived signal is critical for completion of flowering and can be considered to be the basis of a plant-wide floral commitment that is achieved without accompanying meristem autonomy. It has been proposed that cytokinins can be involved in floral induction as the leaf-generated signal that produces completion of the flowering process making it irreversible [66,67]. These cytokinin fluxes during floral induction in LD plants could be altered in the pSAG12::ipt transgenic plants during senescence due to the continued production of cytokinin and this fact could be influencing the reversion process in some inflorescences. To elucidate if there was a correlation between the inflorescence reversion phenotype and the expression level of the exogenous gene in these plants, we carried out real-time RT-PCR experiments. Our results indicated that the expression level of the transgene is higher in those transgenic lines showing inflorescence reversion. However, this phenotype could be considered as an undesirable collateral effect from a commercial point of view and transgenic lines showing occasional inflorescence reversion were discarded.
All the pSAG12::ipt P. zonale plants cultivated in the greenhouse exhibited delayed senescence when compared with WT control plants, especially in the basal leaves. A high number of adult leaves of control plants exhibited an evident senescence phenotype while similar leaves at the same positions in the transgenic plants remained green and fully expanded. To better characterize and determine the delay of senescence in the transgenic plants, young and healthy leaves of similar age from both transgenic and control plants were detached and their petioles were placed in glass tubes with water at 28°C in darkness. The analysis of these leaves over time showed that leaves from the pSAG12::ipt transgenic plants remained green longer than the controls. While the WT leaves exhibited evident symptoms of chlorophyll degradation after 6 days of incubation in the darkness, the transgenic leaves exhibited similar symptoms after 22 days of incubation, indicating a delay in the senescence process. Likewise, necrotic symptoms appeared early in the WT leaves than in the transgenic ones. Quantification of chlorophyll content of detached WT and pSAG12::ipt leaves indicated that the decline of chlorophyll was higher in the WT leaves when compared with the transgenic ones. Moreover, the loss of water in leaves from transgenic plants was minor over the time course analyzed. These data reinforce the idea that the chimaeric pSAG12::ipt construct could be useful in Pelargonium spp. to delay the senescence process and to produce long-lived plants.
We have obtained engineered male sterile plants of P. zonale to prevent undesirable lateral gene flow of the introduced transgenes to related species. The PsEND1 promoter specifically directed expression of the barnase gene to different anther tissues involved in anther architecture (epidermis, endothecium, middle layer, connective tissue). Expression of the barnase gene under control of this promoter caused specific ablation of these tissues at early stages of anther development in the transgenic plants. We readily observed small structures that developed instead of normal anthers in the third floral whorl of transgenic flowers, which displayed premature senescence and collapse of the pollen sacs, microspores and tapetum and a lack of pollen at anthesis. Ablation of the structural anther tissues also produces the improper formation of the tapetum tissue and the subsequent degeneration of the microspores is accompanied by a change in anther wall thickness, by a size reduction and by a change in the epidermal cell types [38]. Since this phenotype is unlikely to be due to expression of barnase in structural tissues, it is likely to be an indirect effect of the loss of the tapetum and microspore cells.
No pollen grains were observed in any section of the ablated anthers from the male sterile plants, indicating that barnase effectively ablates specific cell lines that will form the structural tissues of the anther, preventing pollen development. Transgenic anthers appeared to show effects of barnase expression at every stage examined. This is likely due to the developmentally earlier expression of the PsEND1 promoter. The anther filaments of the transgenic plants were shorter than WT filaments. The formation of short filaments is commonly associated with male sterility or reduced fertility [43,68-70]. These observations reinforce our previous results in other crop species using the same chimaeric construct [38].
