Abstract
We investigated the molecular and kinetic properties of two acetylcholinesterases (AmAChE1 and AmAChE2) from the Western honey bee, Apis mellifera. Western blot analysis revealed that AmAChE2 has most of catalytic activity rather than AmAChE1, further suggesting that AmAChE2 is responsible for synaptic transmission in A. mellifera, in contrast to most other insects. AmAChE2 was predominately expressed in the ganglia and head containing the central nervous system (CNS), while AmAChE1 was abundantly observed not only in the CNS but also in the peripheral nervous system/non-neuronal tissues. Both AmAChEs exist as homodimers; the monomers are covalently connected via a disulfide bond under native conditions. However, AmAChE2 was associated with the cell membrane via the glycophosphatidylinositol anchor, while AmAChE1 was present as a soluble form. The two AmAChEs were functionally expressed with a baculovirus system. Kinetic analysis revealed that AmAChE2 has approximately 2,500-fold greater catalytic efficiency toward acetylthiocholine and butyrylthiocholine than AmAChE1, supporting the synaptic function of AmAChE2. In addition, AmAChE2 likely serves as the main target of the organophosphate (OP) and carbamate (CB) insecticides as judged by the lower IC50 values against AmAChE2 than against AmAChE1. When OP and CB insecticides were pre-incubated with a mixture of AmAChE1 and AmAChE2, a significant reduction in the inhibition of AmAChE2 was observed, suggesting a protective role of AmAChE1 against xenobiotics. Taken together, based on their tissue distribution pattern, molecular and kinetic properties, AmAChE2 plays a major role in synaptic transmission, while AmAChE1 has non-neuronal functions, including chemical defense.
Introduction
Acetylcholinesterase (AChE, EC 3.1.1.7) is a critical enzyme in the cholinergic synapses and neuromuscular junctions of both vertebrates and invertebrates that regulates the level of the neurotransmitter acetylcholine and terminates nerve impulses [1]. AChE is a key enzyme in the insect nervous system, in which the cholinergic system is essential [2], and is the target of organophosphate (OP) and carbamate (CB) insecticides. Reduced sensitivity of AChE has been reported as one of the major resistance mechanisms against OP and CB insecticides in many arthropods [3], including the two-spotted spider mite Tetranychus urticae [4], the house mosquito Culex pipiens [5], the German cockroach Blattella germanica [6] and the Colorado potato beetle Leptinotarsa decemlineata [7].
Two cholinesterases (ChE), AChE and butyrylcholinesterase (BChE, EC 3.1.1.8), have been characterized in vertebrates, while insects possess only AChE and not BChE [8]. Studies on the evolution of ChE, including AChE and BChE, suggest that true ChEs, with highly selective substrate specificity, appear in the early bilaterians [9]. Genes for both AChE and BChE are usually present in most lineages of vertebrates, whereas duplications of the ace gene encoding AChE are observed in a few lineages such as nematodes, arachnids and insects. Recent studies have shown that two different ace loci (ace1, encoding AChE1, which is paralogous to Drosophila ace; ace2, encoding AChE2, which is orthologous to Drosophila ace) have been cloned from various insect species, such as the cotton aphid Aphis gossypii [10], the greenbug Schizaphis graminum [11], the diamondback moth Plutella xylostella [12] and the German cockroach B. germanica [13], while only one type of AChE has been discovered in cyclorrhaphan flies [14], including Drosophila melanogaster [15] and Musca domestica [16]. As indicated by phylogenetic analysis, the two ace genes were derived from a duplication that occurred long before the differentiation of insects, whereas the ace1 copy was lost in Cyclorrhapha during the course of evolution [15]. Of the two insect ace genes, the expression of AChE1 is much greater than that of AChE2 in insects with both genes [12], [13], [17]–[19]. In addition, insects that are resistant to OP and CB insecticides possess point mutations in the ace1 gene that are responsible for target site insensitivity [20], [21]. Based on these findings, it was proposed that AChE1 is likely the major AChE involved in synaptic transmission in insects possessing both AChE1 and AChE2 [19], [22]–[24].
In addition to the different ace loci expressing functionally distinct AChEs, multiple molecular forms of each AChE contribute to the functional diversification of AChEs. Several structurally distinct forms of AChE, which can be differentiated by the number and types of subunits, have been reported in both vertebrates and invertebrates [22], [25]–[29]. Insect AChE exists in three different molecular forms. The main native form is an amphiphilic dimer (G2m) that is attached to the plasma membrane via a glycophosphatidylinositol (GPI)-anchor [22], [28], [29]. A hydrophilic water-soluble dimer (G2s) can be generated from the amphiphilic dimer by proteolysis. The third form is a monomer (G1) that is thought to originate from the reduction of the two dimers. In addition, in D. melanogaster, the 55 and 18 kDa components are generated by the proteolytic cleavage of a 75 kDa precursor of AChE [30].
The Western honey bee, Apis mellifera L, is the most important pollinator in natural and commercial agriculture [31], [32]. In the United States, honey bees work to pollinate over 90 varieties of fruits and vegetables, including apples, avocados, blueberries, cherries, citrus crops, vine crops and almonds; these crops are valued at more than $15 billion per year. Honey bees also produce approximately $150 million in honey annually [33]. Recently, the mysterious disappearance of honey bees, called colony collapse disorder (CCD), has been reported since 2006 in the United States [34], and the global economic costs of bee decline, including lower crop yields and increased production costs, have been estimated at as high as $75 billion per year [35]. Because honey bee colonies are constantly at risk of exposure to various pesticides, including OP and CB insecticides, exposure to these insecticides may be one of the factors contributing to pollinator decline and CCD [36].
To understand the toxicity of OP and CB insecticides against honey bees, it is essential to determine whether A. mellifera AChE1 (AmAChE1) or AChE2 (AmAChE2) is primarily responsible for synaptic function and serves as the major target of OP and CB insecticides. In this study, the tissue distribution patterns and molecular characteristics of the two AmAChEs were investigated by native-PAGE and Western blot analysis with AChE1- and AChE2-specific antibodies. Furthermore, we expressed the two AmAChEs in Sf9 cells with a baculovirus expression system and characterized their kinetic and inhibitory properties.
Materials and Methods
Insects
The colonies of the Western honey bee, A. mellifera, that were used as sources for experimental specimens were maintained at Seoul National University. According to their behaviors and ages [37], we collected forager bees, which were older than 3 weeks of age and returned to the hive with clearly visible pollen loads on their hind legs. The collected honey bees were frozen directly with liquid nitrogen and stored at −75°C until protein extraction.
Protein Sample Preparation
Soluble proteins were extracted from the forager heads with 0.1 M Tris-HCl (pH 7.8) buffer, while membrane-bound proteins were extracted with the same buffer supplemented with 0.5% Triton X-100. To observe the tissue distribution of AmAChE1 and AmAChE2, protein samples were prepared from six various tissues (ganglia, head, thorax, abdomen, leg, and gut) of forager bees with buffer containing Triton X-100. Proteins from various samples were extracted with an appropriate amount of buffer using a micro tissue grinder (Radnoti, Monrovia, CA, USA). The homogenates were centrifuged at 12,000×g for 15 min at 4°C. The supernatant was filtered through glasswool to remove excess lipid and stored at −75°C until use.
