Abstract
Arabidopsis thaliana contains two genes encoding farnesyl diphosphate (FPP) synthase (FPS), the prenyl diphoshate synthase that catalyzes the synthesis of FPP from isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). In this study, we provide evidence that the two Arabidopsis short FPS isozymes FPS1S and FPS2 localize to the cytosol. Both enzymes were expressed in E. coli, purified and biochemically characterized. Despite FPS1S and FPS2 share more than 90% amino acid sequence identity, FPS2 was found to be more efficient as a catalyst, more sensitive to the inhibitory effect of NaCl, and more resistant to thermal inactivation than FPS1S. Homology modelling for FPS1S and FPS2 and analysis of the amino acid differences between the two enzymes revealed an increase in surface polarity and a greater capacity to form surface salt bridges of FPS2 compared to FPS1S. These factors most likely account for the enhanced thermostability of FPS2. Expression analysis of FPS::GUS genes in seeds showed that FPS1 and FPS2 display complementary patterns of expression particularly at late stages of seed development, which suggests that Arabidopsis seeds have two spatially segregated sources of FPP. Functional complementation studies of the Arabidopsis fps2 knockout mutant seed phenotypes demonstrated that under normal conditions FPS1S and FPS2 are functionally interchangeable. A putative role for FPS2 in maintaining seed germination capacity under adverse environmental conditions is discussed.
Introduction
All isoprenoids are derived from the C5 building blocks isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP). In plants, IPP and DMAPP are synthesized via two independent pathways: the mevalonic acid (MVA) pathway in the cytosol [1] and the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway in the plastids [2]. IPP and DMAPP are subsequently used as substrates by distinct short-chain prenyl diphosphate synthases that catalyze the head-to-tail condensation of one molecule of DMAPP with one, two or three IPP units to produce geranyl diphosphate (GPP; C10), farnesyl diphosphate (FPP; C15) and geranylgeranyl diphosphate (GGPP; C20), respectively [3], [4]. Pathway specialized branches starting from these prenyl diphosphates lead ultimately to the production of the astonishing number of isoprenoid end products synthesized by plants. GPP serves as a precursor for monoterpenoids, GGPP is a precursor of diterpenoids, gibberellins, carotenoids and abscisic acid, side chains of chlorophyll, phylloquinone, plastoquinone and tocopherols, and geranylgeranylated proteins, and FPP serves as a precursor for sesquiterpenoids, sterols, brassinosteroids, triterpenoids, polyprenols, side chains of ubiquinone, and farnesylated proteins [5]. It is generally accepted that the intracellular levels of these prenyl diphosphates and their precursors IPP and DMAPP must be strictly controlled to avoid deleterious effects on the metabolic flux through the pathway branches competing for these intermediates [6]. Indeed, overexpression of FPP synthase (FPS) in Arabidopsis [7], [8], [9] and phytoene synthase in tomato [10] results in altered levels of specialized isoprenoid end products that negatively affect plant performance.
Plants contain small FPS (EC 2.5.1.1/EC 2.5.1.10) isozyme families [11], [12], [13], [14], [15], [16], [17], [18]. This, together with the key position of FPP at a node of the isoprenoid pathway to which many branches leading to mitochondrial and cytosolic isoprenoids are connected [19], [20], has fuelled interest in deciphering the role of individual FPS isozymes in the isoprenoid pathway. Arabidopsis contains two genes, FPS1 (At5g47770) and FPS2 (At4g17190), encoding three FPS isozymes: FPS1L, FPS1S and FPS2. The FPS1 gene encodes FPS1S and FPS1L, which differ only by an N-terminal extension of 41 amino acid residues that targets FPS1L into mitochondria [10], [13] whereas the FPS2 gene encodes FPS2 that shares 90.6% amino acid identity with FPS1 isozymes [12]. Although the intracellular localization of Arabidopsis FPS1S and FPS2 remains to be experimentally established, both isozymes are predicted to localize in the cytosol since they do not bear any obvious subcellular targeting signal [12]. However, the possibility that one or both Arabidopsis short FPS isozymes might reside in a different compartment cannot be excluded. In fact, a Catharanthus roseus short FPS lacking a canonical peroxisomal targeting sequence partially localizes to the peroxisomes of C. roseus cells [21]. This observation would support the hypothesis for a partial localization of the early steps of the plant isoprenoid pathway in peroxisomes [22], [23] as reported to occur in mammals [24].
Studies in transgenic Arabidopsis expressing chimeric FPS::GUS genes [25] and analysis of microarray expression data [26] have shown that FPS genes are expressed in all organs throughout plant development, albeit at greatly different levels. FPS1 is widely expressed in all tissues throughout plant development whereas expression of FPS2 is mainly concentrated in floral organs, seeds and the early stages of seedling development. Characterization of Arabidopsis fps1 and fps2 single knockout mutants demonstrated that a single functional FPS gene is enough to sustain normal plant growth and development, thereby indicating that FPS1 and FPS2 can almost fully complement each other. However, simultaneous knockout of both FPS genes is embryo-lethal and severely impairs male genetic transmission [26]. Thus, the small Arabidopsis FPS gene family seemingly constitutes a redundant two-locus genetic network in which as long as one gene functions, no noticeable loss of plant fitness occurs. Nevertheless, FPS1 and FPS2 functions are not completely redundant. FPS activity measurements and analysis of sterol and ubiquinone levels, the major cytosolic and mitochondrial FPP-derived isoprenoids, have shown that FPS1S has a major role during most of the plant life cycle, whereas FPS2 has a predominant role in seeds and during the early stages of seedling development. In fact, FPS2 is by far the major contributor to total FPS activity in mature seeds, though shortly after germination FPS1 replaces FPS2 as the major provider of FPS activity. Hence, lack of FPS2, but not of FPS1, leads to a marked reduction of sitosterol content in mature seeds concomitant to a positive feedback regulatory response of 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR), the enzyme that catalyzes the main regulatory step in the MVA pathway and downstream isoprenoid pathways [27]. The elevated levels of HMGR activity become essential to sustaining a flux through the isoprenoid pathway that is high enough to produce sufficient sterols and likely other isoprenoids needed for normal seed viability, as revealed by the hypersensitivity of fps2 mutant seeds to the HMGR inhibitor mevastatin [26]. Correct quantitative and qualitative sterol composition is essential for proper seed development and viability, not only because sterols have well established roles in maintaining membrane structure and function [28] and as precursors of brassinosteroids [29], but also because they are involved in signalling pathways that are essential for normal embryogenesis [30], [31].
Biochemical characterization of individual FPS isozymes can also greatly contribute to the understanding of their physiological functions. So far, studies on the biochemical properties of plant FPSs have been conducted in a very limited number of species using purified native [32], [33] and recombinant [18], [34] enzyme preparations and, to the best of our knowledge, a comparative biochemical analysis between individual members of FPS isozyme families has only been reported in Artemisia tridentata. This plant contains two FPS isozymes that share 83% of amino acid identity. In spite of this, FDS-1 and FDS-2 exhibit different functional properties, which led the authors to propose specific cellular functions for each of the two isozymes [18]. In the present study, we report the results of a detailed functional and structural characterization of the two Arabidopsis short FPS isozymes FPS1S and FPS2. We also expand our previous FPS gene expression analysis [25] by establishing the spatial and temporal pattern of expression of the FPS genes during seed development, and present the results of functional complementation studies of the fps2 single knockout mutant phenotypes [26] with chimeric FPS gene constructs. Altogether, findings reported herein provide new clues to understand the biological role of FPS isozymes in Arabidopsis.
Materials and Methods
Chemicals
Unlabelled IPP, DMAPP, GPP and FPP were purchased from Echelon Biosciences and [4-14C]IPP (60 mCi/mmol) was from GE Healthcare Life Sciences. Mevastatin (Calbiochem, www.merck-chemicals. com) was dissolved in EtOH to prepare a 5 mM stock solution.
Plant Material and Growth Conditions
Arabidopsis thaliana wild-type (ecotype Col-3 qrt1/qrt1) and fps2 mutant plants were grown in vitro on Murashige and Skoog (MS) medium (Duchefa, http://www.duchefa.com) solidified using 0.8% w/v agar, or in vivo on soil in a climate-controlled growth chamber, under long-day conditions (16 h light/8 h dark) at 22°C. When required, MS medium was supplemented with 1 µM mevastatin.
Heterologous Expression of GST-FPS1S and GST-FPS2 in E. coli and Recombinant Protein Purification
Arabidopsis FPS proteins were expressed as N-terminal GST fusion proteins using a modified version of pGEX-3X plasmid (Pharmacia Biotech). The polylinker of pGEX-3X was modified by introducing a NotI restriction site upstream from the BamHI site that allows to obtain native proteins having an N-terminal end without extra residues after cleavage with Factor Xa protease [35]. The cDNAs coding for FPS1S and FPS2 were amplified by PCR using, respectively, the forward primers 5′-ATGGAGACCGATCTCAAGTCAACC-3′ and 5′-ATGGCGGATCTGAAATCAACC-3′, the common reverse primer 5′-CGCGGATCC CTACTTCTGCCTCTTGTAG-3′ (translation start and stop codons are shown in bold, and BamHI restriction site is underlined) and plasmids pcNC3 [13] and pcNC2 [12] as templates, respectively. The resulting PCR products were digested with BamHI, phosphorylated with T4 Polynucleotide kinase (Promega) and cloned into the NotI (blunt ended by nuclease S1 treatment) and BamHI restriction sites of the modified pGEX-3X, yielding plasmids pGEX-3X-NotI-FPS1 and pGEX-3X-NotI-FPS2. These expression plasmids were transformed into the E. coli strain BL21 (DE3), harbouring pUBS520 encoding E.coli tRNAArg AGA/AGG [36], and transformed cells were grown overnight on LB plates supplemented with ampicillin (100 µg/mL). Plasmids were isolated from overnight cultures and their sequences were verified. To express the recombinant proteins, 30 mL of LB medium supplemented with ampicillin (100 µg/mL) and kanamycin (50 µg/mL) was inoculated with 0.5 mL of an overnight culture of BL21 (DE3) cells harbouring either pGEX-3X-NotI-FPS1 or pGEX-3X-NotI-FPS2 and grown at 37°C to an OD600 of 0.5–0.6. After induction with 0.4 mM isopropyl thio-β-D-galactopyranoside (IPTG) the cultures were shifted to 20°C and incubated for 16 additional hours at 200 rpm. E.coli cells were collected by centrifugation at 7,000 g for 5 min at 4°C, resuspended in 3 mL of PBS (80 mM Na2HPO4, 20 mM NaH2PO4, 100 mM NaCl, pH 7.5) and disrupted by sonication (0.5 min/mL suspension) while being chilled in a −10°C bath. Cell debris was removed by centrifugation at 15,000 g for 30 min at 4°C, and the resulting supernatant was loaded onto a 1 mL Glutathione-Sepharose 4B affinity chromatography column (GE Healthcare) pre-equilibrated with PBS. All procedures for enzyme purification were performed at 4°C unless otherwise indicated. The column was washed with a minimum volume of 10 mL of PBS and re-equilibrated with Factor Xa cleavage buffer (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM CaCl2). GST-FPS fusion proteins were digested by adding to the column 40U of Factor Xa (Amersham Biosciences) in cleavage buffer to the column. After overnight incubation at 20–22°C the resulting native FPS proteins were eluted with cleavage buffer. Fractions of 250 µL were collected and analyzed by 12,5% SDS-PAGE [37] after a quick estimation of protein concentration with a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc.). Fractions enriched in FPS protein were pooled and the remaining Factor Xa protease was removed by treatment with Factor Xa removal resin (Qiagen) according to the manufacturer’s instructions. Glycerol was then added to a final concentration of 15% (v/v) and samples were stored frozen at −80°C. The purity of each FPS preparation was checked by SDS-PAGE. Protein concentration was determined by the method of Lowry [38] using BSA as a standard.
FPS Enzyme Activity Assay and Kinetic Studies
FPS activity of purified recombinant FPS isozymes was measured in a total volume of 100 µL containing 30 mM PIPES (pH 7.0), 6 mM NaCl, 4 mM MgCl2, 150 mM sucrose, 10 µg/mL aprotinin, 2 µg/mL E64, 1 µg/mL pepstatin, 0.35 mM PMSF, 0.30 mg/mL bovine serum albumin (BSA), 100 µM [4-14C]IPP (6.97 µCi/µmol), 100 µM GPP and the appropriate amount of enzyme (between 10 and 40 ng). The reaction was initiated by the addition of the substrates after preincubation of the enzyme for 5 min at 37°C. The reaction was carried out at 37°C for 15 min and terminated by the addition of 585 µl of 2 M HCl pre-chilled at 0°C. Solid NaCl was added to saturation and the reaction products were acid hydrolysed by incubation for 30 min at 37°C. The mixture was extracted with 1 mL of n-hexane and the radioactivity in 500 µL of the hexanic phase was quantified by liquid scintillation counting. To measure FPS activity in extracts from plant tissues, shoots (between 200 and 250 mg) or seeds (between 25 and 40 mg) were mixed with extraction buffer (2 µL per mg of seedlings and 20 µL per mg of seeds) containing 50 mM PIPES, pH 7.0, 250 mM sucrose, 10 mM NaCl, 5 mM MgCl2, 5 mM DTT, 15 µg/mL aprotinin, 3 µg/mL E64, 1.5 µg/mL pepstatin, and 0.5 mM PMSF pre-chilled at 4°C and ground to a fine powder with mortar and pestle. The slurry was centrifuged at 200 g for 10 min at 4°C to remove cell debris and the resulting supernatant was collected and centrifuged again at 16,000 g for 20 min at 4°C. The supernatant was recovered and FPS activity (between 100 and 200 µg of protein) was assayed for 30 min at 37°C as described above. One unit of FPS is defined as the amount of enzyme that incorporates one nanomol of IPP into acid-labile products per minute and mg of protein at 37°C.