Due to the extremely toxic nature of barnase, other researchers have reported a general lack of vigour and a decline in plant growth in transgenic plants carrying the barnase gene [71-73]. The lack of significant effects on growth characteristics is important to know when considering the use of barnase for male sterility in landscaping plants. To prevent the possible effects of ectopic barnase expression, Gardner et al. [40] proposed the introduction of the male and/or female sterility genes in combination with a gene protecting against inappropriate barnase expression (enhanced 35S::barstar). In all the lines of transgenic P. zonale plants expressing barnase under control of the PsEND1 promoter, we did not observe differences with respect to wild type plants in vegetative growth, flowering time or inflorescence number. Morphological analysis of the transgenic plants showed that, under greenhouse conditions, the expression of the PsEND1::barnase construct does not significantly affect the vegetative and floral development, thus confirming the anther specificity of the PsEND1 promoter region previously observed by means of the GUS expression studies in different dicots and monocots [38,39,41,42]. The potential biotechnological applications of the PsEND1 promoter largely depend on both its spatial and temporal expression pattern, since the ectopic expression of the cytotoxic agent would damage other plant tissues and organs, decreasing the agronomic value of hybrid plants.
Expression of the barnase gene in ornamental plants under control of the anther-specific PsEND1 promoter may be used to create efficient male sterile versions of existing popular cultivars without adversely affecting their respective phenotypes. Therefore, this technology would be especially useful to produce environmentally friendly transgenic plants carrying new traits by preventing gene flow between the genetically modified ornamentals.
Conclusions
We have developed a simple and reliable in vitro regeneration protocol for the genetic transformation of Pelargonium spp. By using this methodology, we introduced two new traits in P. zonale cv. 370, one to produce long-life plants and the other to produce male sterility. The resulting phenotypes would be of interest both for consumers and producers.
The chimaeric pSAG12::ipt construct may be useful in Pelargonium spp. to delay the senescence process and to produce long-lived plants, which could have commercial interest. Transgenic pSAG12::ipt plants showed delayed leaf senescence, increased branching and reduced internodal length as compared to non-transformed plants. Moreover, the transgenic pSAG12::ipt plants showed a more compact architecture than the WT. In some pSAG12::ipt inflorescences, flowers coexist with new vegetative structures which are produced at the same time as flowers or new inflorescences. This occasional phenomenon might be due to a change in the determination of the floral meristem leading to inflorescence reversion as a collateral effect of the increased expression levels of the transgene in some transgenic lines. The lines showing inflorescence reversion were discarded because this effect is undesirable from a commercial point of view.
The PsEND1 promoter specifically directed expression of the barnase gene to different anther tissues involved in anther architecture. Expression of the barnase gene under control of this promoter caused specific ablation of these tissues, which become necrotic at early stages of anther development in the transgenic pelargonium plants. No pollen grains were observed in the ablated anthers from the male-sterile plants, indicating that barnase effectively destroys specific cell lines that form the structural tissues of the anther, thus preventing pollen development. The use of engineered male sterility would be especially useful to eliminate pollen allergens and to produce environmentally friendly transgenic plants carrying new traits by preventing gene flow between the genetically modified ornamentals and related plant species.
Abbreviations
AS: Acetosyringone; BA: 6-benzylaminopurine; CEF: Cefotaxime; Gfp: Green fluorescent protein gene; Ipt: Isopentenyl phosphotransferase gene; IAA: Indole-3-acetic acid; KAN: Kanamycin sulphate; KIN: Kinetin; LB: Luria Bertani medium; MES: 2-(N-morpholino)ethane sulfonic acid; MS: Murashige and Skoog medium; NAA: α-naphthalene acetic acid; nptII: Neomycin phosphotransferase gene; PCR: Polymerase chain reaction; TDZ: Thidiazuron; TIM: Timentin (ticarcillin/clavulanic acid); uidA: β-glucuronidase gene.
Competing interests
In the past five years we have received funding from the Spanish Ministry of Science and Innovation (MICINN) and the article-processing charge will be pay with funds from two granted projects. The authors received salaries from two different institutions: The Polytechnic University of Valencia (UPV) or the High Spanish Council of Scientific Research (CSIC). We are not currently applying for a patent related with the content of this manuscript. All the mentioned organisms/institutions do not gain or lose financially from the publication of this manuscript either now or in the future.