Antibody Generation, PAGE and Western blotting
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), native-PAGE, Western blotting and AChE activity staining were conducted as previously described with some modifications [22]. Electrophoresis was performed with a vertical electrophoresis unit (Novex® mini cell, Invitrogen, Carlsbad, CA, USA). Protein preparations from various tissues (20 µg) were separated by native-PAGE gel (7.5%) in triplicate at 120 V for 90 min in a cold chamber with a continuous Tris-glycine buffer system. The gel and running buffers contained 0.5% Triton X-100 (v/v). Following native-PAGE, one set of gels was stained for AChE activity according to previously described methods [38], while the remaining two sets of gels were analyzed by Western blotting as described below.
To determine the multimer formation of AmAChEs, 20 µg of protein samples extracted from forager heads were treated with or without 14 mM ß-mercaptoethanol and separated by SDS-PAGE (4–12% gradient gel). To investigate the anchor properties of AmAChEs, proteins extracted from forager heads (20 µg) were incubated with 0.13 U of phospholipase C (PIPLC) for 20 min at 20°C. Non-treated control samples were incubated in the absence of PIPLC. After digestion, protein samples were separated on a native-PAGE gel in triplicate. After PAGE, one set of gels was stained for AChE activity, while the remaining two sets of gels were analyzed by Western blotting; one set of gels was analyzed with an AChE1-specific antibody, while the other was analyzed with an AChE2-specific antibody, as described below.
Proteins separated on the gels were transferred to Hybond-N nitrocellulose membranes (GE Healthcare, Pittsburgh, PA, USA) by electroblotting. After blocking in PBS buffer containing 0.1% Tween-20 (PBST) and 5% fat-free dry milk for 1 h at room temperature, the nitrocellulose membrane sheets were incubated for 3 h at room temperature or overnight at 4°C with primary antibodies (anti-AChE1 or anti-AChE2) [22]. Anti-AChE1 and anti-AChE2 polyclonal antibodies were generated as described previously [22]. The membranes were then incubated with horseradish-peroxidase conjugated anti-rabbit IgG secondary antibody (Pierce Bio-Technology, Rockford, IL, USA) for 1 h. The antigen-antibody complex on the bands was visualized with a chemiluminescence kit according to the manufacturer's instructions (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
In vitro Expression of AmAChE1 and AmAChE2 with a Baculovirus Expression System
Total RNA was extracted from forager heads with TRI reagent (MRC, Cincinnati, OH, USA) as described by the manufacturer. Following extraction, the total RNA was treated with DNaseI (TAKARA Korea Biomedical Inc., Seoul, Korea) at 37°C for 30 min and concentrated with 3 M sodium acetate. First strand cDNA was synthesized from the DNaseI-treated total RNA with Superscript III reverse transcriptase (Invitrogen) at 55°C for 1 h by priming with oligo dT, and the RNA strand was then removed by incubation with RNase H (Invitrogen) at 37°C for 20 min. The complete cDNA fragments encoding A. mellifera ace1 and ace2 (AmAce1 and AmAce2, respective GenBank accession numbers XM393751 and AF213012) were amplified by Advantage Taq (Clontech, Palo Alto, CA, USA) with gene-specific primers (Table S1) and directly cloned into the pGEM®-T easy vector (Promega, Madison, MU, USA). Partial fragments of AmAce1 and AmAce2 with truncated C-terminal hydrophobic regions were amplified by ExTaq (Takara, Japan) at 95°C for 2 min, (95°C for 30 s, 65°C for 30 s, 72°C for 2 min) × 5 cycles, (95°C for 30 s, 60°C for 30 s, 72°C for 2 min) × 30 cycles and 72°C for 2 min with gene-specific primers containing restriction enzyme sites (Table S1) from respective full ORF clones. The amplified AmAce1 and AmAce2 DNA fragments were digested with XbaI and SacI (Koschem, Korea) and inserted into pBacPAK8 (Clontech) that had been digested with the same restriction endonucleases. Recombinant baculoviruses expressing AmAChE1 and AmAChE2 in SF9 cells were generated as described previously [22]. Virus-infected cells were incubated for 84 h at 27°C. Protein samples were collected by centrifugation and concentrated with an Ultra Amicon YM-30 (Millipore, Bedford, MA, USA). Protein concentrations were determined by the Bradford method with bovine serum albumin as the standard protein [39]; the proteins were then stored at −75°C until use.
Kinetics and Inhibition of AmAChEs
The enzyme assay for AmAChEs expressed in Sf9 cells was performed with 8 different concentrations (0.05 to 1 mM) of acetylthiocholine iodide (ATChI) and butyrylthiocholine iodine (BTChI) according to previously described methods with some modifications [40], [41]. To measure enzyme kinetics, 15 µl of culture supernatant containing 5 µg of one of the two AmAChEs as enzyme sources was added to each well containing 85 µl of substrate mixture in the presence of 0.4 mM 5,5'-dithiobis-(2-nitrobenzoic acid) (DTNB); the wild type virus was used as a blank control. The reaction was monitored at 412 nm for 5 min with 10-sec intervals with a Soft Max® Pro5 microplate reader (Molecular Devices, Menlo Park, CA) at 30°C. Michaelis-Menten constants (Km) and maximal velocity (Vmax) values for each substrate were determined by Lineweaver-Burk plot.
AmAChE inhibition was assayed at 7 different concentrations (10−9 to 10−3 M) of each of three inhibitors (BW284C51, eserine and Iso-OMPA), three organophosphates (DDVP, malaoxon and paraoxon) and four carbamates (aldicarb, carbaryl, carbofuran and propoxur), according to previously described methods with some modifications [42]. Due to high sensitivity of AmAChEs to chlorpyrifos-oxon, 10−12 to 10−6 M of chlorpyrifos-oxon was used for inhibition assay. Five µg of each AmAChE as enzyme source was added to each well containing the substrate mixture of 1 mM ATChI and 0.4 mM DTNB and various concentrations of inhibitors to initiate enzyme reaction. Upon initiation of reaction, AmAChE activities were recorded at 412 nm for 3 min with 10-sec intervals using a microplate reader as above. To investigate the scavenger effects of AmAChE1 on the inhibition of AmAChE2, an AmAChE mixture was inhibited with 7 different concentrations of each of two OP (chlorpyrifos -oxon and malaoxon) and CB (carbofuran and propoxur) according to previously described methods with some modification [43]; the molar ratio of AmAChE1:AmAChE2 was 2∶1 (4∶2 µg) based on the their expressed protein levels in forager bees. The inhibitors were pre-incubated with AmAChE1 for 30 min at 30°C, and AmAChE2 was then added. The same molar amount of BSA instead of AmAChE1 was used as a control. AmAChE activities in the presence of 1 mM ATChI, 0.4 mM DTNB and various concentrations of inhibitors were recorded as above. Five consecutive concentrations of each inhibitor exhibiting a relatively linear range of inhibition were selected for the calculation of median inhibition concentration (IC50). The IC50 for each inhibitor was determined based on log-concentration versus probit (% inhibition) regression analysis using SPSS for Windows version 20.0 K (SPSS Inc., Chicago, IL).
3D Structure Modeling, Hydrophobicity and GPI-anchor Prediction
The 3D structure analyses of AmAChE1 and AmAChE2 were performed with the Automated Comparative Protein Modeling Server of SWISS-MODEL (http://swissmodel.expasy.org/) using human BChE and D. melanogaster AChE as the respective templates. Structure comparisons between AmAChEs were performed with UCSF Chimera MatchMaker ver. 1.4 (University of California, CA). The models were visualized and modified with Swiss PDB viewer 4.0.1 (Swiss Institute of Bioinformatics, Lausanne, Switzerland). The hydrophobicities of AmAChE1 and AmAChE2 were predicted with ProtScale of the ExPASy Proteomics Server (http://expasy.org/cgi-bin/protscale.pl). The potential GPI-anchor sequences of two AmAChEs were predicted with GPI-SOM of the ExPASy Proteomics Server (http://gpi.unibe.ch/).