For pH dependence analysis, enzyme activity assays were carried out using MES (5.5, 6.0), PIPES (6.5, 7.0) and Tris-HCl (7.5, 8.0, 8.5, 9.0). The apparent K m values for the substrates IPP, DMAPP and GPP were calculated from Lineweaver-Burk plots of FPS activity. The K m values for DMAPP and GPP were determined with substrate concentrations in the range of 6.25 to 200 µM with a fixed IPP concentration of 100 µM. The K m value for IPP was determined with concentrations in the range between 1 to 100 µM and a fixed DMAPP concentration of 200 µM. A nonlinear regression analysis in Sigma Plot 7.0 was used to calculate the kinetic parameters.
Determination of HMGR Enzyme Activity and Sitosterol Content in Seeds
For HMGR activity measurements, seeds (50 mg) were frozen in liquid nitrogen, ground to a fine powder with mortar and pestle, and mixed with 0.4 mL of pre-chilled extraction buffer (40 mM sodium phosphate, pH 7.5, 100 mM sucrose, 30 mM EDTA, 50 mM NaCl, 10 mM DTT, 10 µg/mL aprotinin, 1 µg/mL E64, 0.5 µg/mL leupeptin, 1 µg/mL pepstatin, 0.5 mM PMSF and 0.25% (w/v) Triton X-100). The slurry was centrifuged at 200 g for 10 min at 4°C to remove cell debris and HMGR activity was immediately measured in the supernatant as previously described [39]. One unit of HMGR activity is defined as the amount of enzyme that converts one picomol of 3-hydroxy-3-methylglutaryl coenzyme A into MVA per min and mg of protein at 37°C. Sitosterol levels in seeds were analyzed by GC-MS as previously described [26].
Western Blot Analysis
Aliquots (40–50 µg of protein) of the same seed extracts used for FPS activity measurements (16,000 g supernatant) were fractionated by 10% SDS-PAGE and electrotransferred onto Hybond-P polyvinylidene difluoride membranes (Amersham, Buckinghamshire, UK) at a constant intensity of 125 mA for 3 h at 4°C. The membrane was blocked in PBS pH 7.5, 0.5% (v/v) Tween 20 and 5% (v/v) Blotto non-fat dry milk (Santa Cruz Biotechnology inc.) for 16 h at 4°C, and incubated with rabbit polyclonal anti-FPS1S antibody [8] (1∶8000 dilution in blocking solution) for 1 h at room temperature. The membrane was then incubated with goat anti-rabbit IgG conjugated to peroxidase (Amersham) (1∶60000 dilution in blocking solution) for 1 h at room temperature. The FPS1S-antibody complex was visualized using the ECL Advance Western blotting system (GE Healthcare) according to the manufacturer's instructions. Protein loading was assessed by Coomassie blue staining of the membranes.
Differential Scanning Fluorimetry
The difference in thermal stability between FPS1S and FPS2 was analyzed by differential scanning fluorimetry (DSF) [40]. In brief, 20 µL reactions were set up on a 96-well thin-wall plate (Bio-Rad) containing 3–14 µM each protein in assay buffer (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM CaCl2, 15% (v/v) glycerol and 5× Sypro Orange (Invitrogen). Assay buffer was added instead of protein in the control samples. The plates were sealed with optical-quality sealing tape (Bio-Rad) and heated on a iQ5 Real Time (RT)-PCR instrument (Bio-Rad) from 20–80°C in increments of 0.2°C. Fluorescence was monitored with a charge-coupled device (CCD) camera using 490 and 575 nm as emission and excitation wavelengths, respectively. The mid-point temperature of the unfolding transition or melting temperature (T m) was calculated by fitting a Boltzmann model to the fluorescence imaging data after eliminating data beyond the fluorescence intensity maximum.
Homology Modelling
To construct homology models of Arabidopsis FPS1S and FPS2 that could be directly comparable with respect to their sequence differences, we chose as template the crystal structure of unliganded human FPS (PDB 2F7M) [41]. The sequence identity of human FPS and FPS1S was 45% over 99% of its length (339 out of 343 amino acids) and between human FPS and FPS2 was 46% over 99% of its length (338 out of 342 amino acids). The phosphate ion and the four water molecules found in the active site of FPS in this structure were kept to maintain a stable conformation of the active site loops during modelling. First, we threaded the correct FPS1S and FPS2 sequences onto the template structure using Modeller 9.10 [42] and then selected the 10 best models out of 1000 independent models on the basis of the Z-DOPE normalized scores, which ranged from –1.916 to –1.337 for FPS1S and from –1.949 to –1.529 for FPS2 (a Z-DOPE of less than –1 indicates a plausible model with 80% of the Cα atoms lying within 3.5 Å of their correct positions). Next, the top ten models for each protein were refined and minimized using the Rosetta force field and Monte Carlo sampling methods [43], [44] and the improved models were clustered and analyzed to obtain a final model. The final models for FPS1S and FPS2 showed correct stereochemistry as assessed by MolProbity [45].
In Silico Evaluation of Free Energy and Structure Changes Upon Mutation
Free energy (DDG) and structure changes upon mutation were calculated using two established methods, the Rosetta DDG application [46] and the publicly available CC/PBSA web server (http://ccpbsa.biologie.uni-erlangen.de/ccpbsa/) [47]. The changes in stability predicted by the two independent calculations were analyzed in the context of the predicted structural changes. The Rosetta DDG protocol corresponds to row 16 of a recent benchmarking study [46]. Row 16 protocol first repacks all residues according to Rosetta standard sidechain sampling procedures while keeping the backbone fixed, and then minimizes all backbone and sidechain degrees of freedom. Energies are calculated for 50 wild-type and mutant sequence contexts and the predicted DDG is the difference in the free energy between the mutant and wild-type protein. The CC/PBSA method is accessed via a web interface where the coordinates are submitted along with a string describing the desired mutation. In a nutshell, the CC/PBSA method generates two random conformational ensembles each one of them consisting of 300 wild-type and mutant minimized structures for which energies are evaluated using a custom energy function. Those energy contributions are scaled to reproduce experimentally measured free energies using five-fold cross validation.
Gene Constructs for Mutant Complementation
To construct plasmid pCAM-FPS2p::FPS1S, a 1388-bp fragment including 1329 bp of the FPS2 gene promoter, the 5′ untranslated region and the ATG translation start codon, was amplified by PCR using genomic DNA as a template, forward primer 5′-GCGTCGACAGCTTGGAGCATAAGAAG-3′ and reverse primer 5′-TCCGCCATGGATAGGATCAAGG-3′. A SalI restriction site was added at the 5′ end of forward primer and an NcoI site encompassing the ATG start codon of FPS2 was created in the reverse primer by introducing a G (shown in bold) instead of a C. A 2353-bp fragment including the region encoding FPS1S and 339 bp of the 3′-non coding region was amplified by PCR using genomic DNA as a template, forward primer 5′-AGCTCTTCCATGGAGACCGATC-3′ and reverse primer 5′-TTGGAGCTCTTTGGAATGGAATGTAGG-3′. An NcoI restriction site encompassing the ATG start codon of FPS1S was created by introducing a C (shown in bold) instead of a G in the forward primer and a SacI restriction site was added at the 5′ end of reverse primer. Both genomic fragments were cloned into pGEM-T Easy vector (Promega), excised by digestion with either SalI and NcoI or NcoI and SacI, and cloned into the SalI and SacI sites of pBluescript KS+ in a three-piece ligation yielding pBFPS2p::FPS1S. The entire FPS2p::FPS1S fragment was then excised by digestion with SalI and SacI and cloned into pCAMBIA2300 yielding plasmid pCAM-FPS2p::FPS1S.
To create plasmid pCAM-FPS2p::FPS1S-mutdis, a 1375-bp fragment including 1329 bp of the FPS2 gene promoter was amplified by PCR using genomic DNA as a template, forward primer 5′-GCGTCGACAGCTTGGAGCATAAGAAG-3′ and reverse primer 5′-CCCAAGCTTGATAGGATCAAGGAAGGTGT-3′. Restriction sites for SalI and HindIII (underlined) were added at the 5′ end of forward and reverse primers, respectively. The amplified fragment was cloned into pGEM-T Easy vector yielding pGEM-FPS2p. A 2472-bp fragment including the entire coding region of the FPS1 gene and 339 bp of the 3′-non coding region was amplified by PCR using genomic DNA as a template, forward primer 5′-GGGATATCAGTGTGAGTTGTTGTTGT-3′ and reverse primer 5′-TTGGAGCTCTTTGGAATGGAATGTAGG-3′. Restriction sites for EcoRV and SacI (underlined) were added at the 5′ end of forward and reverse primers, respectively. In the forward primer the third base of the ATG codon corresponding to the translation start codon of FPS1L isoform was changed to C (shown in bold). The amplified fragment was cloned into the EcoRV and SacI sites of pBluescript SK+ yielding pBFPS1S-mutdis. The FPS1 gene fragment was then excised from pBFPS1S-mutdis with EcoRV and SacI and cloned into plasmid pGEM-FPS2p, which had been previously digested with HindIII, treated with nuclease S1 to produce blunt ends and digested again with SacI (in the pGEM-T polylinker) yielding pGEM-FPS2p::FPS1S-mutdis. The entire FPS2p::FPS1S-mutdis fragment was then excised by digestion with SalI and SacI and cloned into pCAMBIA2300 yielding plasmid pCAM-FPS2p::FPS1S-mutdis.
To construct plasmid pCAM-FPS1p::FPS2, a 1526-bp fragment including 1338 bp of the FPS1 gene promoter and 185 bp of the 5′ leader region up to the ATG translation start codon of FPS1S was amplified by PCR using DNA from genomic clone pgNC241 as a template [12], forward primer 5′-GCGTCGACATAGTAGTTAATGTTGGGG-3′ and reverse primer 5′-TCTCCATGGAAGAGCTTTGGATACG-3′. A SalI site was added at the 5′ end of forward primer and an NcoI site encompassing the ATG start codon of FPS1S was created by introducing a G (shown in bold) instead of a T in the reverse primer. A 2553-bp fragment including the entire coding region of the FPS2 gene and 438 bp of the 3′-non coding region was amplified by PCR using genomic DNA as a template, forward primer 5′-GATCCTATCCATGGCGGATCTG-3′ and reverse primer 5′-AGCGAGCTCATTTCCACTAATCTTCTCG-3′. An NcoI restriction site encompassing the ATG start codon of FPS2 was created by introducing a C (shown in bold) instead of an A in the forward primer. A SacI restriction site was added at the 5′ end of reverse primer. Both genomic fragments were cloned into pGEM-T Easy vector, excised by digestion with either SalI and NcoI or NcoI and SacI, and cloned into the SalI and SacI sites of pBluescript KS+ in a three-piece ligation yielding pBFPS1p::FPS2. The entire FPS1p::FPS2 fragment was then excised by digestion with SalI and SacI and cloned into pCAMBIA1300-T-Nos yielding plasmid pCAM-FPS1p::FPS2. Plasmid pCAMBIA1300-T-Nos was generated by introducing the T-Nos sequence from pBI221 into the SacI and EcoRI sites of pCAMBIA1300.
To create plasmid pCAM-FPS1mutdisp::FPS2, a 1526-bp fragment of the FPS1 flanking region was amplified by PCR using as a template a chimeric translational FPS1S::GUS gene fusion in which the ATG start codon of FPS1L had been converted to an ATC codon by site directed mutagenesis [25] and the same forward and reverse primers used to construct pCAM-FPS1p::FPS2. The amplified fragment was cloned into pGEM-T Easy yielding pGEM-FPS1mutdisp. The FPS1 gene fragment was then excised with SalI and NcoI and cloned into pBFPS1p::FPS2 to replace the corresponding non-mutated region of FPS1 promoter, yielding plasmid pBFPS1mutdisp::FPS2. The entire pBFPS1mutdisp::FPS2 fragment was then excised with SalI and SacI and cloned into pCAMBIA1300-T-Nos yielding plasmid pCAM-FPS1mutdisp::FPS2.
All PCR fragments used to construct plasmids described above were sequenced to exclude amplification artifacts. The correct fusion of the genomic fragments was also confirmed by sequencing.
Plant Transformation and Transgene Expression Analysis
Agrobacterium tumefaciens strain GV3101 harbouring plasmids described above was used to transform Arabidopsis fps2-1 mutant plants by the floral dip method [48]. Seeds from infiltrated plants were surface sterilized and sown in Petri dishes containing solid MS medium supplemented with 50 µg/mL kanamycin. Antibiotic-resistant seedlings (T1) were transplanted into soil and grown to maturity. Lines homozygous for the transgenes containing a single insertion were selected by segregation analysis of the kanamycin resistance trait.
To analyze the expression of the transgenes introduced into the fps2-1 mutant, total RNA was isolated from 12-days-old seedlings form fps2-1 lines harbouring FPS2p::FPS1S, FPS2p::FPS1-mutdis, FPS1p::FPS2, or FPS1mutdisp::FPS2. Total RNA (1,5 µg) was treated with DNAse (Ambion) and single-stranded cDNA pools were synthesized using oligo-dT primer and SuperScript III reverse transcriptase (Invitrogen) according to standard protocols. PCR reactions were carried out by 35 cycles of amplification (45 s at 94°C, 60 s at 50°C and 90 s at 72°C with a 5 min final extension at 72°C) using as template 7.5 µL of a 1∶10 dilution of the corresponding single-stranded cDNA pools and 1 unit of Taq polymerase (Biotools). Primers 5′- GGCTTTGCACACCTTCCTTG-3′ and 5′-CCTGTGGATGTGATTGCGAAG-3′ were used for expression analysis of FPS2p::FPS1S and FPS2p::FPS1-mutdis genes, and primers 5′-GGTGGGAGTCTCTATCGTCGTCGTATCCAA-3′ and 5′-CGGAGAGGCCCGAGTATG-3′ were used for expression analysis of FPS1p::FPS2 and FPS1mutdisp::FPS2 genes. The expression of the ACT2 (At3g18780) gene was analyzed using primers 5′-GATCTGGCATCACACTTTCTAC-3′ and 5′- GCCTTGGAGATCCACATCTGCTG-3′. The expression of the PP2AA3 (At1g13320) gene encoding the 65 kDa regulatory subunit of protein phosphatase 2A (PP2A) was analyzed using primers 5′-TAACGTGGCCAAAATGATGC-3′ and 5′-GAAGCCAACATTAACATTAGTAGC-3′.