Authors’ contributions
BG-S and BP performed the transformation experiments of P. zonale, chlorophyll quantification and loss of water experiments, both authors contributed equally to this paper. TA contributed to the production of in vitro regenerated and greenhouse P. zonale plants. AA performed the ploidy level analyses of transgenic lines. MB and VM designed the experiments related to the production of pSAG12::ipt transgenic plants. ER performed the molecular analyses (PCR and real-time RT-PCR) of transgenic plants. JPB and LAC designed the experiments related to the production of male sterile PsEND1::barnase transgenic plants. All authors read and approved the final version of the manuscript.
Contributor Information
Begoña García-Sogo, Email: bgarcia@btc.upv.es.
Benito Pineda, Email: bpineda@btc.upv.es.
Edelín Roque, Email: edroque@ibmcp.upv.es.
Teresa Antón, Email: maanmar2@doctor.upv.es.
Alejandro Atarés, Email: aatares@ibmcp.upv.es.
Marisé Borja, Email: marise.borja@plantresponse.com.
José Pío Beltrán, Email: jbeltran@ibmcp.upv.es.
Vicente Moreno, Email: vmoreno@ibmcp.upv.es.
Luis Antonio Cañas, Email: lcanas@ibmcp.upv.es.
Acknowledgements
This work was funded by grants AGL2009-13388-C03-01 and BIO2009-08134 from the Spanish Ministry of Science and Innovation (MICINN). We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
References
- Winkelmann T, Kaviani K, Serek M. Development of a shoot regeneration protocol for genetic transformation in Pelargonium zonale and Pelargonium peltatum hybrids. Plant Cell Tiss Org Cult. 2005;80:33–42. doi: 10.1007/s11240-004-5788-5. [DOI] [Google Scholar]
- Cassells AC, Carney BF. Adventitious regeneration in Pelargonium x domesticum. Acta Hort. 1987;212:419–423. [Google Scholar]
- Dunbar KB, Stephens CT. Shoot regeneration of hybrid seed geranium (Pelargonium x hortorum) and regal geranium (Pelargonium x domesticum) from primary callus cultures. Plant Cell Tiss Org Cult. 1989;19:13–21. doi: 10.1007/BF00037772. [DOI] [Google Scholar]
- Boase MR, Deroles SC, Winnefield CS, Butcher SM, Borst NK, Butler RC. Genetic transformation of regal pelargonium (Pelargonium x domesticum ‘Dubonnet’) by Agrobacterium tumefaciens. Plant Sci. 1996;121:47–61. doi: 10.1016/S0168-9452(96)04508-6. [DOI] [Google Scholar]
- Robichon MP, Renou JP, Jalouzot R. Plant regeneration of ivy leaved geranium through shoot organogenesis. Plant Cell Tiss Org Cult. 1997;49:209–212. doi: 10.1023/A:1005843901764. [DOI] [Google Scholar]
- Agarwal PK, Ranu RS. Regeneration of plantlets from leaf and petiole explants of Pelargonium x hortorum. In Vitro Cell Dev Biol Plant. 2000;36:392–397. doi: 10.1007/s11627-000-0070-y. [DOI] [Google Scholar]
- Mithila J, Murch SJ, KrishnaRaj S, Saxena PK. Recent advances in Pelargonium in vitro regeneration system. Plant Cell Tiss Org Cult. 2001;67:1–9. doi: 10.1023/A:1011601517200. [DOI] [Google Scholar]
- Cassells AC. The effect of 2,3,5-triiodobenzoic acid on caulogenesis in callus culture of tomato and pelargonium. Physiol Plant. 1979;46:159–164. doi: 10.1111/j.1399-3054.1979.tb06550.x. [DOI] [Google Scholar]
- KrishnaRaj S, Bi YM, Saxena PK. Somatic embryogenesis and Agrobacterium-mediated transformation system for scented geraniums (Pelargonium sp. ‘Frensham’) Planta. 1997;201:434–440. doi: 10.1007/s004250050086. [DOI] [Google Scholar]
- Haensch KT. Somatic embryogenesis in vitro from adult plants of pelargonium: Influence of genotype and basal medium. Gartenbauwissenschaft. 1999;64:193–200. [Google Scholar]