Results
Expression Patterns of AmAChEs in Various Tissues
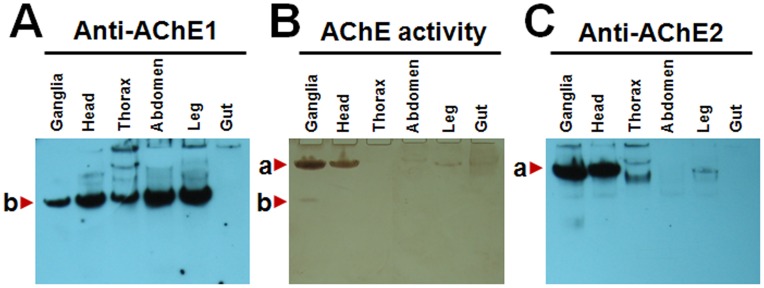
To determine the tissue-specific expression profiles of two AmAChEs, native-PAGE was performed on proteins extracted from six tissues (thoracic ganglia, head, thorax, abdomen, legs and gut) of forager bees, and their AChE activities were visualized by activity staining (Fig. 1B). The AmAChEs were concentrated in the ganglia and heads containing the central nervous system (CNS) (band ‘a’), but little activity was detected in the peripheral nervous system (PNS) such as in the thorax, abdomen, legs and gut.
Figure 1. Tissue distribution of AmAChE1 and AmAChE2 as assessed by native polyacrylamide gel electrophoresis and Western blot analysis.
Protein samples (20 µg) from various tissues were loaded onto a 7.5% polyacrylamide gel and run for 90 min at 120 V. After electrophoresis, one gel was stained for activity with acetylthiocholine iodide as a substrate (B). The other gels were analyzed by Western blot with anti-AChE1 (A) or anti-AChE2 (C) polyclonal antibodies.
Western blot analysis revealed that the main AChE activity is actually associated with AmAChE2 (Fig. 1C, band ‘a’), and AmAChE2 was more predominantly expressed in the CNS than in other tissues. By contrast, although AmAChE1 activity was faintly detected in ganglia (Fig. 1B, band ‘b’), AmAChE1 was not detectable in other tissues. Nevertheless, AmAChE1 seemed to be abundantly expressed not only in the CNS but also in the PNS such as in the thorax, abdomen and legs, as judged by Western blot analysis (Fig. 1A, band ‘b’).
Molecular Characterization of AmAChEs
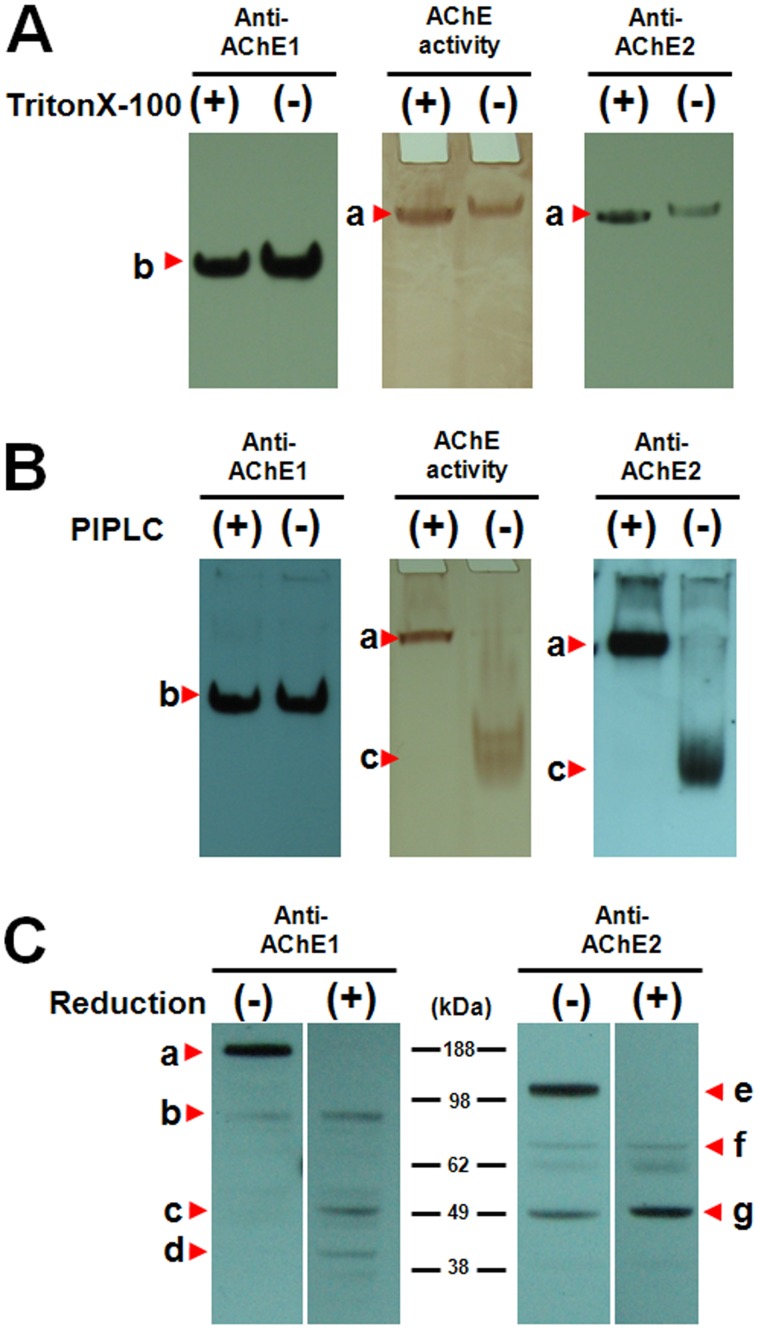
Crude protein was extracted from forager heads with 0.1 M Tris-HCl buffer in the presence or absence of Triton X-100 to determine the molecular formations and soluble nature of AmAChE1 and AmAChE2. When the protein was extracted with Triton X-100-containing buffer, both AmAChE1 and AmAChE2 were strongly detected (Fig. 2A, see the Triton X-100 (+) lanes). In the absence of Triton X-100, the AmAChE1 band was still clearly observed, while AmAChE2 was faintly detected (Fig. 2A, see the Triton X-100 (−) lanes), suggesting the membrane-anchored nature of AmAChE2. To confirm the membrane-anchored properties of AmAChEs, AmAChEs extracted from forager heads were digested with PIPLC. After PIPLC treatment, no molecular changes were observed in AmAChE1, whereas amphiphilic AmAChE2 was completely converted to the hydrophilic form (Fig. 2B). This result indicates that AmAChE2 is associated with the membrane via a GPI-anchor, while AmAChE1 is present in a soluble form due to the absence of a GPI-anchor, as indicated by PIPLC treatment as well as protein extraction in the absence of Triton X-100.
Figure 2. Molecular characterization of AmAChE1 and AmAChE2 by polyacrylamide gel electrophoresis and Western blot analysis.
Protein samples were extracted from the heads of forager honey bees with 0.1 M Tris-HCl buffer in the presence or absence of 0.5% Triton X-100 to determine the soluble nature of the AmAChEs (A). To investigate the GPI-anchor properties of the AmAChEs, protein samples were treated with PIPLC (B). Protein samples were mixed with or without β-mercaptoethanol and separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis to determine the multimer formation of AmAChE1 and AmAChE2 (C).