GUS Assay in Arabidopsis Seeds
Siliques were harvested, opened longitudinally, placed in GUS assay buffer (50 mM phosphate buffer, pH 7.0, 0.2% (v/v) Triton X-100, 20 mM X-Gluc and 2 mM potassium ferricyanide) and subjected to vacuum for 10 min. After incubation for 24 hours at 37°C, siliques were placed in ethanol:acetic acid (1∶1) and incubated for either 4 hours (young seeds with embryos at the globular and heart stages) or 8 hours (mature seeds with embryos at torpedo and cotyledon stages). GUS-stained seeds were cleared in Hoyer’s medium for 3–4 days in darkness [49]. Dissected seeds were placed on a slide covered with a coverslip, stored in darkness for 24 hours and observed under a Zeiss Axiophot microscope equipped with Nomarski optics. Photographs were taken using the same microscope equipped with an Olympus DP70 photo camera.
Expression of GFP-FPS1S, GFP-FPS2, FPS1S-GFP and FPS2-GFP in Agroinfiltrated Leaves of N. benthamiana Plants
The FPS1S coding sequence (1029 bp) was amplified by PCR using forward primer 5′-ACGCGTCGACAATGGAGACCGATCTCAAGTCAACC-3′, reverse primer 5′-CTGTCGATATCCCTTCTGCCTCTTGTAGATCTTAGC-3′ and plasmid pcNC3 [13] as a template. The sequence coding for FPS2 (1039 bp) was amplified using forward primer 5′-ACGCGTCGACAATGGCGGATCTGAAATCAACCTTC-3′, reverse primer 5′-GAGTATGATATCCCTTCTGCCTCTTGTAGATCTTAGC-3′ and plasmid pcNC2 [12] as a template. Translational start codons are shown in bold. SalI and EcoRV restriction sites (underlined) were added at the 5′ end of the forward and reverse primers, respectively. The amplified fragments were cloned into the corresponding sites of plasmid pENTR3C (Gateway®, Invitrogen) yielding plasmids pENTR-FPS1S and pENTR-FPS2. Both cDNA fragments were subsequently transferred from pENTR-FPS1S and pENTR-FPS2 to pMDC43 and pMDC83 (Gateway®, Invitrogen) yielding plasmids pMDC43FPS1S, pMDC43FPS2, pMDC83FPS1S and pMDC83FPS2, in which the FPS coding sequences were fused in-frame to the 5′- or 3′-ends of the green fluorescent protein (GFP) coding sequence. In all cases the coding sequences were under the transcriptional control of the CaMV35S gene promoter. All constructs were sequenced to confirm the in-frame fusions. Plasmids coding for the different protein fusions were transformed into Agrobacterium tumefaciens strain EHA105. The recombinant A. tumefaciens strains were grown overnight at 28°C in YEB liquid medium supplemented with 100 µg/mL rifampicine and 25 µg/mL kanamycin. Cells were harvested by centrifugation and resuspended to an OD600 of 0.150 in a solution containing 10 mM MgCl2, 10 mM HEPES, pH 5.6, and 200 µM acetosyringone (3,5-dimethoxy-4′-hydroxy-acetophenone). Prior infiltration, bacterial suspensions were incubated at room temperature for 3 h. For co-expression experiments, suspensions of A. tumefaciens harbouring the FPS-GFP expression constructs were mixed with A. tumefaciens cultures harbouring constructs for expression of the cyan fluorescent protein (CFP)-peroxisome marker (CFP-SKL) [50] and the tobacco etch polytovirus helper component protein (HC-Pro) silencing suppressor [51] in a 1∶1:1 ratio. Leaves of 2–4 week old N. benthamiana plants were infiltrated by gently appresing a 2-ml syringe without a needle to the abaxial surface of fully expanded leaves. Following a 2–3 day incubation of infiltrated plants under long-day conditions at 25°C and 60% humidity, abaxial epidermis of leaf tissue was examined by confocal laser-scanning microscopy using a Leica SP5 microscope (Leica Microsystems). GFP and CFP were excited by using 488 and 458 laser lines, respectively. Images were acquired sequentially to avoid crosstalk between channels. LAS-AF Lite 2.6.0 software was used for image capture and for merging false-coloured images of transiently co-transformed cells.
Results
Biochemical Characterization of Recombinant FPS1S and FPS2 Isozymes
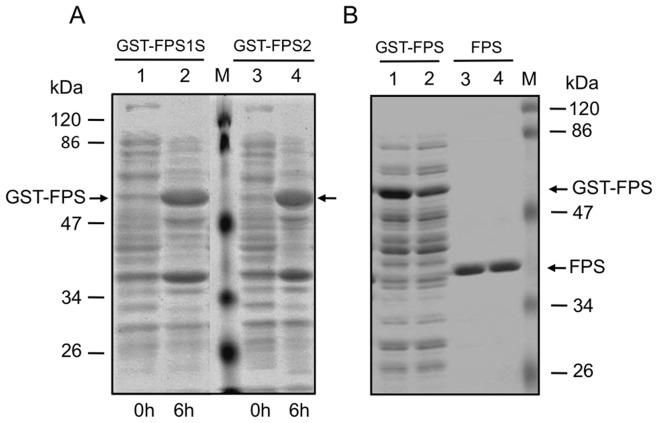
Arabidopsis FPS1S and FPS2 isozymes were expressed in E. coli as soluble N-terminal GST fusions (Figure 1A). The resulting recombinant proteins were purified through Glutathione-Sepharose 4B affinity chromatography, digested with Factor Xa protease to release native FPS1S and FPS2 enzymes, and further purified to remove both the excised GST moiety and Factor Xa protease. SDS-PAGE analysis showed that this purification procedure yielded highly purified preparations of both enzymes (Figure 1B) that were used for biochemical characterization.
Figure 1. Expression in E. coli and purification of recombinant FPS1S and FPS2 proteins.
(A) Total protein extracts from E. coli cells harbouring either pGEX-3X-NotI-FPS1 or pGEX-3X-NotI-FPS2 before (lanes 1 and 3) and after induction (lanes 2 and 4) of GST-FPS1S and GST-FPS2 expression with 0.4 mM IPTG for 6 hours at 22°C. (B) Soluble protein extracts of IPTG-induced E. coli cells harbouring either pGEX-3X-NotI-FPS1 (lane 1) or pGEX-3X-NotI-FPS2 (lane 2), and purified native FPS1S (lane 3) and FPS2 (lane 4) protein preparations after Glutathione-Sepharose 4B affinity column chromatography, proteolytic digestion with Factor Xa and protease removal. Arrows indicate the position of GST-FPS protein fusions and purified native FPS proteins. Molecular masses of standards (M) are indicated in kDa.
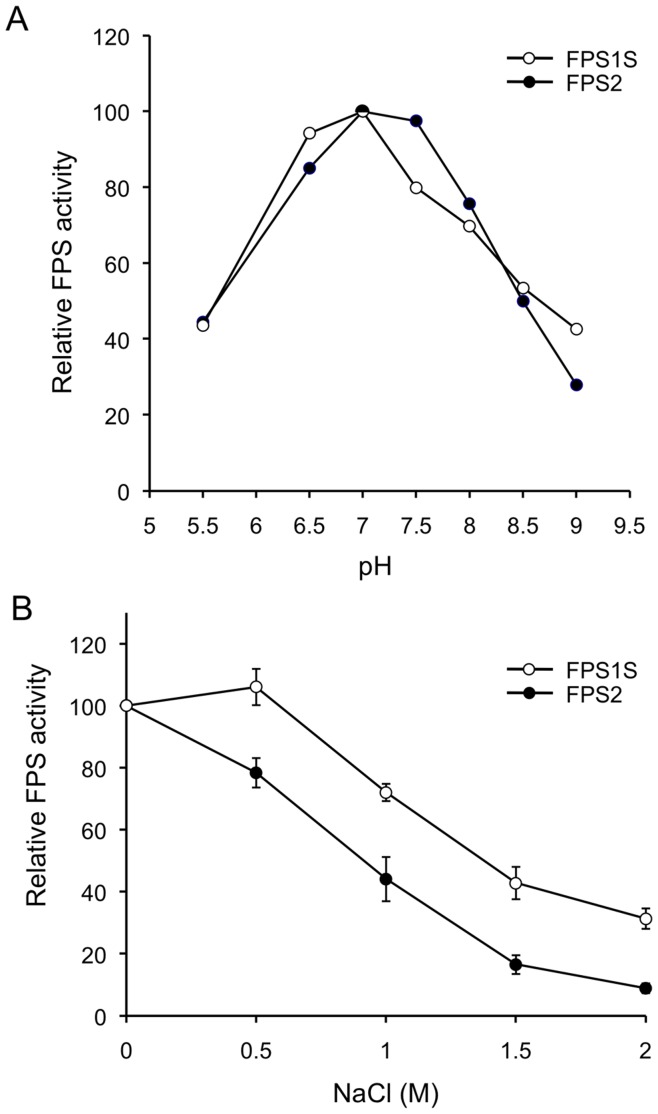
Prenyltransferases are known to require a divalent metal ion for catalytic activity. Thus, we first analyzed the effect of different Mg2+ concentrations on FPS1S and FPS2 activity. Similar values of activity were obtained when the enzyme activity was measured in the presence of MgCl2 concentrations ranging between 1 and 5 mM (data not shown). The optimal pH range for FPS1S and FPS2 was also determined. Both enzymes showed a similar pH-rate profile with only minor differences (Figure 2A). Maximal activity for both enzymes was observed at a pH value of 7.0, albeit the pH-rate profile of FPS1S was slightly shifted toward more acidic pH values compared to FPS2. We next investigated the effect of NaCl on the activity of FPS1S and FPS2 using concentrations in the range from 0 to 2 M. As shown in Figure 2B, the activity of both enzymes progressively declined as the concentration of NaCl increased, though in the case of FPS1S this effect was observed only at NaCl concentrations higher than 0.5 M. At all NaCl concentrations tested, FPS2 was markedly more sensitive to the inhibitory effect of NaCl than FPS1S. Purified FPS1S and FPS2 were also subjected to kinetic analyses and the resulting steady-state kinetic constants are shown in Table 1. Both enzymes displayed typical Michaelis-Menten behaviour, as observed for other FPSs, and exhibited similar K m values in the µmolar range (from 8.3 to 31.5 µM) for both IPP and the allylic substrates DMAPP and GPP. The affinity (K m) of FPS1S and FPS2 for the reaction intermediate GPP was approximately 2.7-fold higher than for DMAPP, thus indicating a preference of both enzymes for the allylic intermediate. Similarly, comparison of the specificity constants (kcat/K m) indicated a 2-fold higher catalytic efficiency for GPP compared to DMAPP. The inhibitory effect of the reaction product FPP on FPS1S and FPS2 activity was analyzed using concentrations in the range from 0 to 1 mM. The activity inhibition profile was nearly identical for both enzymes, with a maximal 30% reduction of activity at the highest FPP concentration assayed.
Figure 2. Effect of pH and NaCl on FPS1S and FPS2 enzyme activity.
(A) FPS activity of purified FPS1S and FPS2 was determined at pH values ranging from 5.5 to 9.0 as described under Materials and Methods. Enzyme activities are expressed relative to the maximal activity values for FPS1S and FPS2. (B) Effect of NaCl on FPS1S and FPS2 enzyme activity. FPS activity of purified FPS1S and FPS2 was determined at the indicated NaCl concentrations. Enzyme activities are expressed relative to the activity values for FPS1S and FPS2 measured without NaCl. The mean values and SE were calculated from three independent experiments.
Table 1. Steady-state kinetic constants for FPS1S and FPS2.
| Variable substrate | Kinetic parameter | FPS1S | FPS2 |
| IPP | K m (μM) | 23.1±0.4 | 31.3±0.5 |
| kcat (min−1) | 62.5 | 269.0 | |
| kcat/K m (min−1μM−1) | 2.7 | 8.6 | |
| DMAPP | K m (μM) | 22.7±2.1 | 31.5±0.5 |
| kcat (min−1) | 52.2 | 197.3 | |
| kcat/K m (min−1μM−1) | 2.3 | 6.2 | |
| GPP | K m (μM) | 8.3±1.2 | 11.3±1.0 |
| kcat (min−1) | 36.4 | 132.8 | |
| kcat/K m (min−1μM−1) | 4.4 | 11.7 |
The K m mean values and SE were calculated from three independent experiments.
kcat values were calculated for the dimeric form of the enzyme.