- Haensch KT. Morpho-histological study of somatic embryo-like structures in hypocotyls culture of Pelargonium x hortorum Bailey. Plant Cell Rep. 2004;22:376–381. doi: 10.1007/s00299-003-0726-2. [DOI] [PubMed] [Google Scholar]
- Boase MR, Bradley JM, Borst NK. An improved method for transformation of regal pelargonium (Pelargonium x domesticum) by Agrobacterium tumefaciens. Plant Sci. 1998;1998(139):59–69. [Google Scholar]
- Pellegrineschi A, Davolio-Mariani O. Agrobacterium rhizogenes-mediated transformation of scented geranium. Plant Cell Tiss Org Cult. 1996;47:79–86. doi: 10.1007/BF02318969. [DOI] [Google Scholar]
- Bi YM, Cammue BPA, Goodwin PH, KrishnaRaj S, Saxena PK. Resistance to Botrytis cinerea in scented geranium transformed with a gene encoding the antimicrobial protein Ace-AMP1. Plant Cell Rep. 1999;18:835–840. doi: 10.1007/s002990050670. [DOI] [Google Scholar]
- Van Staden J, Cook EL, Nooden LD. In: Senescence and Aging in Plants. Nooden LD, Leopold AC, editor. Academic, London; 1998. Cytokinins and senescence; pp. 282–328. [Google Scholar]
- Singh S, Letham DS, Palni LMS. Cytokinin biochemistry in relation to leaf senescence. Physiol Plant. 1992;86:388–406. doi: 10.1111/j.1399-3054.1992.tb01334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino RM. Cytokinins in plant senescence: from spray and pray to clone and play. Bioessays. 1996;18:557–565. doi: 10.1002/bies.950180707. [DOI] [Google Scholar]
- Gan S, Amasino RM. Making sense of senescence: molecular genetics regulation of leaf senescence. Plant Physiol. 1997;113:313–319. doi: 10.1104/pp.113.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V. The molecular biology of leaf senescence. J Exp Bot. 1997;48:181–199. doi: 10.1093/jxb/48.2.181. [DOI] [Google Scholar]
- Nam HG. The molecular genetic analysis of leaf senescence. Curr Opin Biotechnol. 1997;8:200–207. doi: 10.1016/S0958-1669(97)80103-6. [DOI] [PubMed] [Google Scholar]
- Skene KGM. In: The Developmental Function of Roots. Torrey JG, Clarkson DT, editor. Academic, New York; 1975. Cytokinin production by roots as a factor in the control of plant growth; pp. 365–396. [Google Scholar]
- Nooden LD, Singh S, Letham DS. Correlation of xylem sap cytokinin levels with monocarpic senescence in soybean. Plant Physiol. 1990;93:33–39. doi: 10.1104/pp.93.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kays SJ. Postharvest Physiology of Perishable Plant Products. AVI/Van Nostrand Reinhold, New York; 1991. [Google Scholar]
- McGaw BA, Burch LR. In: Plant Hormones: Physiology, Biochemistry and Molecular Biology. 2. Davies PJ, editor. Kluwer Academic Publishers, Dordrecht, The Netherlands; 1995. Cytokinin biosynthesis and metabolism; pp. 98–117. [Google Scholar]
- Mok DWS, Mok MC. Cytokinin: Chemistry, Activity, and Function. CRC Press, Boca Raton, FL; 1994. [Google Scholar]
- Medford JI, Horgan R, El-Sawi Z, Klee HJ. Alterations of endogenous cytokinins in transgenic plants using a chimeric isopentenyl transferase gene. Plant Cell. 1989;1:403–413. doi: 10.1105/tpc.1.4.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart CM, Scofield SR, Bevan MW, Dyer TA. Delayed leaf senescence in tobacco plants transformed with tmr, a gene for cytokinin production in Agrobacterium. Plant Cell. 1991;3:647–656. doi: 10.1105/tpc.3.7.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hagen G, Guilfoyle TJ. Altered morphology in transgenic tobacco plants that over-produce cytokinins in specific tissues and organs. Dev Biol. 1992;153:386–395. doi: 10.1016/0012-1606(92)90123-X. [DOI] [PubMed] [Google Scholar]