The molecular masses of AmAChEs were analyzed by SDS-PAGE in the presence or absence of β-mercaptoethanol reduction (Fig. 2C). Under non-reducing conditions, bands of approximately 185 kDa (band ‘a’) and 140 kDa (band ‘e’) were strongly visualized as putative dimers, while bands of 88 kDa (band ‘b’) and 79 kDa (band ‘f’) were faintly detected as putative monomers by the respective AmAChE1- and AChE2-specific antibodies (see reduction (-) lanes). In addition to the putative dimeric form, a band of approximately 50 kDa representing cleaved monomer (band ‘g’) was also clearly detected for AmAChE2. This finding suggests that dimers are the predominant forms of both AmAChEs under native conditions and that the cleaved monomer is also abundantly present in AmAChE2. After reduction with β-mercaptoethanol, the relative quantities of the other forms, such as the monomers (88 kDa band ‘b’ for AmAChE1 versus 70 kDa band ‘f’ for AmAChE2) and cleaved forms (48 kDa band ‘c’ and 40 kDa band ‘d’ for AmAChE1 vs. 50 kDa band ‘g’ for AmAChE2), increased, while the dimeric forms of both AmAChEs disappeared, supporting the presence of a disulfide bond in the dimer conformation. As indicated by the amino acid sequence alignment with Drosophila and German cockroach AChEs (Fig. S1B), the presence of cysteine at the C-terminal region of several insect AChEs also supports the role of a disulfide bond connection in dimer formation for both AmAChE1 and AmAChE2.
Kinetic Properties of AmAChEs
As judged by Western blotting with AChE1- and AChE2-specific antibodies, two types of AmAChEs were successfully expressed with the recombinant baculovirus expression system. In vitro expressed AmAChE2 demonstrated strong AChE activity, whereas AmAChE1 activity was barely detectable, as observed in the protein samples from the heads of forager honey bees (Fig. S2A). In addition, the estimated molecular masses of both AmAChEs (approximately 70 kDa) were confirmed by Western blotting following SDS-PAGE (Fig. S2B).
Two cholinesterase substrates, ATChI and BTChI, were used to study the kinetic properties of the two AmAChEs. The Km and Vmax values were calculated by double-reciprocal plots (Fig. S3). The kinetic parameters for the two substrates are presented in Table 1. AmAChE1 and AmAChE2 exhibited 3.8- and 4-fold higher catalytic efficiency [Maximal velocity/Michaelis-Menten constants (Vmax/Km)], respectively, toward ATChI than BTChI, confirming the typical substrate specificities of AChEs. In a cross-enzyme comparison of AmAChE1 and AmAChE2, AmAChE2 showed lower Km values than AmAChE1 (1.28- and 12.5-fold for ATChI and BTChI, respectively), demonstrating higher substrate affinity compared to AmAChE1. AmAChE2 also exhibited approximately 200-fold higher Vmax values for both ATChI and BTChI than did AmAChE1. Taken together, AmAChE2 exhibited approximately 2,500- and 2,400-fold higher catalytic efficiencies toward ATChI and BTChI, respectively, than did AmAChE1, indicating that AmAChE2 is a much more efficient enzyme than AmAChE1. The ratio Vmax(BTChI)/Vmax(ATChI) was 0.51 and 0.5 for AmAChE1 and AmAChE2, respectively, indicating that the substrate spectrums of AmAChE1 and AmAChE2 were similar.
Table 1. Kinetic properties of recombinant two AmAChEs in hydrolyzing various substrates*.
| Substrate | Kinetic property | AmAChE1 | AmChE2 | Ratio† |
| ATChI | Vmax(mM/min/mg protein) | 0.308±0.081 | 60.8±0.686 | 197 |
| Km (mM) | 1.14±0.381 | 0.089±0.011 | 0.078 | |
| Vmax/Km (Ratio) | 0.269 | 684 | 2543 | |
| BTChI | Vmax (mM/min/mg protein) | 0.157±0.029 | 30.4±3.91 | 194 |
| Km (mM) | 2.23±0.279 | 0.179±0.029 | 0.08 | |
| Vmax/Km (Ratio) | 0.07 | 170 | 2429 | |
| Substrate specificity | Vmax (BTChI)/Vmax (ATChI) | 0.51 | 0.5 |
Results are reported as the mean ± SD (n = 3).
AmAce2/AmAce1.
Inhibitory Properties of AmAChEs
The inhibitory properties of AmAChEs were determined with various concentrations of three reversible cholinesterase-specific inhibitors, four OPs and four CBs (Fig. S4 and Table 2). Both AmAChEs were effectively inhibited by BW284C51 and eserine but not by Iso-OMPA, a BChE-specific inhibitor, suggesting that both enzymes retain typical features of AChEs [44]. BW284C51 similarly inhibited both AmAChEs, while AmAChE2 was 16-fold more sensitive to eserine than was AmAChE2, as judged by IC50 values. In the inhibition assay with OPs and CBs, AmAChE2 was, in general, much more sensitive to these insecticides than was AmAChE1 (Table 2). As indicated by the IC50 values of the OPs, AmAChE2 was approximately 4-, 7-, 3- and 45-fold more sensitive to chlorpyrifos, DDVP, malaoxon and paraoxon, respectively. The CBs also inhibited AmAChE2 more effectively than did AmAChE1 (3,500-, 19- and 3-fold for carbaryl, carbofuran and propoxur, respectively), whereas neither AmAChE was inhibited by aldicarb. In a cross-inhibitor comparison of OPs and CBs, OPs generally exhibited much lower IC50 values than CBs, suggesting that OPs are more efficient inhibitors of both AmAChE1 and AmAChE2 than CBs.
Table 2. IC50 (M) values of different inhibitors of two recombinant AmAChEs*.
| Inhibitor | AmAce1 | AmAce2 | Ratio of IC50 † |
| BW284C51 | (4.33±0.58) × 10−9 | (2.67±0.21) × 10−9 | 1.68 |
| Eserin | (1.73±0.41) × 10−5 | (1.09±0.03) × 10−6 | 15.89 |
| Iso_OMPA | n/a | n/a | n/a |
| Chlorpyrifos oxon | (6.12±0.25) × 10−7 | (1.53±0.19) × 10−7 | 3.99 |
| DDVP | (3.53±0.15) × 10−4 | (5.03±0.40) × 10−5 | 7.01 |
| Malaoxon | (9.19±0.43) × 10−6 | (3.26±0.29) × 10−6 | 2.82 |
| Paraoxon | (4.58±0.11) × 10−5 | (1.01±0.04) × 10−6 | 45.4 |
| Aldicarb | n/a | n/a | n/a |
| Carbaryl | (1.20±0.51) × 10−3 | (3.43±0.69) × 10−7 | 3.49 × 103 |
| Carbofuran | (1.69±0.24) × 10−5 | (9.02±0.19) × 10−7 | 18.78 |
| Propoxur | (1.85±0.12) × 10−4 | (5.33±0.49) × 10−5 | 3.48 |
Results are reported as the mean ± SD (n = 3).
AmAce1/AmAce2.