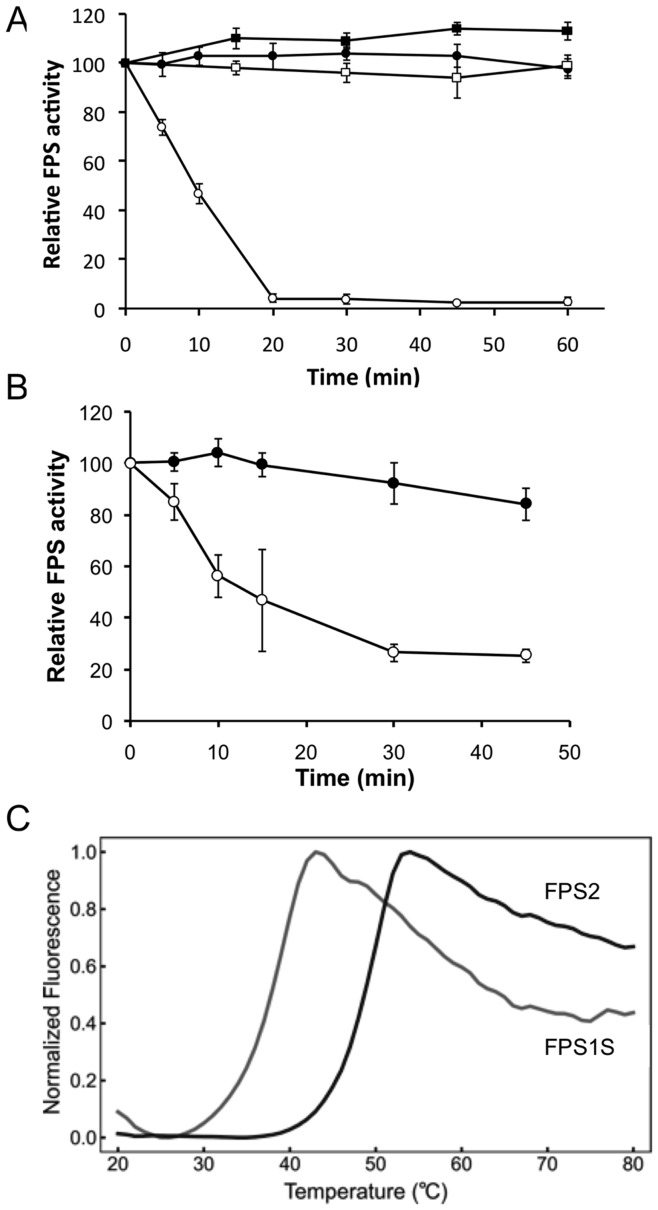
Interestingly, FPS2 preparations consistently showed higher specific activity than the FPS1S ones. In fact, FPS2 showed catalytic rate constant (kcat) values for IPP, DMAPP, and GPP that were 3.2-, 2.7- and 2.7-fold higher, respectively, than FPS1S, thus indicating that FPS2 is a more efficient catalyst than FPS1S. These differences cannot be attributed to differential stability of the enzymes either under storage conditions or during the purification procedure since both retained more than 90% of their initial activity after 2 months of storage at −80°C and their activity remained virtually unaffected after incubation for 1 hour at 37°C (Figure 3A). In contrast to this later observation, we observed a markedly different response of FPS1S and FPS2 to treatment at 45°C for different time-periods. As shown in Figure 3A, FPS2 activity remained unaltered after incubation for 1 hour at 45°C whereas FPS1S activity was completely abolished after 20 minutes at the same temperature. A similar result was obtained when protein extracts from the Arabidopsis fps1-1 and fps2-1 single knockout mutants bearing only one functional FPS gene (FPS2 and FPS1 respectively) were incubated at 45°C for different time-periods and assayed for FPS activity. Again, incubation at 45°C had almost no effect on FPS2-derived activity in extracts from fps1-1 plants, whereas FPS1-derived activity in extracts from fps2-1 plants was severely diminished upon incubation at the same temperature (Figure 3B). These differences in thermal stability were corroborated by differential scanning fluorimetry (DSF) [40]. The melting temperature (T m) derived from the DSF data is a useful diagnostic tool because greater thermal stability is associated with a positive shift in T m with respect to a reference T 0 (T m – T 0 = DT m >0) and vice versa. DSF experiments with 3–14 µM FPS1S or FPS2 in FPS activity assay buffer yielded T m values of 37°C and 48.3°C, respectively, a difference in T m of 11.3°C (Figure 3C). Based on these results, it can be concluded that FPS2 is more resistant to thermal inactivation than FPS1S because FPS2 protein is thermodynamically more stable than FPS1S.
Figure 3. Thermal stability of FPS1S and FPS2 enzyme activity.
(A) Activity of purified FPS1S (open symbols) and FPS2 (closed symbols) was measured after incubation at either 37°C (squares) or 45°C (circles) for the indicated time periods. (B) FPS activity in 16,000 g protein extracts from fps1-1 (FPS2 activity, closed circles) and fps2-1 (FPS1 activity, open circles) mutants was determined after incubation at 45°C for the indicated times. In both cases enzyme activities are expressed relative to the FPS activity values at time 0 min and the mean values and SE were calculated from three independent experiments. (C) Differential scanning fluorimetry (DSF) results plotted as change in fluorescence emission intensity (normalized to unity at its maximum) with increasing temperature (20–80°C). The FPS1S and FPS2 curves correspond to 6 µM enzyme.
Structural Basis of the Enhanced Thermal Stability of FPS2
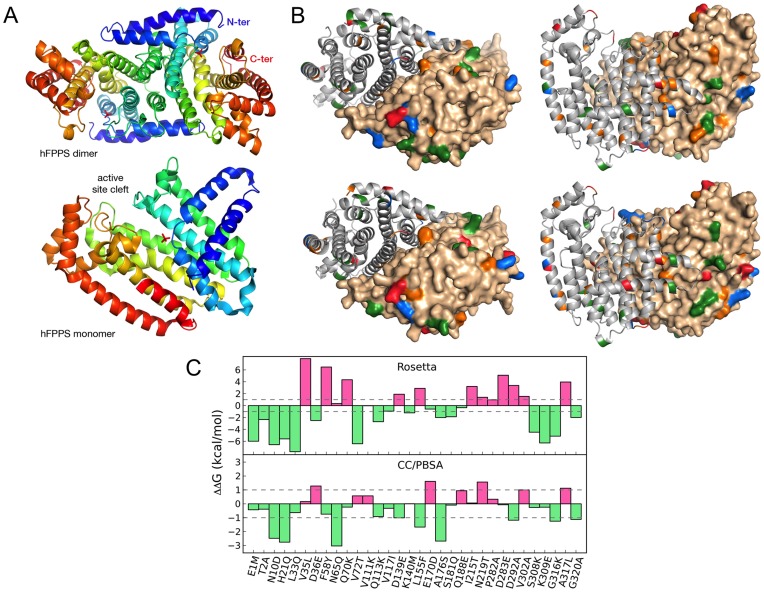
To further our understanding of the differential thermal stability between FPS1S and FPS2, homology models for both proteins were built using as a template the crystal structure of unliganded human FPS (PDB 2F7M) [41] (Figure 4A) and carefully assessed with respect to their stereochemistry. Best models for both enzymes showed correct stereochemical parameters and native dimer interfaces using various stringent criteria [45], [52]. The root-mean-square (r.m.s.) deviation between each model and the template structure was 0.26 Å and between the two models was 0.29 Å. Mapping of the sequence substitutions between FPS1S and FPS2 onto their respective ribbon structures and molecular surfaces (Figure 4B) showed that the immense majority of these substitutions occur at the outer surface of the enzyme.
Figure 4. Homology modelling of FPS1S and FPS2 proteins and in silico DDG calculations.
(A) Ribbon representation of the dimeric and monomeric structures of human FPS (PDB 2F7M), which was used to template the threading of Arabidopsis FPS1S and FPS2. The active site cleft is labelled and a bound phosphate ion is shown in sticks. (B) Sequence substitutions between FPS1S (top) and FPS2 (bottom) were mapped onto the ribbon structure (left monomer) or the molecular surface (right monomer) of the homology modeled dimers. Chemical character is colour coded as follows: red, acidic (Asp, Glu); blue, basic (Arg, Lys, His); green, polar (Ser, Thr, Asn, Gln, Tyr); orange, apolar (Met, Phe, Pro, Trp, Val, Leu, Ile, Ala). (C) Histogram of DDG (kcal/mol) upon single-site substitution calculated using Rosetta DDG application (top) or CC/PBSA (bottom). Mutations predicted to occur with a decrease in DDG are coloured green and those expected to increase DDG are coloured pink. Horizontal dashed lines at –1 to +1 kcal/mol bound the neutral area where DDG is supposed to contribute little to the overall stabilization or destabilization of the mutated protein.
Detailed analysis of the thirty-two substitutions that separate FPS2 from FPS1S revealed that the increase in surface polarity and the greater capacity to form surface salt bridges of FPS2 compared with FPS1S appear to be determining factors of the enhanced thermostability of FPS2. Indeed, there are several discernable trends in amino acid substitution between FPS2 and FPS1S that lends support to this hypothesis. First, FPS2 has four charged residues more than FPS1S of which three are basic and one is acidic. Secondly, polar and charged sidechains in FPS2 tend to be longer on average than in FPS1S (e.g., substitutions follow Asp = > Glu, Asn = > Gln), which could afford greater rotameric freedom and thereby facilitate formation of polar interactions on the protein’s surface. In summary, inspecting the FPS1S and FPS2 homology models around each mutated residue suggested that as many as three additional salt bridges (and several strong hydrogen bonds) could be established in FPS2 compared to FPS1S. Other metrics of structure stability, such as sidechain hydrogen bonds (31 in all/4 strong versus 30 in all/2 strong) and salt bridges across the dimer interface (7 versus 5) interface did also favour FPS2 with respect to FPS1S. Interestingly, a number of aromatic-aromatic, aromatic-sulfur and pi-cation interactions seem to be more prevalent in FPS2 than in FPS1S, and could also make a sizable contribution to the overall thermal stability of the two enzymes.
To accurately calculate the changes in free energy and structure induced by single residue substitutions we applied the Rosetta DDG [46] and CC/PBSA [47] methods to predict the free energy and structure changes underwent by FPS1S as single FPS2-mimicking substitutions are introduced. Results indicated that the cumulative free energy change summed over all mutated residues was energetically very favourable regardless of the method used, in qualitative agreement with the results from DSF experiments. Moreover, the Rosetta DDG and CC/PBSA protocols flag 58% (15 versus 11) and 69% (9 versus 4) of the non-neutral substitutions as stabilizing (DDG < –1 kcal/mol). Of the non-stabilizing mutations, Rosetta DDG brands more mutations than CC/PBSA as destabilizing (DDG > +1 kcal/mol) than neutral (DDG between –1 and +1 kcal/mol). Figure 4C shows this pattern for both the Rosetta DDG and the CC/PBSA protocols. Of the 21 out of 32 substitutions that change any residue in FPS1S to a polar/charged residue in FPS2, 85% (18) are predicted as non-destabilizing (the three exceptions are Q70K, E170D and D283E) thereby providing corroborative evidence for the important role of surface electrostatic interactions for FPS2 thermal stability. Perhaps one of the larger discrepancies between the two prediction methods involves substitutions F58Y, N65Q, Q70K and V72T, for which DDG estimates are reversed. These four substitutions, which are located in three consecutive helices, cluster tightly together around a 6-Å sphere and therefore it is plausible that the true structure of FPS2 had undergone compensatory changes that cannot be accurately captured by our single-site mutation calculations. Future structural studies of FPS1S and FPS2 should provide more accurate rationales for the observed stability differences.
Subcellular Localization of FPS1S and FPS2
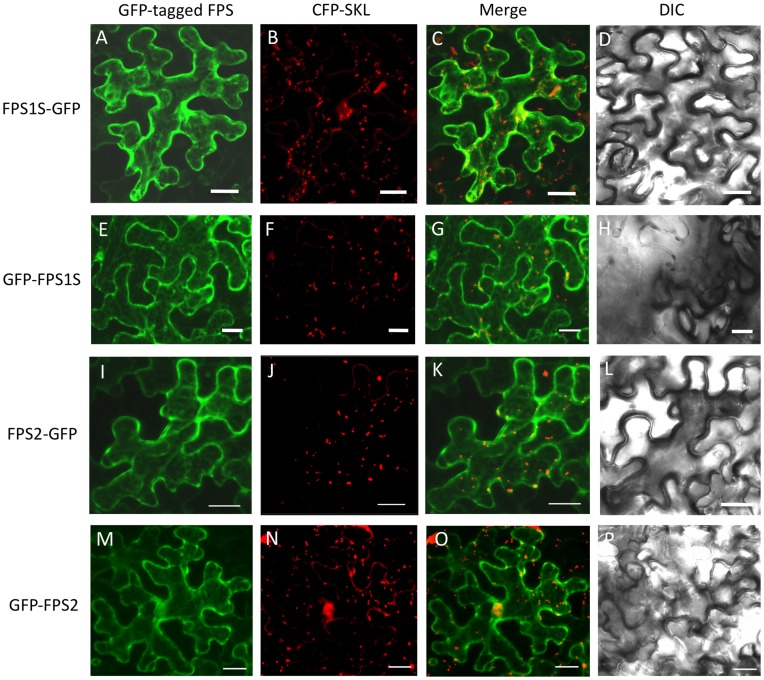
Arabidopsis FPS1S and FPS2 isoforms have long been considered to localize in the cytosol, mainly because no obvious organellar targeting signals have been detected in their primary structure. To provide experimental data supporting this assumption, the subcellular localization of Arabidopsis FPS1S and FPS2 was investigated by transiently expressing N- and C-terminal fusions of both FPS isoforms to the GFP in agroinfiltrated N. benthamiana leaf cells (Figure 5). Confocal laser microscopy analysis of the transfected cells revealed that all four tested proteins (GFP-FPS1S, FPS1S-GFP, GFP-FPS2, and FPS2-GFP) showed a diffuse pattern of fluorescence throughout the cytosol that was completely different from the punctuate fluorescence pattern characteristic for peroxisomal proteins, as revealed by comparison with the fluorescence signal of the peroxisomal marker CFP-SKL. These results strongly suggested that both FPS1S and FPS2 localize in the cytosol and not in the peroxisomes.
Figure 5. Subcellular localization of GFP-tagged FPS1S and FPS2 proteins.
Confocal laser scanning micrographs showing the distribution of fluorescence in agroinfiltrated N. benthamiana epidermal cells transiently co-expressing FPS1S-GFP (A), GFP-FPS1S (E), FPS2-GFP (I) or GFP-FPS2 (M) with the peroxisome protein marker CFP-SKL (B, F, J and N). Co-localization evaluation of GFP-tagged FPS proteins with the peroxisome protein marker (C, G, K and O). Differential interference contrast (DIC) images showing the morphology of transformed cells (D, H, L and P). Scale bars = 20 µm.