- Wang J, Letham DS, Cornish E, Stevenson KR. Studies of cytokinin action and metabolism using tobacco plants expressing either the ipt or the gus gene controlled by a chalcone synthase promoter: I. Developmental features of the transgenic plants. Aust J Plant Physiol. 1997;24:661–672. doi: 10.1071/PP96101. [DOI] [Google Scholar]
- Wang J, Letham DS, Cornish E, Wei K, Hocart CH, Michael M, Stevenson KR. Studies of cytokinin action and metabolism using tobacco plants expressing either the ipt or gus gene controlled by a chalcone synthase promoter: II. ipt and gus gene expression, cytokinin levels and metabolism. Aust J Plant Physiol. 1997;24:673–683. doi: 10.1071/PP96102. [DOI] [Google Scholar]
- Gan S, Amasino RM. Inhibition of leaf senescence by autoregulated production of cytokinin. Science. 1995;270:1986–1988. doi: 10.1126/science.270.5244.1986. [DOI] [PubMed] [Google Scholar]
- Lohman KN, Gan S, John MC, Amasino RM. Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol Plant. 1994;92:322–328. doi: 10.1111/j.1399-3054.1994.tb05343.x. [DOI] [Google Scholar]
- Hidekamp F, Dinkse WG, Hille J, Van Ormondt H. Nucleotide sequence of the Agrobacterium tumefaciens octopine Ti plasmid-encoded tmr gene. Nucleic Acids Res. 1983;11:6211–6233. doi: 10.1093/nar/11.18.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Ding Y, Liu X, Sun C, Cao S, Wang D, He S, Wang X, Li L, Tian W. Rice transformation with a senescence inhibition chimeric gene. Chin Sci Bull. 1998;43:1810–1815. doi: 10.1007/BF02883378. [DOI] [Google Scholar]
- Nguyen KHT, Kane EJ, Dix PJ. In: Plant Biotechnology and In Vitro Biology in the 21st Century. Altman A, Ziv M, Izhar S, editor. IX International Congress Plant Tissue Culture. Kluwer Academic Publishers, Dordrecht, The Netherlands; 1998. Hormonal regulation of senescence in cauliflower (Brassica oleracea var. Botrytis). Abstract no. 96; p. 164. [Google Scholar]
- McCabe MS, Mohapatra U, Schepers F, van Dun K, Power JB, Davey M. Delayed senescence in transgenic lettuce using an autoregulated ipt gene. J Exp Bot Suppl. 1998;49:49. [Google Scholar]
- McCabe MS, Garratt LC, Schepers F, Jordi WJRM, Stoopen GM, Davelaar E, van Rhijn JHA, Power JB, Davey MR. Effects of PSAG12-IPT gene expression on development and senescence in transgenic lettuce. Plant Physiol. 2001;127:505–516. doi: 10.1104/pp.010244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque E, Gómez MD, Ellull P, Wallbraun M, Madueño F, Beltrán JP, Cañas LA. The PsEND1 promoter: a novel tool to produce genetically engineered male-sterile plants by early anther ablation. Plant Cell Rep. 2007;26:313–325. doi: 10.1007/s00299-006-0237-z. [DOI] [PubMed] [Google Scholar]
- García-Sogo B, Pineda B, Castelblanque L, Antón T, Medina M, Roque E, Torresi C, Beltrán JP, Moreno V, Cañas LA. Efficient transformation of Kalanchoe blossfeldiana and production of male-sterile plants by engineered anther ablation. Plant Cell Rep. 2010;29:66–77. doi: 10.1007/s00299-009-0798-8. [DOI] [PubMed] [Google Scholar]
- Gardner N, Felsheim R, Smith AG. Production of male- and female-sterile plants through reproductive tissue ablation. J Plant Physiol. 2009;166(8):871–881. doi: 10.1016/j.jplph.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Gómez MD, Beltrán JP, Cañas LA. The pea END1 promoter drives anther-specific gene expression in different plant species. Planta. 2004;219:967–981. doi: 10.1007/s00425-004-1300-z. [DOI] [PubMed] [Google Scholar]
- Pistón F, García C, de la Viña G, Beltrán JP, Cañas LA, Barro F. The pea PsEND1 promoter drives the expression of GUS in transgenic wheat at the binucleate microspores stage and during pollen tube development. Mol Breed. 2008;21:401–405. doi: 10.1007/s11032-007-9133-7. [DOI] [Google Scholar]