Reduction of AmAChE2 Inhibition by the Presence of AmAChE1
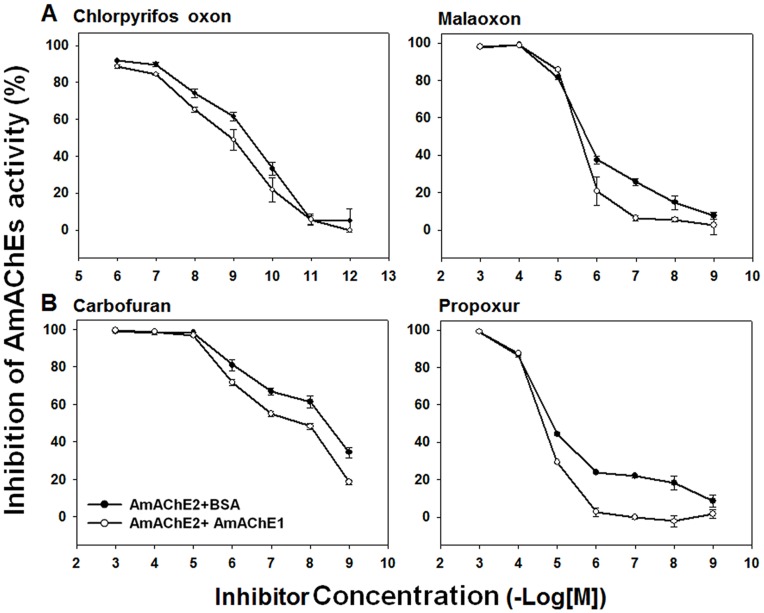
To examine the physiological function of AmAChE1, the inhibition rates of two OPs and two CBs against AmAChE2, the major catalytic enzyme in the honey bee, were measured in the presence or absence of AmAChE1 (Fig. 3). As judged by the IC50 values (Table S2), the overall inhibition was reduced 2.3- to 4.5-fold when inhibitors were pre-incubated with AmAChE1 and then added to AmAChE2 compared to pre-incubation with BSA, suggesting that one of the physiological functions of AmAChE1 may be chemical defense against xenobiotics, perhaps by sequestering inhibitors.
Figure 3. Inhibition of AmAChE2 by two organophosphates (A) and two carbamates (B) in the presence (○) or absence of AmAChE1 (.
•). Inhibitors were pre-incubated with AmAChE1 for 30 min at 30°C, and AmAChE2 was then added. The same molar amount of BSA in place of AmAChE1 was used as a control.
Three-dimensional Structure Modeling
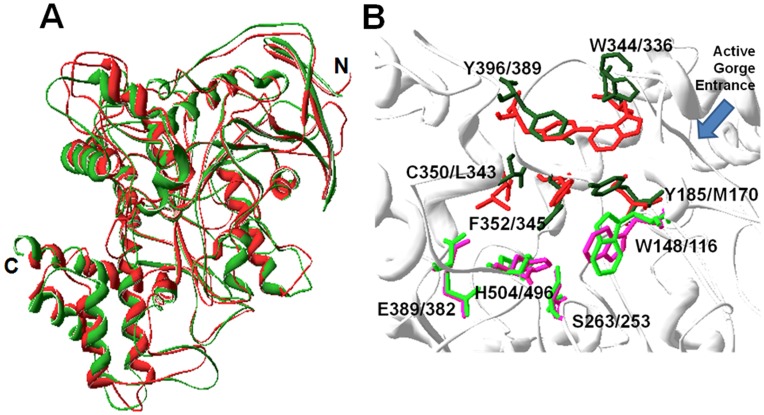
The 3D structures of AmAChE1 and AmAChE2 were predicted with Swiss Model using the structures of human BChE and D. melanogaster AChE as the respective templates (Fig. 4). In the structure comparison between AmAChE1 and AmAChE2, most of the α-helix and β-stranded sheets were highly overlapped, demonstrating that the overall structures of the two AmAChEs were similarly folded (Fig. 4A). When the catalytic gorge structures of the two AmAChEs were merged, the choline-binding site (W148) and the catalytic triad (S263, E389 and H504) of AmAChE1 closely overlapped those of AmAChE2 (Fig. 4B). However, the shape of the peripheral anionic site (PAS) changed because of differences in two amino acid residues: Y185 in AmAChE1 versus M170 in AmAChE2 and C350 in AmAChE1 versus L343 in AmAChE2. In addition, the angles of the choline-binding site (W344/336) at the entrance to the active gorge in AmAChE1 and AmAChE2 differed by approximately 90°, suggesting that the differences in the PAS conformation may be responsible for the differences in the substrate and inhibition kinetics of AmAChE1 and AmAChE2.
Figure 4. Comparison of the predicted three-dimensional (3D) structures of AmAChE1 (green color) and AmAChE2 (red color).
The superimposed 3D structures (A) and zoom-in views (B) of the two AmAChEs were compared. The positions of amino acid residues involved in the formation of the catalytic triads, choline-binding sites and peripheral anionic sites are indicated (AmAChE1/AmAChE2) (B).
Discussion
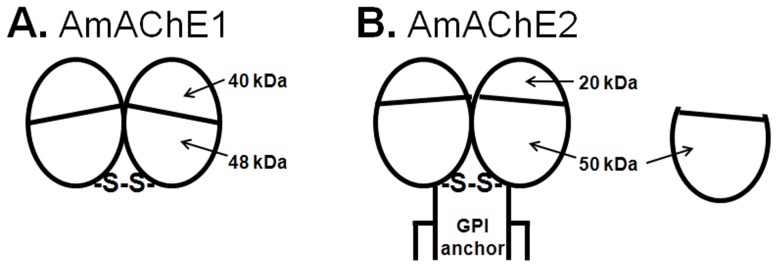
In this study, we determined the molecular polymorphisms of AmAChEs and the identity of the major catalytic enzyme by native-PAGE and Western blot analyses with AChE1- and AChE2-specific antibodies. The most predominant molecular form of both AmAChEs was a dimer formed by an intersubunit disulfide bridge (Fig. 2C), as observed in Drosophila [30], German cockroach [22] and Torpedo AChEs [45]. In addition, amphiphilic dimers of AmAChE2 were completely converted to hydrophilic dimers by PIPLC treatment, whereas the molecular migration pattern of AmAChE1 was unchanged (Fig. 2B). According to the hydrophobicity plot (Fig. S1A) and GPI-anchor prediction (Fig. S1B), AmAChE2 is predicted to have GPI-anchor sequences at the hydrophobic C-terminal region. By contrast, no GPI-anchor sequence was predicted in the AmAChE1 sequence, and the C-terminal region of AmAChE1 is predicted to be hydrophilic, supporting the soluble and membrane-anchored properties of AmAChE1 and AmAChE2, respectively. Based on the molecular characteristics of AmAChE1 and AmAChE2, schematic models of the structures of these enzymes were generated (Fig. 5). The monomer of both AmAChE1 and AmAChE2 is composed of two small subunits (40 and 48 kDa for AmAChE1 vs. 20 and 50 kDa for AmAChE2), and two monomers are linked by a disulfide bond at the C-terminal regions. AmAChE1 predominantly exists as a soluble dimer, while the most abundant molecular form of AmAChE2 appeared to be attached to the membrane via a GPI-anchor. In addition, a small fraction of AmAChE2 is present as a 50-kDa subunit under native conditions (Fig. 5B).
Figure 5. Schematic diagrams of the molecular structures of AmAChE1 (A) and AmAChE2 (B).
One type of AmAChE1 is predominately expressed (A), while two different molecular polymorphisms of AmAChE2 were abundantly observed (B).