Expression Pattern of Arabidopsis FPS1 and FPS2 Genes during Seed Development
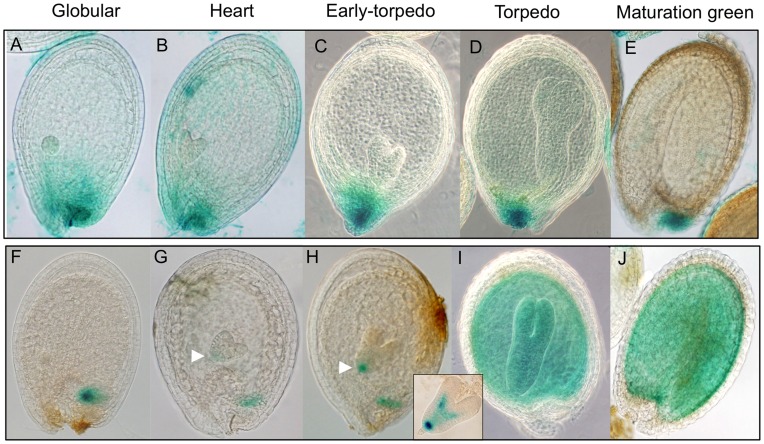
Characterization of Arabidopsis fps single knockout mutants revealed a differential contribution of FPS isoforms to total FPS activity in mature seeds [26]. In order to investigate whether this effect could be due to differential expression of FPS1 and FPS2 in seed tissues, we analyzed the spatial and temporal pattern of expression of GUS activity in seeds at different developmental stages harvested from Arabidopsis lines harbouring FPS1S::GUS and FPS2::GUS transgenes (Figure 6). These chimeric gene fusions included approximately 1350 bp of the corresponding FPS promoter region [25]. In FPS1S::GUS plants, the primary site of GUS activity detection at all stages of development was the maternal chalazal seed coat (Figure 6A–E). At the globular, heart and early-torpedo stages of embryo development some variable and weaker GUS staining could also be detected in surrounding tissues (Figure 6A–C) although it is difficult to establish whether this expression reflected diffusion of the substrate or true expression of the transgene. At latter stages of embryo development GUS activity was restricted to the chalazal seed coat (Figure 6D and E). Interestingly, no expression of GUS activity in the embryo could be detected at any developmental stage. The pattern of GUS expression driven by the FPS2 promoter was completely different (Figure 6F–J). At the globular stage of embryo development GUS staining was detected only in the chalazal endosperm (Figure 6F), but from the heart stage onward GUS activity could also be detected in the embryo (Figure 6G–J). At the heart and early torpedo stage GUS expression in the embryo was primarily localized to the root apical meristem region (Figure 6G and H, arrowhead) and the pro-vascular tissue of embryo (Figure 6, inset). Up to this developmental stage, a very faint staining could also be detected in the endosperm of some seeds when they were subjected to intensive staining. At later developmental stages GUS activity was clearly detected in the whole embryo as well as in the surrounding endosperm (Figure 6I). In mature seeds, strong GUS activity was also present in the cotyledonary embryo (Figure 6J), which is in sharp contrast to the absence of GUS activity in the embryo of mature seeds expressing FPS1S::GUS (Figure 6E). In conclusion, analysis of FPS1S::GUS and FPS2::GUS expression analysis demonstrated that FPS genes are differentially regulated during Arabidopsis seed development, showing highly complementary expression patterns particularly at late stages of seed formation.
Figure 6. Histochemical analysis of GUS activity during seed development in Arabidopsis lines harbouring transgenes FPS1S::GUS (panels A to E) and FPS2::GUS (panels F to J).
Seeds containing embryos at the globular (A and F), heart (B and G), early-torpedo (C and H), torpedo (D and I) and maturation green (E and J) developmental stages were analyzed for GUS expression as described under Material and Methods. GUS expression in the root meristematic region of the embryo at heart (G) and early-torpedo (H) stages in FPS2::GUS seeds is indicated by arrowheads. The inset between panels (H) and (I) shows expression of FPS2::GUS in the root meristematic region and the pro-vascular tissue of an embryo at the early-torpedo stage.
Functional Complementation of fps2 Mutant Seed Phenotypes by FPS1S
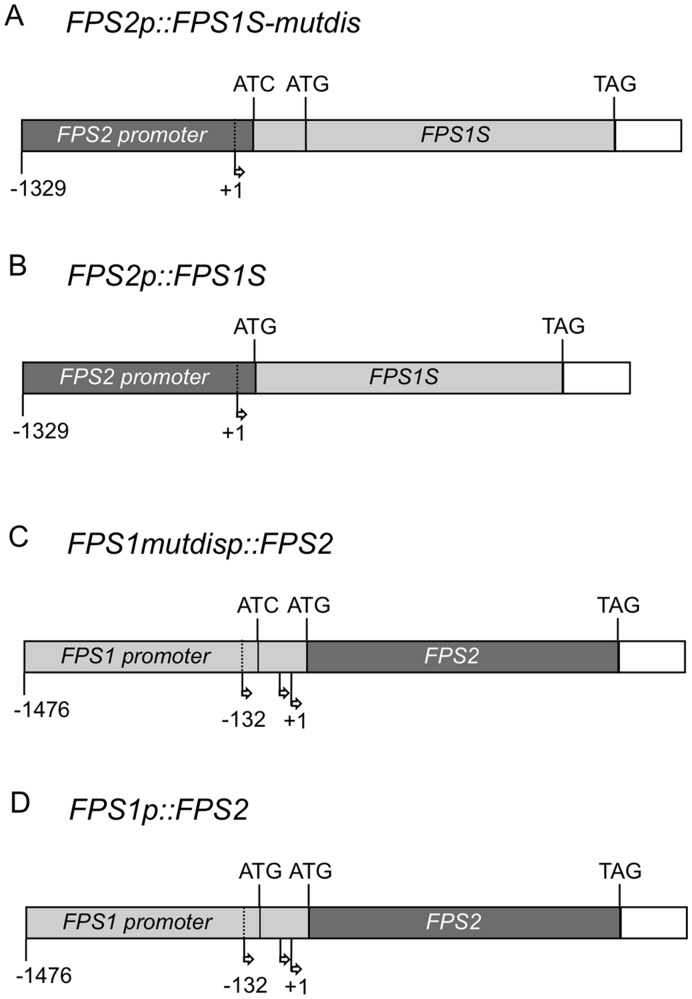
Mature seeds lacking FPS2 activity display several phenotypes including reduced levels of sitosterol, the main sterol found in plant tissues, increased HMGR activity and hypersensitivity to mevastatin compared to both fps1 and wild-type seeds [26]. To determine whether FPS1S could complement these phenotypes, transgenic fps2-1 plants expressing FPS1S under the control of the FPS2 gene promoter were obtained. To this end we created two different gene constructs that were able to produce only isoform FPS1S (Figure 7A and B). The FPS2p::FPS1S-mutdis gene consisted of 1375 bp of the FPS2 5′-flanking region, including 1329 bp of the promoter and the entire 5′-untranslated region, fused to the genomic coding region of FPS1 in which the distal ATG start codon was converted into an ATC codon to ensure that only FPS1S isoform could be produced. The FPS2p::FPS1S gene differed from FPS2p::FPS1S-mutdis gene in that the region comprised between the two ATG codons was removed.
Figure 7. Schematic representation of chimeric genes FPS2p::FPS1-mutdis (A), FPS2p::FPS1S (B), FPS1mutdisp::FPS2 (C) and FPS1p::FPS2 (D).
Dark gray boxes represent FPS2 genomic sequences whereas sequences corresponding to FPS1 gene are represented by light gray boxes. In all constructs the 3′-untranslated regions of both FPS genes is represented by an empty box. The position of the translational start codons is indicated as ATG and that of stop codons is indicated as TAG. In the FPS2p::FPS1S-mutdis (A) and FPS1mutdisp::FPS2 (C) constructs, the 5′-most ATG codon in exon 1 of the FPS1 gene was mutated to an ATC codon (encoding Ile). Arrows indicate the transcription start sites. In the FPS1 gene, position +1 was assigned to the most internal transcription start site.
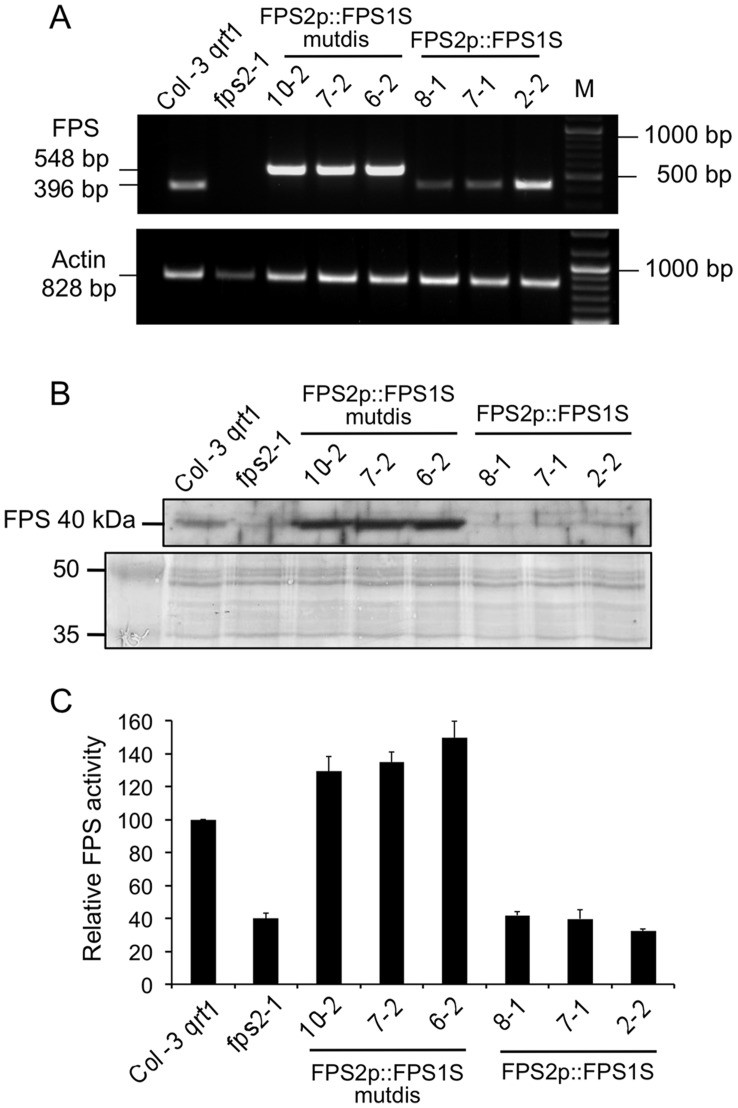
Several independent lines of these two kinds of transgenic plants were obtained and characterized. Among them, lines 6.2, 7.2 and 10.2 for FPS2p::FPS1S-mutdis and lines 2.2, 7.1 and 8.1 for FPS2p::FPS1S were selected for further characterization. The expression of FPS2p::FPS1S-mutdis and FPS2p::FPS1S was analyzed in young seedlings using semiquantitative RT-PCR and a primer set consisting of a forward primer specific for the FPS2 5′-untranslated sequence and a reverse primer located in the FPS1 coding region. The three selected FPS2p::FPS1S-mutdis lines showed very similar levels of expression of the FPS1 mRNA that were even higher than those detected in wild-type plants (Figure 8A). Accordingly, protein extracts from seeds of FPS2p::FPS1S-mutdis lines also contained both higher levels of FPS1S protein, as demonstrated by Western blot analysis (Figure 8B), and higher values of FPS activity (Figure 9C) compared to wild-type seeds. On the contrary, FPS protein and enzyme activity levels in extracts from seeds of the three FPS2p::FPS1S transgenic lines were comparable to those found in extracts of fps2-1 mutant seeds, which indeed were much lower that those detected in wild-type seed extracts (Figure 8B and C). The inability of FPS2p::FPS1S to restore wild-type levels of FPS protein and enzyme activity was not due to a lack of expression of the transgene, since its mRNA was detected in the seedlings of the three selected lines. In two of them (lines 7.1 and 8.1), expression of the transgene was less intense than in wild-type plants whereas in the third one (line 2.2) expression was slightly stronger than in control plants (Figure 8A). These observations suggested that translation of the chimeric FPS2::FPS1S mRNA expressed by FPS2p::FPS1S was severely impaired, which is most likely due to the different structural environment around the AUG start codon of the FPS2::FPS1S mRNA compared to that of the FPS2 and FPS2::FPS1S-mutdis mRNAs (Figure S1). Among the different structural features known to be involved in translational control of specific mRNAs [53], base-pairing involving nucleotides within the AUG codon has been recognized as an important structural determinant that may influence translation initiation [54], [55].
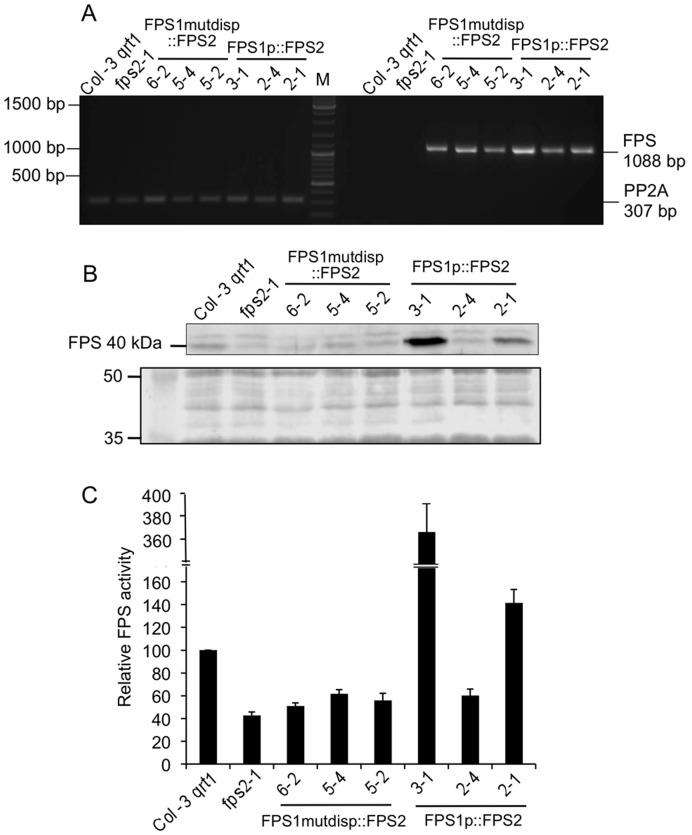
Figure 8. Characterization of fps2-1 mutant lines harbouring FPS2p::FPS1S and FPS2p::FPS1-mutdis genes.