- Mariani C, DeBeuckeleer M, Truettner J, Leemans J, Goldberg RB. Induction of male sterility in plants by a chimaeric ribonuclease gene. Nature. 1990;347:737–741. doi: 10.1038/347737a0. [DOI] [Google Scholar]
- Mariani C, Gossele V, De Beuckeleer M, De Block M, Goldberg RB, De GW, Leemans J. A chimaeric ribonuclease inhibitor gene restores fertility to male sterile plants. Nature. 1992;357:384–387. doi: 10.1038/357384a0. [DOI] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio-assay with tobacco tissue cultures. Physiol Plantarum. 1962;75:325–332. [Google Scholar]
- Shahin EA. Totipotency of tomato protoplasts. Theor Appl Genet. 1985;69:235–240. doi: 10.1007/BF00662431. [DOI] [PubMed] [Google Scholar]
- Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/S0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Hartley RW. Barnase and barstar: expression of its cloned inhibitor permits expression of a cloned ribonuclease. J Mol Biol. 1968;202:913–915. doi: 10.1016/0022-2836(88)90568-2. [DOI] [PubMed] [Google Scholar]
- Atarés A, Moyano E, Morales B, Schleicher P, García-Abellán JO, Antón T, García-Sogo B, Pérez-Martín F, Lozano R, Flores FB, Moreno V, Bolarín MC, Pineda B. An insertional mutagenesis programme with an enhancer trap for the identification and tagging of genes involved in abiotic stress tolerance in the tomato wild-related species Solanum pennellii. Plant Cell Rep. 2011;30:1865–1879. doi: 10.1007/s00299-011-1094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of total cellular DNA from plants, algae and fungi. Plant Mol Biol Manual. 1994;D1:1–8. [Google Scholar]
- He J, Gray J, Leisner S. A Pelargonium ARGONAUTE4 gene shows organ-specific expression and differences in RNA and protein levels. J Plant Physiol. 2010;167:319–325. doi: 10.1016/j.jplph.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotenoids: Pigments of photosynthetic biomenbranes. Met Enzymol. 1987;148:350–382. [Google Scholar]
- Elliot AR, Campbell JA, Dugdale B, Brettell RIS, Grof CPL. Green fluorescent protein facilitates rapid in vivo detection of genetically transformed plant cells. Plant Cell Rep. 1999;18:707–714. doi: 10.1007/s002990050647. [DOI] [Google Scholar]
- Escobar MA, Park JI, Polito VS, Leslie CA, Uratsu SL, Mc Granahan GH, Dandekar AM. Using GFP as a scorable marker in walnut somatic embryo transformation. Ann Bot. 2000;85(6):831–835. doi: 10.1006/anbo.2000.1143. [DOI] [Google Scholar]
- Ghorbel R, Juárez J, Navarro L, Peña L. Green fluorescent protein as a screenable marker to increase the efficiency of generating transgenic woody fruit plants. Theor Appl Genet. 1999;99:350–358. doi: 10.1007/s001220051244. [DOI] [Google Scholar]
- Pérez-Clemente RM, Pérez A, García L, Beltrán JP, Cañas LA. Transformation and regeneration of peach plants (Prunus persica L.) from embryo sections using the green fluorescent protein (GFP) as a vital marker. Mol Breed. 2004;14:419–427. doi: 10.1007/s11032-004-0506-x. [DOI] [Google Scholar]
- Rakosy-Tican E, Aurori CM, Dijkstra C, Thieme R, Aurori A, Davey MR. The usefulness of the gfp reporter gene for monitoring Agrobacterium-mediated transformation of potato dihaploid and tetraploid genotypes. Plant Cell Rep. 2007;26(5):661–671. doi: 10.1007/s00299-006-0273-8. [DOI] [PubMed] [Google Scholar]