Contrary to the general idea that AChE1 is the major synaptic enzyme in most insects having both AChE1 and AChE2 studied to date [19], [22], AmAChE2 appears to function as the main synaptic enzyme in the honey bee, as judged by the migration patterns of the AChE activity bands and the protein bands detected by Western blotting. In addition, the distribution of AmAChE2 was greater in tissues associated with the CNS, such as the ganglia and head, while AmAChE1 was strongly detected not only in the CNS but also in the PNS/non-neuronal tissues, such as the thorax, abdomen and legs (Fig 1). In Drosophila [1], [29], [46], the membrane-bound form of AChE exhibits greater AChE activity than does the soluble AChE. Moreover, the AChE activities of the fruit fly and house fly were reduced after treatment with PIPLC or β-mercaptoethanol to convert membrane-bound AChE to the soluble form [16], [28]. Likewise, as judged by native-PAGE in conjunction with Western blotting, the enzymatic activity of membrane-bound AmAChE2 is much greater than that of the soluble AmAChE1, which has negligible enzymatic activity. In fact, a comparison of the catalytic properties of in vitro-expressed AmAChE1 and AmAChE2 also strongly confirmed that AChE activity in the honey bee primarily originates from AmAChE2, which possesses approximately 2,500-fold higher catalytic efficiency than AmAChE1 (Table 1). Furthermore, in the inhibition assays, AmAChE2 was much more sensitive to inhibitors in general, as indicated by the IC50 values (Table 2), implying that the major target molecule of OP and CB insecticides is AmAChE2, which functions in neurotransmission. Compared to other insecticides, chlropyrifos-oxon exhibited much lower IC50 values to AmAChE2 (Table 2). Interestingly, it was reported that LD50 value of chlorpyrifos to the honey bee was generally much lower than other insecticides [32]. Taken together, the high sensitivity of AmAmChE2 to chlorpyrifos-oxon may be mainly responsible for the high toxicity of chlorpyrifos to the honey bee. In a previous study, the AmAChE responsible for the majority of AChE activity was cloned, and its expression pattern was evaluated as a function of the worker bee’s development state, although the major AChE in the honey bee was not assigned to one of the two different types of AChEs [47]. The AmAChE cDNA sequence (GenBank accession number AF213012) used in that study is the same as that of AChE2, also supporting that A. mellifera expresses AChE2 as the major catalytic enzyme.
As shown by 3D structure comparison, the catalytic triad conformations of the two AmAChEs were similar, with the exception of some predicted structural differences at the active gorge entrance (Fig. 4). Most notably, the aromatic side chain angles of the choline-binding sites (W344 in AmAChE1 vs. W336 in AmAChE2) differed by approximately 90°. In addition, Y185 and C350 in AmAChE1 were replaced by M170 and L343 in AmAChE2, respectively, altering the topology of the gorge entrance. These amino acid replacements were highly similar to those of B. germanica, and the catalytic efficiency of cockroach AChE2 was also higher than that of AChE1 [22]. As observed in B. germanica, these conformational differences between the two AmAChEs appear to be responsible for the kinetic and inhibitory properties.
Considering that a large fraction of AmAChE1 exists as a soluble form in the PNS (Fig. 1) and the enzymatic activity of AmAChE1 is lower than that of AmAChE2 (Table 1), AmAChE1 seems to play other non-neuronal functions, in contrast to AmAChE2. As in the case of vertebrate BChE [48] and nematode AChE [43], we investigated the hypothesis that AmAChE1 plays a role in chemical defense against xenobiotics. When OPs and CBs were pre-incubated with AmAChE1 for 30 min prior to the addition of AmAChE2, the inhibition of AmAChE2 by the insecticides was reduced significantly (Fig. 3), implying that AmAChE1 functions as a ubiquitous sequestration protein rather than as a specific hydrolase by binding nonspecifically to a variety of xenobiotics, particularly OP and CB insecticides. This physiological property of AmAmChE1 highly resembles that of pinewood nematode AChE3 (BxAChE3) [43]. Among three BxAChEs, BxAChE3 showed the highest expression level and lowest catalytic efficiency toward substrates [49]. In addition, a significant reduction in the inhibition of AChE by insecticides was observed in the presence of BxAChE3, indicating that BxAChE3 protects BxAChE1 and BxAChE2 [43] from potential inhibitors. The bio-scavenging function of ChE has been broadly investigated in vertebrate BChEs. In pigs and monkeys, administration of BChE increases LD50 values against nerve agents, demonstrating that BChE can function as a biological scavenger to provide significant protection against the behavioral and lethal effects of nerve agent intoxication before these agents arrive at their target sites [50]–[52]. This soluble enzyme is also abundantly observed in non-neuronal tissues, such as plasma, liver, lung and intestine [53]. These results support the physiological function of soluble AmAChE1 as a bio-scavenger that is predominantly expressed in both neuronal (most likely the extracellular space in the blood-brain barrier) and non-neuronal tissues (most likely the hemocoel) without high catalytic efficiency. In fact, although the honey bee is considered to be particularly sensitive to insecticides, it was no more sensitive than other insect species to various insecticides including CBs, OPs, nicotinoids, organochlorines, pyrethroids and others [32], which may be due to the bio-scavenging function of AmAChE1. In addition to such putative bio-scavenging function, a recent study suggested that a non-neuronal AChE is likely involved in various physiological functions, such as female reproduction, embryo development and growth of offspring in Tribolium castaneum [23], [24].
In summary, AmAChE2 was present at high concentrations in the CNS, such as the head and ganglia, and has high catalytic efficiency. Based on its tissue distribution, membrane-anchoring properties and high catalytic efficiency, AmAChE2 appears to play a major role in postsynaptic transmission. Inhibition studies with OPs and CBs demonstrated that AmAChE2 is more sensitive to inhibitors than AmAChE1, supporting the role of AmAChE2 as the major target molecule of OP and CB insecticides. Although AmAChE1 has low enzyme activity, soluble dimeric AmAChE1 was also abundant in non-neuronal tissues. In particular, the ability of AmAChE1 to reduce the inhibition of AmAChE2 by insecticides strongly suggests a physiological function of AmAChE1 as a bio-scavenger that provides a chemical defense against xenobiotics.
Supporting Information
Hydrophobicity prediction (A) and GPI-anchor prediction (B) of AmAChE1 and AmAChE2. Red ovals indicate the C-terminal region of each of the AmAChEs (A). The predicted GPI-anchor sites and the cysteine residues that form the disulfide bond in each of the AChEs are represented by red arrows and red circles, respectively (B).
(TIF)
Expression of recombinant AmAChE1 and AmAChE with a baculovirus expression system. The expression of AmAChE1 and AmAChE2 was confirmed by AChE activity staining and Western blotting following native polyacrylamide gel electrophoresis (A) and sodium dodecyl sulfate polyacrylamide gel electrophoresis (B).
(TIF)
Double-reciprocal plots for AmAChE1 (A) and AmAChE2 (B) for the calculation of Km and Vmax. The straight lines were generated by plotting 1/v versus 1/[S]; the slope indicates Km/Vmax, and the intercept of the x-axis (1/[S]) indicates −1/Km.
(TIF)
Inhibition of AmAChE1 (•) and AmAChE2 (○) by cholinesterase-specific inhibitors (A), organophosphates (B) and carbamates (C). The results are the mean of three determinations (n = 3). Vertical bars indicate standard deviations.
(TIF)
Primers used for the in vitro expression of AmAChE1 and AmAChE2 with a baculovirus expression system.
(DOCX)
IC50 (M) values of different inhibitors of recombinant AmAChE2 with or without pre-incubation with AmAChE1.
(DOCX)
Acknowledgments
The authors thank Prof. Y.H. Je and Dr. J.Y. Choi for their technical assistance to generate recombinant baculovirus.