(A) The expression of FPS2p::FPS1S and FPS2p::FPS1-mutdis was investigated using total RNA from 12-day-old seedlings of Arabidopsis wild-type, fps2-1 and the indicated lines of the fps2-1 mutant harbouring FPS2p::FPS1S-mutdis and FPS2p::FPS1S chimeric genes (upper panel). PCR products were electrophoresed in a 1% agarose gel. The size in bp of the amplified cDNA fragments corresponding to FPS2p::FPS1-mutdis, FPS2p::FPS1S and ACT2 (actin) genes is indicated on the left. The size of the fragment amplified from FPS2p::FPS1S-mutdis lines (548 bp) was larger than that amplified from both FPS2p::FPS1S lines and wild-type plants (396 bp) because the FPS2::FPS1-mutdis mRNA contains the region between the two ATG translation start codons of the FPS1 gene, which is not present in the FPS2::FPS1S mRNA. Numbers on the right indicate the sizes in bp of DNA markers shown in lane M. (B) Western blot analysis of total FPS protein in 16,000 g extracts from seeds of plant lines indicated above (upper panel). The lower panel shows the Coomassie blue-stained electrophoretic protein patterns in the 35 to 50 kDa range of extracts used for FPS protein level determinations. Images show the results of one representative experiment. (C) FPS activity in the 16,000 g protein extracts used for Western blot analysis. FPS activity in fps mutants is expressed relative to that in the wild-type, which was assigned a value of 100. The mean values and SE were calculated from three independent experiments.
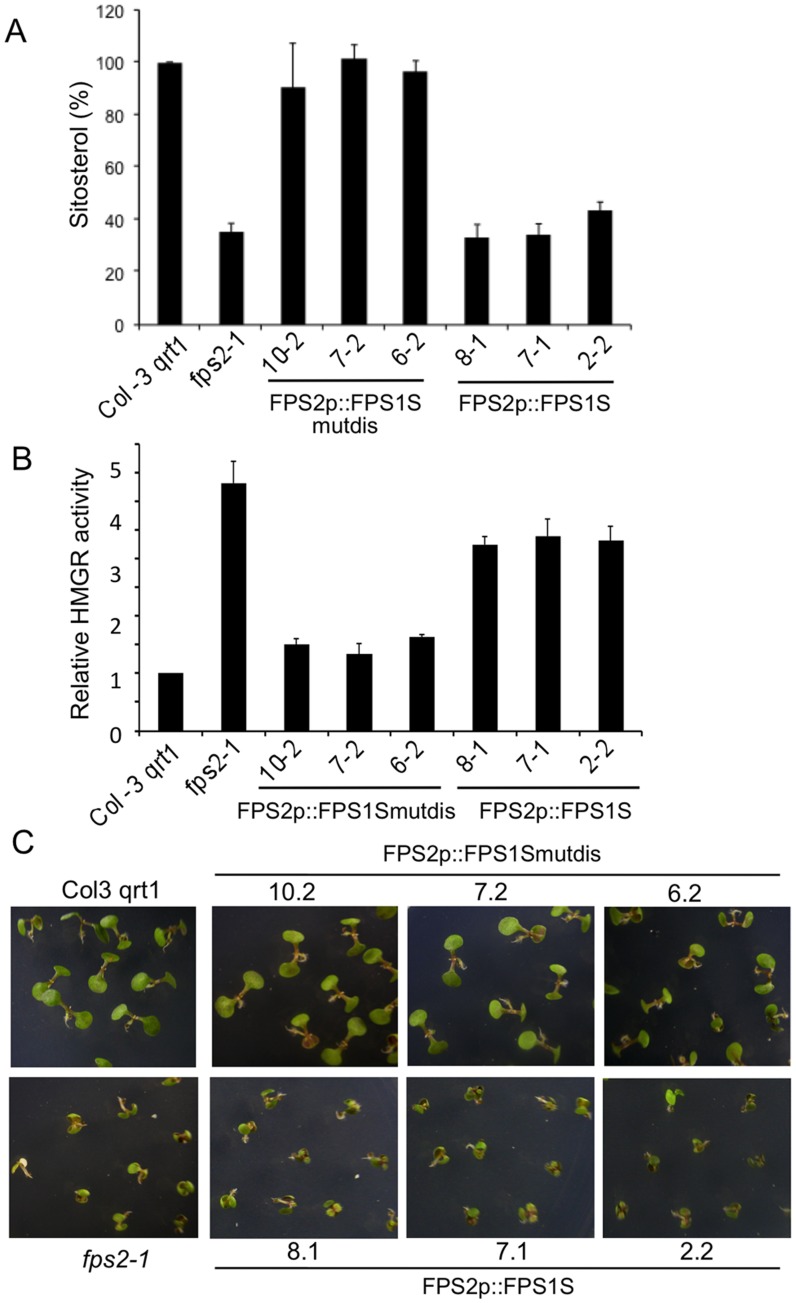
Figure 9. Reversion of fps2-1 phenotypes by expressing FPS1S under control of the FPS2 promoter.
Sitosterol (A) and HMGR activity (B) levels in seeds from Arabidopsis wild-type, fps2-1 and the indicated lines of the fps2-1 mutant harbouring FPS2p::FPS1S-mutdis and FPS2p::FPS1S chimeric genes. Values are expressed relative to the wild-type values. Data represent the mean values and SE of three independent experiments. (C) Reversion of mevastatin hypersensitivity. Representative seedlings of the same lines were grown for 12 days under long-day conditions (16 h light/8 h dark) on MS plates supplemented with 1 µM mevastatin.
To complete the characterization of the transgenic lines we next assessed whether expression of FPS1S driven by the FPS2 promoter could complement the characteristic phenotypes of fps2-1 seeds. Sitosterol and HMGR activity levels were quantified in seeds of FPS2p::FPS1S-mutdis and FPS2p::FPS1S lines. None of the fps2-1 seed phenotypes could be rescued by FPS2p::FPS1S expression (Figures 9A–C), a result that was fully consistent with the lack of FPS1S protein production and enzyme activity recovery (Figure 8B and C). On the contrary, sitosterol and HMGR activity levels were both restored to those of wild-type seeds in lines expressing FPS2p::FPS1S-mutdis (Figures 9A and B). Recovery of wild-type levels of HMGR activity in seeds also restored normal mevastatin sensitivity to these lines (Figure 9C). Altogether, these results demonstrated that FPS1S is able to functionally replace FPS2.
Rescue of fps2 Mutant Seed Phenotypes by Increasing Total FPS Activity in Seeds
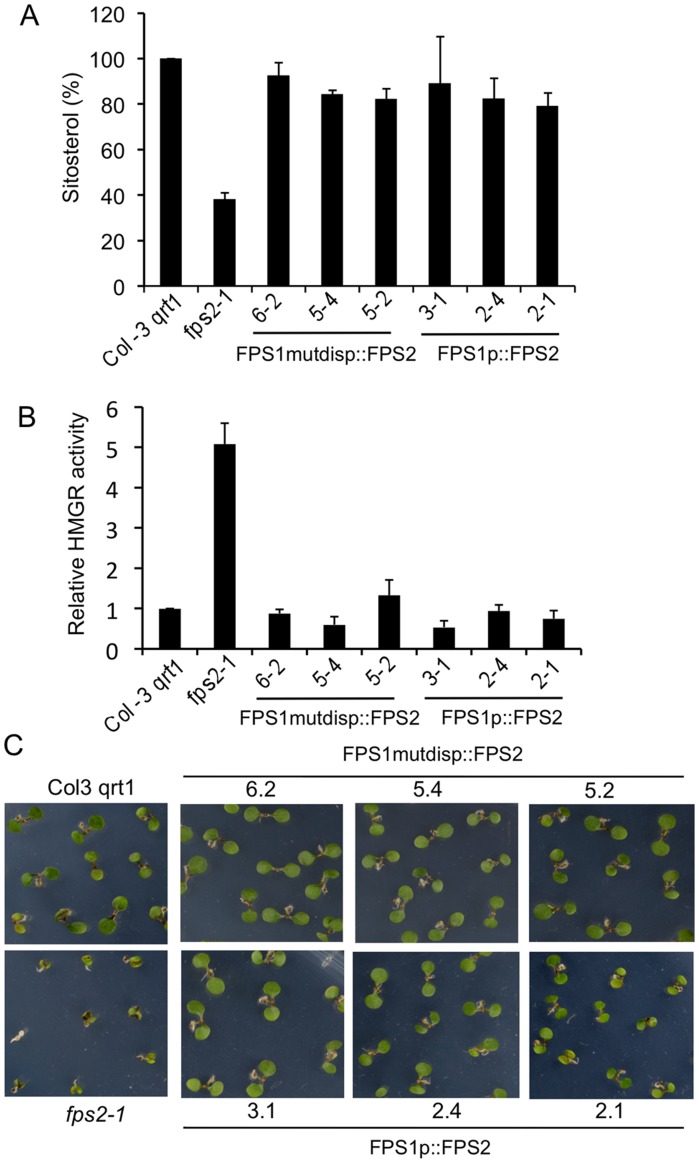
Once we had demonstrated that expression of FPS1S driven by the FPS2 promoter was able to rescue the phenotypes caused by loss of function of FPS2, we set out to determine whether expression of FPS2 driven by the FPS1 promoter could also rescue the fps2-1 phenotypes. To this end fps2-1 plants harbouring genes FPS1mutdisp::FPS2 and FPS1p::FPS2 (Figure 7C and D) were created and characterized. FPS1mutdisp::FPS2 consisted of 1526 bp of the FPS1 5′-flanking region, including 1338 bp of the FPS1 gene promoter and the entire 5′-untranslated region in which the distal ATG start codon was converted into an ATC codon, fused to the genomic coding region of FPS2 so that only FPS2 isoform could be produced from this transgene. In the FPS1p::FPS2 gene, the distal ATG codon was not disrupted and therefore this transgene could potentially express both FPS2 and a long version of FPS2 equivalent to FPS1L. Among the transgenic lines obtained, lines 6.2, 5.4 and 5.2 harbouring FPS1mutdisp::FPS2, and lines 3.1, 2.4 and 2.1 harbouring FPS1p::FPS2 were selected for further characterization. The expression of the transgenes was analyzed in young seedlings of these lines using semiquantitative RT-PCR and a primer set consisting of a forward primer specific for the FPS1 5′-untranslated sequence and a reverse primer specific for the FPS2 coding region. All selected lines showed comparable expression levels of the chimeric FPS2 mRNAs that, as expected, were not detected either in wild-type plants or in fps2-1 plants (Figure 10A). However, striking differences in FPS protein and activity levels were observed between the two groups of transgenic plants. FPS protein content in seeds of the three FPS1mutdisp::FPS2 lines was comparable and only slightly above that found in fps2-1 seed extracts. By contrast, total FPS protein levels in FPS1p::FPS2 seeds varied greatly. The amount of FPS protein in line 2.4 was pretty similar to that found in FPS1mutdisp::FPS2 plants whereas line 3.1 had strikingly elevated levels of FPS protein. Line 2.1 showed intermediate levels of FPS that were, nevertheless, higher than in wild-type plants (Figure 10B). These differences could be due to the production of different relative amounts of short and long versions of FPS2 in FPS1p::FPS2 seeds. Unfortunately, it is not possible to determine the relative contributions of the individual short and long FPS2 forms to the total amount of FPS2 protein since they both have the same size after processing of the long version of FPS2 [9]. In any case, a close correlation between FPS protein content and enzyme activity levels was observed in the seed extracts of all lines (Figure 10C). Analysis of the characteristic fps2-1 phenotypes in seeds of the transgenic lines revealed that the phenotypes associated to loss of function of FPS2 were fully rescued in all transgenic lines regardless of the degree of FPS activity enhancement in the mature mutant seeds (Figure 11A–C). In fact, a small increase of FPS activity of only 1.2-fold (line 6.2) was sufficient to restore wild-type sitosterol and HMGR activity values as well as normal Mst sensitivity to the same extent than a drastic enhancement of FPS activity (approximately 9-fold in line 3.1). These results suggested that impaired synthesis of FPP in the mature embryo due to the lack of FPS2 activity could be fully compensated just by increasing slightly the synthesis of FPP in the seed maternal tissues.
Figure 10. Characterization of fps2-1 mutant lines harbouring FPS1mutdisp::FPS2 and FPS1p::FPS2 genes.
(A) The expression of FPS1mutdisp::FPS2 and FPS1p::FPS2 was investigated using total RNA from 12-day-old seedlings of Arabidopsis wild-type, fps2-1 and the indicated lines of the fps2-1 mutant harbouring FPS1mutdisp::FPS2 and FPS1p::FPS2 chimeric genes. PCR products were electrophoresed in a 1% agarose gel. The size in bp of the amplified cDNA fragments corresponding to FPS1mutdisp::FPS2 and FPS1p::FPS2 (1088 bp) and PP2A genes (307 bp) is indicated on the right. Numbers on the left indicate the sizes in bp of DNA markers shown in lane M. (B) Western blot analysis of total FPS protein in 16,000 g extracts from seeds of plant lines indicated above (upper panel). The lower panel shows the Coomassie blue-stained electrophoretic protein patterns in the 35 to 50 kDa range of extracts used for FPS protein level determinations. Images show the results of one representative experiment. (C) FPS activity in the 16,000 g protein extracts used for Western blot analysis. FPS activity in mutants is expressed relative to that in the wild-type, which was assigned a value of 100. The mean values and SE were calculated from three independent experiments.
Figure 11. Reversion of fps2-1 phenotypes by expressing FPS2 under the control of the FPS1 promoter.