- Yancheva SD, Shlizerman LA, Golubowicz S, Yabloviz Z, Perl A, Hanania U, Flaishman MA. The use of green fluorescent protein (GFP) improves Agrobacterium-mediated transformation of ‘Spadona’ pear (Pyrus communis L.) Plant Cell Rep. 2006;25:183–189. doi: 10.1007/s00299-005-0025-1. [DOI] [PubMed] [Google Scholar]
- Baranski B, Klocke E, Schumann G. Green fluorescent protein as an efficient selection marker for Agrobacterium rhizogenes mediated carrot transformation. Plant Cell Rep. 2006;25:190. doi: 10.1007/s00299-005-0040-2. [DOI] [PubMed] [Google Scholar]
- Mercuri A, Sacchetti A, De Benedetti A, Schiva T, Alberti S. Green fluorescent flowers. Plant Sci. 2001;161:961–968. doi: 10.1016/S0168-9452(01)00497-6. [DOI] [Google Scholar]
- Hraska M, Rakousky S, Curn V. Green fluorescent protein as a vital marker for nondestructive detection of transformation events in transgenic plants. Plant Cell Tiss Org Cult. 2006;86:303–318. doi: 10.1007/s11240-006-9131-1. [DOI] [Google Scholar]
- Domínguez A, Cervera M, Pérez RM, Romero J, Fagoaga C, Cubero J, López MM, Juárez JA, Navarro L, Peña L. Characterisation of regenerants obtained under selective conditions after Agrobacterium-mediated transformation of citrus explants reveals production of silenced and chimeric plants at unexpected high frequencies. Mol Breed. 2004;14:171–183. [Google Scholar]
- Hempel FD, Patricia C, Zambryski PC, Feldman LJ. Photoinduction of flower identity in vegetatively biased primordia. Plant Cell. 1988;10:1663–1676. doi: 10.1105/tpc.10.10.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Took F, Ordidge M, Chiurugwi T, Battey N. Mechanisms and function of flower and inflorescence reversion. J Exp Bot. 2005;56(420):2587–2599. doi: 10.1093/jxb/eri254. [DOI] [PubMed] [Google Scholar]
- Lejeune P, Kinet J-M, Bernier G. Cytokinin fluxes during floral induction in the long day plant Sinapis alba L. Plant Physiol. 1988;86:1095–1098. doi: 10.1104/pp.86.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune P, Bernier G, Requier M-C, Kinet J-M. Cytokinin in phloem and xylem saps of Sinapis alba during floral induction. Physiol Plant. 1994;90:522–528. doi: 10.1111/j.1399-3054.1994.tb08810.x. [DOI] [Google Scholar]
- Theis R, Röbbelen G. Anther and microspore development in different male sterile lines of oilseed rape (Brassica napus L.) Angew Bot. 1990;64:419–434. [Google Scholar]
- Denis M, Delourme R, Gourret JP, Mariani C, Renard M. Expression of engineered nuclear male-sterility in Brassica napus. Plant Physiol. 1993;101:1295–1304. doi: 10.1104/pp.101.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worral D, Hird DL, Hodge R, Paul W, Draper J, Scott R. Premature dissolution of microsporocyte callose wall causes male sterility in transgenic tobacco. Plant Cell. 1992;4:759–771. doi: 10.1105/tpc.4.7.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannath A, Bandyopadhyay P, Arumugam N, Gupta V, Burma PK, Pental D. The use of spacer DNA fragment insulates the tissue-specific expression of a cytotoxic gene (barnase) and allows high-frequency generation of transgenic male sterile lines in Brassica juncea L. Mol Breed. 2001;8:11–23. doi: 10.1023/A:1011916216191. [DOI] [Google Scholar]
- Stanislaus MA, Cheng CL. Genetically engineered self-destruction: an alternative to herbicides for cover crop systems. Weed Sci. 2002;50:794–801. doi: 10.1614/0043-1745(2002)050[0794:GESDAA]2.0.CO;2. [DOI] [Google Scholar]
- Wei H, Meilan R, Brunner AM, Skinner JS, Ma C, Gandhi HT, Strauss SH. Field trial detects incomplete barstar attenuation of vegetative cytotoxicity in Populus trees containing a poplar LEAFY promoter: barnase sterility transgene. Mol Breed. 2007;19:69–85. [Google Scholar]