Funding Statement
This research was supported by the Grant (2012R1A1A2007260) from the Basic Science Research Program through the National Research Foundation of Korea (NRF), which is funded by the Ministry of Education, Science and Technology, and the Grant (308024-05-3-SB010) from the Korea Institute of Planning and Evaluation for Technology of Food. Deok Jea Cha was supported in part by the Brain Korea 21 program. Je Won Jung and Hyung Wook Kwon were supported in part by WCU (World Class University) program through the Korea Science and Engineering Foundation funded by the National Research Foundation of Korea (R31-10056).
References
- 1. Toutant JP, Arpagaus M, Fournier D (1988) Native molecular forms of head acetylcholinesterase from adult Drosophila melanogaster - Quaternary structure and hydrophobic character. J Neurochem 50: 209–218. [DOI] [PubMed] [Google Scholar]
- 2. Fournier D, Bride JM, Hoffmann F, Karch F (1992) Acetylcholinesterase. Two types of modifications confer resistance to insecticide. J Biol Chem 267: 14270–14274. [PubMed] [Google Scholar]
- 3. Fournier D, Mutero A (1994) Modification of acetylcholinesterase as a mechanism of resistance to insecticides. Comp Biochem Phys C 108: 19–31. [Google Scholar]
- 4. Smissaert HR (1964) Cholinesterase inhibition in spider mites susceptible + resistant to organophosphate. Science 143: 129–131. [DOI] [PubMed] [Google Scholar]
- 5. Raymond M, Fournier D, Bride JM, Cuany A, Berge J, et al. (1986) Identification of resistance mechanisms in Culex pipiens (Diptera, Culicidae) from Southern France - Insensitive acetylcholinesterase and detoxifying oxidases. J Econ Entomol 79: 1452–1458. [DOI] [PubMed] [Google Scholar]
- 6. Hemingway J, Small GJ, Monro AG (1993) Possible mechanisms of organophosphorus and carbamate insecticide resistance in German cockroaches (Dictyoptera, Blattelidae) from different geographical areas. J Econ Entomol 86: 1623–1630. [DOI] [PubMed] [Google Scholar]
- 7. Argentine JA, Zhu KY, Lee SH, Clark JM (1994) Biochemical mechanisms of Azinphosmethyl resistance in isogenic strains of Colorado potato beetle. Pestic Biochem Phys 48: 63–78. [Google Scholar]
- 8. Toutant JP (1989) Insect acetylcholinesterase - Catalytic properties, tissue distribution and molecular forms. Prog Neurobiol 32: 423–446. [DOI] [PubMed] [Google Scholar]
- 9. Pezzementi L, Chatonnet A (2010) Evolution of cholinesterases in the animal kingdom. Chem-Biol Interact 187: 27–33. [DOI] [PubMed] [Google Scholar]
- 10. Li F, Han ZJ (2002) Purification and characterization of acetylcholinesterase from cotton aphid (Aphis gossypii Glover). Arch Insect Biochem 51: 37–45. [DOI] [PubMed] [Google Scholar]
- 11. Gao JR, Kambhampati S, Zhu KY (2002) Molecular cloning and characterization of a greenbug (Schizaphis graminum) cDNA encoding acetylcholinesterase possibly evolved from a duplicate gene lineage. Insect Biochemisty and Molecular Biolology 32: 765–775. [DOI] [PubMed] [Google Scholar]
- 12. Baek JH, Kim JI, Lee DW, Chung BK, Miyata T, et al. (2005) Identification and characterization of ace1-type acetylcholine sterase likely associated with organophosphate resistance in Plutella xylostella . Pestic Biochem Phys 81: 164–175. [Google Scholar]
- 13. Kim JI, Jung CS, Koh YH, Lee SH (2006) Molecular, biochemical and histochemical characterization of two acetylcholinesterase cDNAs from the German cockroach Blattella germanica . Insect Mol Biol 15: 513–522. [DOI] [PubMed] [Google Scholar]
- 14. Huchard E, Martinez M, Alout H, Douzery EJP, Lutfalla G, et al. (2006) Acetylcholinesterase genes within the Diptera: takeover and loss in true flies. P R Soc B 273: 2595–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weill M, Fort P, Berthomieu A, Dubois MP, Pasteur N, et al. (2002) A novel acetylcholinesterase gene in mosquitoes codes for the insecticide target and is non-homologous to the ace gene in Drosophila . P Roy Soc Lond B Bio 269: 2007–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fournier D, Berge JB, Dealmeida MLC, Bordier C (1988) Acetylcholinesterases from Musca domestica and Drosophila melanogaster brain are linked to membranes by a glycophospholipid anchor sensitive to an endogenous phospholipase. J Neurochem 50: 1158–1163. [DOI] [PubMed] [Google Scholar]
- 17. Lee DW, Kim SS, Shin SW, Kim WT, Boo KS (2006) Molecular characterization of two acetylcholinesterase genes from the oriental tobacco budworm, Helicoverpa assulta (Guenee). Bba-Gen Subjects 1760: 125–133. [DOI] [PubMed] [Google Scholar]
- 18. Lee SW, Kasai S, Komagata O, Kobayashi M, Agui N, et al. (2007) Molecular characterization of two acetylcholinesterase cDNAs in Pediculus human lice. J Med Entomol 44: 72–79. [DOI] [PubMed] [Google Scholar]
- 19. Seong KM, Kim YH, Kwon DH, Lee SH (2012) Identification and characterization of three cholinesterases from the common bed bug, Cimex lectularius . Insect Mol Biol 21: 149–159. [DOI] [PubMed] [Google Scholar]
- 20. Nabeshima T, Mori A, Kozaki T, Iwata Y, Hidoh O, et al. (2004) An amino acid substitution attributable to insecticide-insensitivity of acetylcholinesterase in a Japanese encephalitis vector mosquito, Culex tritaeniorhynchus . Biochem Bioph Res Co 313: 794–801. [DOI] [PubMed] [Google Scholar]
- 21. Weill M, Malcolm C, Chandre F, Mogensen K, Berthomieu A, et al. (2004) The unique mutation in ace-1 giving high insecticide resistance is easily detectable in mosquito vectors. Insect Mol Biol 13: 1–7. [DOI] [PubMed] [Google Scholar]
- 22. Kim YH, Choi JY, Je YH, Koh YH, Lee SH (2010) Functional analysis and molecular characterization of two acetylcholinesterases from the German cockroach, Blattella germanica . Insect Mol Biol 19: 765–776. [DOI] [PubMed] [Google Scholar]
- 23. Lu YH, Pang YP, Park Y, Gao XW, Yao JX, et al. (2012) Genome Organization, Phylogenies, Expression Patterns, and Three-Dimensional Protein Models of Two Acetylcholinesterase Genes from the Red Flour Beetle. PLoS One 7: e32288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu YH, Park Y, Gao XW, Zhang X, Yao JX, et al. (2012) Cholinergic and non-cholinergic functions of two acetylcholinesterase genes revealed by gene-silencing in Tribolium castaneum. Sci Rep-Uk 2: doi: 10.1038/srep00288. [DOI] [PMC free article] [PubMed]
- 25. Badiou A, Brunet JL, Belzunces LP (2007) Existence of two membrane-bound acetylcholinesterases in the honey bee head. Arch Insect Biochem 66: 122–134. [DOI] [PubMed] [Google Scholar]
- 26. Massoulie J, Bon S (1982) The Molecular forms of cholinesterase and acetylcholinesterase in Vertebrates. Annu Rev Neurosci 5: 57–106. [DOI] [PubMed] [Google Scholar]
- 27. Perrier A, Cousin X, Boschetti N, Haas R, Chatel JM, et al. (2000) Two distinct proteins are associated with tetrameric acetylcholinesterase on the cell surface. Mol Biol Cell 11: 276–276. [DOI] [PubMed] [Google Scholar]
- 28. Fournier D, Cuany A, Bride JM, Berge JB (1987) Molecular polymorphism of head acetylcholinesterase from adult houseflies (Musca domestica L). J Neurochem 49: 1455–1461. [DOI] [PubMed] [Google Scholar]
- 29. Gnagey AL, Forte M, Rosenberry TL (1987) Isolation and characterization of acetylcholinesterase from Drosophila . J Biol Chem 262: 13290–13298. [PubMed] [Google Scholar]
- 30. Fournier D, Bride JM, Karch F, Berge JB (1988) Acetylcholinesterase from Drosophila melanogaster - Identification of 2 subunits encoded by the same gene. Febs Lett 238: 333–337. [DOI] [PubMed] [Google Scholar]
- 31. Allen-Wardell G, Bernhardt P, Bitner R, Burquez A, Buchmann S, et al. (1998) The potential consequences of pollinator declines on the conservation of biodiversity and stability of food crop yields. Conserv Biol 12: 8–17. [Google Scholar]
- 32. Hardstone MC, Scott JG (2010) Is Apis mellifera more sensitive to insecticides than other insects? Pest Manag Sci 66: 1171–1180. [DOI] [PubMed] [Google Scholar]
- 33.Sass J (2011) Why We need bees: Nature's tiny workers put food on our tables. Natural Resources Defense Council March.