Sitosterol (A) and HMGR activity (B) levels in seeds from Arabidopsis wild-type, fps2-1 and the indicated lines of the fps2-1 mutant harbouring FPS1mutdisp::FPS2 and FPS1p::FPS2 chimeric genes. Values are expressed relative to the wild-type values. Data represent the mean values and SE of three independent experiments. (C) Reversion of mevastatin hypersensitivity. Representative seedlings of the same lines were grown for 12 days under long-day conditions (16 h light/8 h dark) on MS plates supplemented with 1 µM mevastatin.
Discussion
Isozymes usually display specific biochemical properties that allow fine-tuning of metabolic pathways to meet the specific needs of tissues and organs at different developmental stages and/or under different environmental conditions. The occurrence in plants of small FPS isozyme families has raised the still-unanswered question about the role of individual FPS isozymes in the cytosol/ER isoprenoid biosynthetic pathway. Most of the information currently available on the biological function of FPS isozyme family members has been obtained from the pattern of expression of the corresponding genes [11], [14], [15], [16], [17], [25] and the characterization of mutants that either overexpress [8], [10] or lack individual FPS isozymes [26]. By contrast, much less attention has been paid to investigate and compare the biochemical properties of individual FPS isozyme family members, despite this knowledge may also greatly contribute to the understanding of their role in the isoprenoid pathway [18].
In this study we report a detailed functional and structural characterization of the Arabidopsis FPS1S and FPS2 isozymes. Both enzymes were efficiently expressed in E. coli as GST-FPS fusion proteins, though the GST tag was removed by proteolytic cleavage during the purification process (Figure 1) to avoid any interference it could have on FPS activity [56]. Biochemical characterization of the purified native FPS1S and FPS2 revealed that both enzymes displayed very similar pH-rate profiles with an optimum at pH 7.0 (Figure 2). This pH preference was consistent with the predicted cytosolic localization of both FPS isozymes because the pH of the cytoplasm of plant cells is kept slightly alkaline at 7.2-7.5 under non-eliciting conditions [57]. The cytosolic localization of both FPS isozymes was demonstrated by transiently expressing N- and C-terminal fusions of FPS1S and FPS2 with GFP in agroinfiltrated N. benthamiana leaf cells. Under our experimental conditions all the four fusion proteins localized exclusively in the cytosol (Figure 5). This result was fully consistent with the absence of canonical subcellular targeting signals in the FPS1S and FPS2 proteins, and argued against the possibility that FPS1S and FPS2 could harbour a cryptic signal for peroxisomal targeting as recently reported to occur in a C. roseus short FPS enzyme. In contrast to our results, an N-terminal fusion of CrFPS with YFP was found to localize both in the peroxisomes and the cytosol of C. roseus cells despite the protein does not contain any classical peroxisomal targeting sequence [21]. It is also worth noting that our finding that none of the Arabidopsis short FPS isoforms reside in the peroxisomes was fully consistent with the results of extensive proteomic studies that have detected both FPS1S and FPS2 in the cytosolic proteome of Arabidopsis [58] and could not detect any FPS protein in purified Arabidopsis peroxisomes [59], [60]. The distinct subcellular localization of Arabidopsis and C. roseus short FPS isozymes could be related to the different profile of isoprenoid compounds produced by these two plant species [3], [61], though the central position of FPS in the isoprenoid pathway and the fundamental role of isoprenoid biosynthesis in plant biochemistry and physiology would argue against this hypothesis. Thus, given the ongoing controversy regarding the subcellular localization of FPS and other MVA pathway enzymes and the potential interference of fluorescent protein tags with proper subcellular targeting of proteins, further studies based on alternative experimental approaches are required in order to establish whether or not short FPS isozymes localize in different subcellular compartments in a plant-species dependent manner.
Comparison of the steady-state kinetic constants of purified FPS1S and FPS2 (Table 1) revealed that both enzymes have similar K m values for their substrates and a clear preference for GPP over DMAPP as the allylic substrate for FPP formation. A similar kinetic behaviour has also been reported for FPS enzymes from both eukaryotic and prokaryotic organisms [33], [18], [56], [62], [63]. The kinetic similarities between FPS1S and FPS2 are consistent with the high degree of sequence conservation (90.6% identity) shared by these isozymes. However, FPS1S and FPS2 also displayed some remarkable differential properties that must be a consequence of the small differences in their primary structure. FPS2 is a more efficient catalyst that FPS1S by a factor of 2.5- to 3-fold (Table 1). All known FPS proteins contain seven highly conserved amino acid motifs [64] that are involved in enzyme catalysis and product chain length determination [5], [65]. Arabidopsis FPS1S and FPS2 differ by only 32 amino acid residues out of a total of 343 and 342 amino acids, respectively. These amino acid changes are scattered throughout the sequence but, interestingly, none of them is located within the conserved motifs involved in catalysis [12]. Thus, it is difficult to attribute the differences in catalytic efficiency between FPS1S and FPS2 to a particular amino acid substitution. Rather it appears that this functional difference is the consequence of subtle changes in the three-dimensional structure of the enzymes due to multiple individual amino acid changes acting together. In fact, FPS is a homodimeric enzyme in which subunits interact with each other to form a shared active site rather than bearing an independent active site in each subunit [66]. The suggested structural differences between FPS1S and FPS2 must also account for the greater thermal stability (Figure 3) and higher sensitivity to the inhibitory effect of NaCl (Figure 2) exhibited by FPS2 compared to FPS1S. In fact, a single amino acid substitution is sufficient to alter temperature and salt sensitivity in the case of malate dehydrogenase [67], [68], [69]. Indeed, inspection of atomic-level homology models of FPS1S and FPS2 and theoretical calculations of the free energy and structural changes that accompany the single-site substitution of FPS2 unique amino acids into the FPS1S structural template indicate that surface salt bridge formation and reduced conformational entropy might largely account for the increased thermostability observed for FPS2. These stabilizing electrostatic effects are reflected in specific amino acid composition biases that have been extensively studied in thermophilic proteins [70], [71].
Our recent characterization of Arabidopsis fps single knockout mutants showing that FPS1S and FPS2 can almost fully compensate each other’s loss throughout most of the plant life cycle demonstrated that FPS1S and FPS2 have largely overlapping physiological functionalities. The only signs of functional specialization were observed in mature seeds and early developing seedlings. At these developmental stages FPS2 becomes the major contributor to total FPS activity, to the point that residual FPS1-derived activity in mature seeds is unable to supply enough FPP for normal sterol production, which triggers a compensatory enhancement of HMGR activity that is crucial for proper seed germination and seedling establishment [26]. Interestingly, our GUS expression analysis during seed development revealed a marked spatial and temporal divergence in the FPS1 and FPS2 expression pattern, which from the torpedo stage onward showed a strong degree of qualitative complementarity (Figure 6), also known as reciprocal expression pattern. This is when only one gene copy is expressed in some organs or tissue types and the other copy is expressed in others [72]. In the mature stages of seed development FPS2 expression clearly dominates over the expression of FPS1. The FPS2 gene is expressed throughout the endosperm and the embryo at the torpedo stage, and throughout the cotyledonary embryo in mature seeds, whereas the expression of FPS1 remains restricted to the maternal chalazal seed coat. The exclusive expression of FPS2 in the whole cotyledonary embryo, which at this stage occupies most of the seed, along with the higher catalytic efficiency of FPS2, explain why this isozyme is the major contributor to total FPS activity in mature seeds [26]. Interestingly, our results from genetic cross-complementation studies of the fps2-1 mutant seed phenotypes showed that expression of FPS1S driven by the FPS2 gene promoter (Figure 8) restored wild-type sitosterol and HMGR activity levels as well as normal sensitivity to mevastatin (Figure 9). This finding demonstrated that under normal conditions FPS1S and FPS2 are completely functionally interchangeable, which is fully consistent with the kinetic similarities exhibited by FPS1S and FPS2, and raises the question as to why FPS2 is the predominant FPS isozyme expressed in mature seeds and during the early stages of seed germination and seedling emergence. Seed viability is essential for survival of higher plants and therefore seeds are well equipped to withstand extended periods of exposure to harsh environmental conditions, as for instance extreme temperatures that may cause protein denaturation and inactivation among other effects. Arabidopsis seeds can complete germination even after treatment for more than 3.5 hours at 45°C [73], a temperature that is lethal for seedlings [74]. It is thus tempting to speculate that FPS2 has been evolutionarily selected as the predominant FPS isozyme in mature seeds of Arabidopsis because of its striking thermotolerance that could contribute to maintain seed germination capability of seeds exposed to high temperatures. It will be interesting to determine whether other isoprenoid biosynthetic enzymes expressed in seeds also display this biochemical feature. Moreover, the higher catalytic efficiency of FPS2 compared to FPS1S would enable early developing seedlings to sustain an active synthesis of isoprenoid precursors until the newly made FPS1S replaces FPS2 in this task.
FPP synthesis has been found to be indispensable for Arabidopsis embryos to progress beyond the pre-globular/globular stage and continue further development [26]. Our GUS expression analysis in seeds suggests that early developing embryos do not synthesize their own FPP since no embryo-specific expression of any of the FPS genes could be detected until the heart stage of embryo development (Figure 6). Rather it seems that early developing embryos import FPP or downstream FPP-derived isoprenoid precursors from other seed tissues like the maternal chalazal seed coat and/or the chalazal endosperm, where FPP can be synthesized as inferred from the expression of FPS1 and FPS2 in these tissues. This is a plausible hypothesis since the maternal chalazal seed coat and the chalazal endosperm are both considered to be specialized seed tissues involved in the uptake, reprocessing and release of metabolites into the endosperm to support embryogenesis [75], [76]. The proposal that an active synthesis of isoprenoid precursors occurs in the chalazal endosperm at the early stages of embryo development is further supported by the results of a previous study showing a strong expression of isopentenyl transferase (IPT) genes AtIPT4 and AtIPT8 in the chalazal zone of Arabidopsis seeds, that disappeared when the embryo progressed to the heart stage [77]. IPTs catalyze the first committed step of the cytokinin biosynthetic pathway, the isopentenylation of AMP, ADP and ATP from DMAPP [78]. Interestingly, the finding that FPS2 gene started to be expressed in embryos at the heart stage of development suggests that the embryo acquires the ability to synthesize its own FPP at this stage, though this biosynthetic capability does not seem to be essential for normal embryo and seed development since embryos in fps2 mutant seeds lacking FPS2 activity are fully viable (Figure S2). These observations support the hypothesis that the maternal chalazal seed coat is able to supply sufficient FPP or FPP-derived precursors to the embryo and the endosperm to sustain normal seed development, though the amount of isoprenoid precursors supplied by this tissue would not be sufficient to sustain normal sterol production in the fps2 seeds (Figure 9). The finding that expression of FPS2 under control of the FPS1 promoter (Figure 10), whose activity is restricted to the chalazal seed coat based on the GUS expression analysis (Figure 6), was also able to restore normal sterols levels to fps2 seeds (Figure 11) lends further support to the hypothesis that the maternal chalazal seed coat can function as a source of FPP for the developing embryo and reinforces the view that maternal seed tissues contribute sterol precursors to developing Arabidopsis embryos [79]. Nevertheless, to the best of our knowledge direct experimental evidence of FPP exchange between cells has yet to be provided. These cross-complementation experiments also indicated that the level of FPS1-derived activity in the chalazal seed coat of fps2 seeds is only slightly below the minimum threshold of total FPS activity needed to sustain a normal flux through the sterol pathway in seeds, since a 1.2-fold increase of FPS activity in this tissue with respect to the activity in fps2 seeds (Figure 10) is enough to prevent all fps2 seed and seedling phenotypes including normal sterols levels (Figure 11). In conclusion, the existence of two potential sources of FPP in seeds would explain why under normal conditions embryos and seeds of fps single knockout mutants develop like their wild-type counterparts.
Supporting Information
Predicted secondary structure of the region containing the AUG translation initiation codon of the FPS2::FPS1S, FPS2, and FPS2::FPS1S-mutdis mRNAs. Secondary structure models were generated by using the RNAfold web server (http://rna.tbi.univie.ac.at/). The AUG start codons are marked with an oval.
(TIF)
Normal embryo development in fps2-1 mutant seeds. Seeds with embryos at the indicated developmental stages were fixed and cleared for visualization as previously described [26].
(TIF)
Acknowledgments
The excellent technical support from Patricia Lordén and members of the Greenhouse and Microscopy facilities at CRAG and the Serveis Científico-Tècnics of the University of Barcelona is greatly appreciated. The authors also thank the Galicia Supercomputing Center (CESGA) for the provision of computational support. V.K. and D.C. were recipients of PhD fellowships from the Spanish Ministerio de Educación y Ciencia.
Funding Statement
This work was supported by grants from the Spanish Ministerio de Ciencia e Innovación (cofinanced by the European Regional Development Fund) (BIO2009-06984 and CSD2007-00036 to A.F., and PET2008_0101 and BFU2010-22260-C02-02 to M.C.V.), the Generalitat de Catalunya (SGR-00914 to A.F.), and the European Commission Framework Programme 7 (FP7 Project ComplexINC No. 279039 to M.C.V.) The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Croteau RB, Kutchan T, Lewis NG (2000) Natural products (secondary metabolites). In: Buchanan B, Gruissem W, Jones R, editors. Biochemistry and Molecular Biology of Plants. Rockville MD: American Society of Plant Biologists. 1250–1268.