- 34. Johnson RM, Evans JD, Robinson GE, Berenbaum MR (2009) Changes in transcript abundance relating to colony collapse disorder in honey bees (Apis mellifera). P Natl Acad Sci USA 106: 14790–14795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Swinton SM, Lupi F, Robertson GP, Hamilton SK (2007) Ecosystem services and agriculture: Cultivating agricultural ecosystems for diverse benefits. Ecol Econ 64: 245–252. [Google Scholar]
- 36. Frazier M, Mullin C, Frazier J, Ashcraft S (2008) What have pesticides got to do with it? Am Bee J 148: 521–523. [Google Scholar]
- 37. Wagener-Hulme C, Kuehn JC, Schulz DJ, Robinson GE (1999) Biogenic amines and division of labor in honey bee colonies. Journal of Comparative Physiology a-Sensory Neural and Behavioral Physiology 184: 471–479. [DOI] [PubMed] [Google Scholar]
- 38. Lewis PR, Shute CCD (1966) The distribution of cholinesterase in cholinergic neurons demonstrated with electron microscope. J Cell Sci 1: 381–390. [DOI] [PubMed] [Google Scholar]
- 39. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 40. Ellman GL, Courtney KD, Andres V Jr, Feather-Stone RM (1961) A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem Pharmacol 7: 88–95. [DOI] [PubMed] [Google Scholar]
- 41. Grant DF, Bender DM, Hammock BD (1989) Quantitative kinetic assays for glutathione S-transferase and general esterase in individual mosquitos using an Eia Reader. Insect Biochem 19: 741–751. [Google Scholar]
- 42. Zhu KY, Brindley WA (1992) Enzymological and inhibitory properties of acetylcholinesterase purified from Lygus hesperus Knight (Hemiptera, Miridae). Insect Biochem Molec 22: 245–251. [Google Scholar]
- 43. Kang JS, Lee DW, Koh YH, Lee SH (2011) A soluble acetylcholinesterase provides chemical defense against xenobiotics in the pinewood nematode. PLoS One 6: e19063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lotti M (1995) Cholinesterase inhibition - Complexities in interpretation. Clin Chem 41: 1814–1818. [PubMed] [Google Scholar]
- 45. Macpheequigley K, Vedvick TS, Taylor P, Taylor SS (1986) Profile of the disulfide bonds in acetylcholinesterase. J Biol Chem 261: 3565–3570. [PubMed] [Google Scholar]
- 46. Zador E (1989) Tissue specific expression of the acetylcholinesterase gene in Drosophila melanogaster . Mol Gen Genet 218: 487–490. [DOI] [PubMed] [Google Scholar]
- 47. Shapira M, Thompson CK, Soreq H, Robinson GE (2001) Changes in neuronal acetylcholinesterase gene expression and division of labor in honey bee colonies. J Mol Neurosci 17: 1–12. [DOI] [PubMed] [Google Scholar]
- 48. Lockridge O, Masson P (2000) Pesticides and susceptible populations: people with butyrylcholinesterase genetic variants may be at risk. Neurotoxicology 21: 113–126. [PubMed] [Google Scholar]
- 49. Kang JS, Lee DW, Choi JY, Je YH, Koh YH, et al. (2011) Three acetylcholinesterases of the pinewood nematode, Bursaphelenchus xylophilus: Insights into distinct physiological functions. Mol Biochem Parasit 175: 154–161. [DOI] [PubMed] [Google Scholar]
- 50. Lenz DE, Maxwell DM, Koplovitz I, Clark CR, Capacio BR, et al. (2005) Protection against soman or VX poisoning by human butyrylcholinesterase in guinea pigs and cynomolgus monkeys. Chem-Biol Interact 157: 205–210. [DOI] [PubMed] [Google Scholar]
- 51. Cerasoli DM, Griffiths EM, Doctor BP, Saxena A, Fedorko JM, et al. (2005) In vitro and in vivo characterization of recombinant human butyrylcholinesterase (Protexia (TM)) as a potential nerve agent bioscavenger. Chem-Biol Interact 157: 363–365. [DOI] [PubMed] [Google Scholar]
- 52. Broomfield CA, Maxwell DM, Solana RP, Castro CA, Finger AV, et al. (1991) Protection by butyrylcholinesterase against organophosphorus poisoning in nonhuman-primates. J Pharmacol Exp Ther 259: 633–638. [PubMed] [Google Scholar]
- 53. Lazari O, Hussein AS, Selkirk ME, Davidson AJ, Thompson FJ, et al. (2003) Cloning and expression of two secretory acetylcholinesterases from the bovine lungworm, Dictyocaulus viviparus . Mol Biochem Parasit 132: 83–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hydrophobicity prediction (A) and GPI-anchor prediction (B) of AmAChE1 and AmAChE2. Red ovals indicate the C-terminal region of each of the AmAChEs (A). The predicted GPI-anchor sites and the cysteine residues that form the disulfide bond in each of the AChEs are represented by red arrows and red circles, respectively (B).
(TIF)
Expression of recombinant AmAChE1 and AmAChE with a baculovirus expression system. The expression of AmAChE1 and AmAChE2 was confirmed by AChE activity staining and Western blotting following native polyacrylamide gel electrophoresis (A) and sodium dodecyl sulfate polyacrylamide gel electrophoresis (B).
(TIF)
Double-reciprocal plots for AmAChE1 (A) and AmAChE2 (B) for the calculation of Km and Vmax. The straight lines were generated by plotting 1/v versus 1/[S]; the slope indicates Km/Vmax, and the intercept of the x-axis (1/[S]) indicates −1/Km.
(TIF)
Inhibition of AmAChE1 (•) and AmAChE2 (○) by cholinesterase-specific inhibitors (A), organophosphates (B) and carbamates (C). The results are the mean of three determinations (n = 3). Vertical bars indicate standard deviations.
(TIF)
Primers used for the in vitro expression of AmAChE1 and AmAChE2 with a baculovirus expression system.
(DOCX)
IC50 (M) values of different inhibitors of recombinant AmAChE2 with or without pre-incubation with AmAChE1.
(DOCX)