- 2. Rodríguez-Concepción M (2006) Early steps in isoprenoid biosynthesis: multilevel regulation of the supply of common precursors in plant cells. Phytochem Rev 5: 1–15. [Google Scholar]
- 3. Wang KC, Ohnuma S (2000) Isoprenyl diphosphate synthases. Biochim Biophys Acta 1529: 33–48. [DOI] [PubMed] [Google Scholar]
- 4. Vandermoten S, Haubruge E, Cusson M (2009) New insights into short-chain prenyltransferases: structural features, evolutionary history and potential for selective inhibition. Cel Mol Life Sci 66: 3685–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tholl D, Sungbeon L (2011) Terpene specialized metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0143 doi:10.1043/tab.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crowell DN, Salaz MS (1992) Inhibition of growth of cultured tobacco cells at low concentrations of lovastatin is reversed by cytokinin. Plant Physiol 100: 2090–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Masferrer A, Arró M, Manzano D, Schaller H, Fernandez-Busquets X, et al. (2002) Overexpression of Arabidopsis thaliana farnesyl diphosphate synthase (FPS1S) in transgenic Arabidopsis induces a cell death/senescence-like response and reduced cytokinin levels. Plant J 30: 123–132. [DOI] [PubMed] [Google Scholar]
- 8. Manzano D, Fernández-Busquets X, Schaller H, González V, Boronat A, et al. (2004) The metabolic imbalance underlying lesion formation in Arabidopsis thaliana overexpressing farnesyl diphosphate synthase (isoform 1S) leads to oxidative stress and is triggered by the developmental decline of endogenous HMGR activity. Planta 219: 982–992. [DOI] [PubMed] [Google Scholar]
- 9. Manzano D, Busquets A, Closa M, Hoyerová K, Schaller H, et al. (2006) Overexpression of farnesyl diphosphate synthase in Arabidopsis mitochondria triggers light-dependent lesion formation and alters cytokinin homeostasis. Plant Mol Biol 61: 195–213. [DOI] [PubMed] [Google Scholar]
- 10. Fray RG, Wallace A, Fraser PD, Valero D, Hedden P, et al. (1995) Constitutive expression of a fruit phytoene synthase gene in transgenic tomatoes causes dwarfism by redirecting metabolites from the gibberellin pathway. Plant J 8: 693–701. [Google Scholar]
- 11. Attucci S, Aitken SM, Gulick PJ, Ibrahim RK (1995) Farnesylpyrophosphate synthase from white lupin: molecular cloning, expression, and purification of the expressed protein. Arch Biochem Biophys 321: 493–500. [DOI] [PubMed] [Google Scholar]
- 12. Cunillera N, Arró M, Delourme D, Karst F, Boronat A, et al. (1996) Arabidopsis thaliana contains two differentially expressed farnesyl-diphosphate synthase genes. J Biol Chem 271: 7774–7780. [DOI] [PubMed] [Google Scholar]
- 13. Cunillera N, Boronat A, Ferrer A (1997) The Arabidopsis thaliana FPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphate synthase isoform. J Biol Chem 272: 15381–15388. [DOI] [PubMed] [Google Scholar]
- 14. Li C-P, Larkins BA (1996) Identification of a maize endosperm-specific cDNA encoding farnesyl pyrophosphate synthetase. Gene 171: 193–196. [DOI] [PubMed] [Google Scholar]
- 15. Pan Z, Herickhoff L, Backhaus RA (1996) Cloning, characterization, and heterologous expression of cDNAs for farnesyl diphosphate synthase from the guayule rubber plant reveals that this prenyltransferase occurs in rubber particles. Arch Biochem Biophys 332: 196–204. [DOI] [PubMed] [Google Scholar]
- 16. Sanmiya K, Ueno O, Matsuoka M, Yamamoto N (1999) Localization of farnesyl diphosphate synthase in chloroplasts. Plant Cell Physiol 40: 348–354. [DOI] [PubMed] [Google Scholar]
- 17. Gaffe J, Bru JP, Causse M, Vidal A, Stamitti-Bert L, et al. (2000) LEFPS1, a tomato farnesyl pyrophosphate gene highly expressed during early fruit development. Plant Physiol 123: 1351–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hemmerlin A, Rivera SB, Erickson HK, Poulter CD (2003) Enzymes encoded by the farnesyl diphosphate synthase gene family in the big sagebrush Artemisia tridentata ssp. spiciformis. J Biol Chem 278: 32132–32140. [DOI] [PubMed] [Google Scholar]
- 19. Suzuki M, Muranaka T (2007) Molecular genetics of plant sterol backbone synthesis. Lipids 42: 47–54. [DOI] [PubMed] [Google Scholar]
- 20. Vranovà E, Coman D, Gruissem W (2012) Structure and dynamics of the isoprenoid pathway network. Mol Plant 5: 318–333. [DOI] [PubMed] [Google Scholar]
- 21. Thabet I, Guirimand G, Courdavault V, Papon N, Godet S, et al. (2011) The subcellular localization of periwinkle farnesyl diphosphate synthase provides insight into the role of peroxisome in isoprenoid biosynthesis. J Plant Physiol 168: 2110–2116. [DOI] [PubMed] [Google Scholar]
- 22. Sapir-Mir M, Mett A, Belausov E, Tal-Meshulam S, Frydman A, et al. (2008) Peroxisomal localization of Arabidopsis isopentenyl diphosphate isomerases suggests that part of the plant isoprenoid mevalonic acid pathway is compartmentalized to peroxisomes. Plant Physiol 148: 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simkin AJ, Guirimand G, Papon N, Courdavault V, Thabet I, et al. (2011) Peroxisomal localisation of the final steps of the mevalonic acid pathway in planta. Planta 234: 903–914. [DOI] [PubMed] [Google Scholar]
- 24. Kovacs WJ, Tape KN, Shackelford JE, Duan X, Kasumov T, et al. (2007) Localization of the pre-squalene segment of the isoprenoid biosynthetic pathway in mammalian peroxisomes. Histochem Cell Biol 127: 273–290. [DOI] [PubMed] [Google Scholar]
- 25. Cunillera N, Boronat A, Ferrer A (2000) Spatial and temporal patterns of GUS expression directed by 5′ regions of the Arabidopsis thaliana farnesyl diphosphate synthase genes FPS1 and FPS2. Plant Mol Biol 44: 745–758. [DOI] [PubMed] [Google Scholar]
- 26. Closa M, Vranovà E, Bortolotti C, Bigler L, Arró M, et al. (2010) The Arabidopsis thaliana FPP synthase isozymes have overlapping and specific functions in isoprenoid biosynthesis, and complete loss of FPP synthase activity causes early developmental arrest. Plant J 63: 512–525. [DOI] [PubMed] [Google Scholar]
- 27. Bach TJ, Boronat A, Campos N, Ferrer A, Vollack KU (1999) Mevalonate biosynthesis in plants. Crit Rev Biochem Mol Biol 34: 107–122. [DOI] [PubMed] [Google Scholar]
- 28. Hodzic A, Rappolt M, Amenitsch H, Laggner P, Pabst G (2008) Differential modulation of membrane structure and fluctuations by plant sterols and cholesterol. Biophys J 94: 3935–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54: 137–164. [DOI] [PubMed] [Google Scholar]
- 30. Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, et al. (2000) FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev 14: 1471–1484. [PMC free article] [PubMed] [Google Scholar]
- 31. Schrick K, Mayer U, Martin G, Bellini C, Kuhnt C, et al. (2002) Interactions between sterol biosynthesis genes in embryonic development of Arabidopsis. Plant J 31: 61–73. [DOI] [PubMed] [Google Scholar]
- 32. Hugueney P, Camara B (1990) Purification and characterization of farnesyl pyrophosphate synthase from Capsicum annuum . FEBS Lett 273: 235–238. [DOI] [PubMed] [Google Scholar]
- 33. Tholl D, Croteau R, Gershenzon J (2001) Partial purification and characterization of the short-chain prenyltransferases, geranyl diphospate synthase and farnesyl diphosphate synthase, from Abies grandis (grand fir). Arch Biochem Biophys 386: 233–242. [DOI] [PubMed] [Google Scholar]
- 34. Schmidt A, Gershenzon J (2007) Cloning and characterization of isoprenyl diphosphate synthases with farnesyl diphosphate and geranylgeranyl diphosphate synthase activity from Norway spruce (Picea abies) and their relation to induced oleoresin formation. Phytochemistry 68: 2649–2659. [DOI] [PubMed] [Google Scholar]
- 35. Montella C, Bellsolell L, Pérez-Luque R, Badía J, Baldomà L, et al. (2005) Crystal structure of an iron-dependent group III dehydrogenase that interconverts l-lactaldehyde and l-1,2-propanediol in Escherichia coli . J Bacteriol 187: 4957–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brinkmann U, Mattes RE, Buckel P (1989) High-level expression of recombinant genes in Escherichia coli is dependent on the availability of the dnaY gene product. Gene 85: 109–114. [DOI] [PubMed] [Google Scholar]
- 37. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 38. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193: 265–75. [PubMed] [Google Scholar]
- 39. Dale S, Arró M, Becerra B, Morrice NG, Boronat A, et al. (1995) Bacterial expression of the catalytic domain of 3-hydroxy-3-methylglutaryl-CoA reductase (isoform HMGR1) from Arabidopsis thaliana, and its inactivation by phosphorylation at Ser577 by Brassica oleracea 3-hydroxy-3-methylglutaryl-CoA reductase kinase. Eur J Biochem 233: 506–513. [DOI] [PubMed] [Google Scholar]
- 40. Niesen FH, Berglund H, Vedadi M (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2: 2212–2221. [DOI] [PubMed] [Google Scholar]
- 41. Rondeau J-M, Bitsch F, Bourgier E, Geiser M, Hemmig R, et al. (2006) Structural basis for the exceptional in vivo efficacy of bisphosphonate drugs. Chem Med Chem 1: 267–273. [DOI] [PubMed] [Google Scholar]
- 42. Eswar N, Eramian D, Webb B, Shen MY, Sali A (2008) Protein structure modeling with MODELLER. Methods Mol Biol 426: 145–159. [DOI] [PubMed] [Google Scholar]
- 43.Kim DE, Chivian D, Baker D (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32 (Web Server issue), W526–531. [DOI] [PMC free article] [PubMed]
- 44. Kaufmann KW, Lemmon GH, Deluca SL, Sheehan JH, Meiler J (2010) Practically useful: what the Rosetta protein modeling suite can do for you. Biochemistry 49: 2987–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, et al.. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35 (Web Server issue), W375–383. [DOI] [PMC free article] [PubMed]
- 46. Kellogg EH, Leaver-Fay A, Baker D (2011) Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins 79: 830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Benedix A, Becker CM, de Groot BL, Caflisch A, Böckmann RA (2009) Predicting free energy changes using structural ensembles. Nat Methods 6: 3–4. [DOI] [PubMed] [Google Scholar]
- 48. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 49. Stangeland B, Salehian Z (2002) An improved clearing method for GUS staining in Arabidopsis endosperm. Plant Mol Biol Rep 20: 107–114. [Google Scholar]
- 50. Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- 51. Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Mallory AC, et al. (1998) A viral suppressor of gene silencing in plants. Proc Natl Acad Sci USA 95: 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797. [DOI] [PubMed] [Google Scholar]
- 53. Adeli K (2011) Translational control mechanisms in metabolic regulation: critical role of RNA binding proteins, microRNAs, and cytoplasmic RNA granules. Am J Physiol Endocrinol Metab 301: E1051–E1064. [DOI] [PubMed] [Google Scholar]
- 54. Reineke LC, Komar AA, Caprara MG, Merrick WC (2008) A small stem-loop element directs internal initiation of the URE2 internal ribosome entry site in Saccharomyces cerevisiae . J Biol Chem 283: 19011–19025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Blaszczyk L, Ciesiolka J (2011) Secondary structure and the role in translation initiation of the 5′-terminal region of p53 mRNA. Biochemistry 50: 7080–7092. [DOI] [PubMed] [Google Scholar]
- 56. Mekkriengkrai D, Sando T, Hirooka K, Sakdapipanich J, Tanaka Y, et al. (2004) Cloning and characterization of farnesyl diphosphate synthase from the rubber-producing mushroom Lactarius chrysorrheus . Biosci Biotechnol Biochem 68: 2360–2368. [DOI] [PubMed] [Google Scholar]
- 57. Felle HH (2001) pH: Signal and messenger in plant cells. Plant Biol 3: 577–591. [Google Scholar]
- 58.Ito J, Batth TS, Petzold CJ, Redding-Johansson AM, Mukhopadhyay A, et al.. (2011) Analysis of the Arabidopsis cytosolic proteome highlights subcellular partitioning of central plant metabolism. J Proteome Res 10, 1571–1582. [DOI] [PubMed]
- 59.Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, et al. (2007) Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19, 3170–3193. [DOI] [PMC free article] [PubMed]
- 60. Reumann S, Quan S, Aung K, Yang P, Manandhar-Shrestha K, et al. (2009) In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol 150: 125–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guirimand G, Courdavault V, St-Pierre B, Burlat V (2010) Biosynthesis and regulation of alkaloids. In Pua EC, Davey M, editors. Plant Developmental Biology. Biotechnological Perspectives. Springer Verlag, Berlin, Heidelberg. vol. 2, 139–160
- 62. Sen SE, Trobaugh C, Béliveau C, Richard T, Cusson M (2007) Cloning, expression and characterization of a dipteran farnesyl diphosphate synthase. Insect Biochem Mol Biol 37: 1198–1206. [DOI] [PubMed] [Google Scholar]
- 63. Ranzer LK, Brück TB, Brück WM, López JV, Kerr RG (2009) A new prokaryotic farnesyldiphosphate synthase from the octocoral Eunicea fusca: differential display, inverse PCR, cloning, and characterization. Mar Biotechnol 11: 62–73. [DOI] [PubMed] [Google Scholar]
- 64. Koyama T, Obata S, Osabe M, Takeshita A, Yokoyama K, et al. (1993) Thermostable farnesyl diphosphate synthase of Bacillus stearothermophilus: molecular cloning, sequence determination, overproduction, and purification. J Biochem (Tokyo) 113: 355–363. [DOI] [PubMed] [Google Scholar]
- 65. Szkopińska A, Plochocka D (2005) Farnesyl diphosphate synthase; regulation of product specificity. Acta Biochim Polon 52: 45–55. [PubMed] [Google Scholar]
- 66. Koyama T, Gotoh Y, Nishino T (2000) Intersubunit location of the active site of farnesyl diphosphate synthase: Reconstruction of active enzymes by hybrid-type heteromeric dimers of site-directed mutants. Biochemistry 39: 463–469. [DOI] [PubMed] [Google Scholar]
- 67. Madern D, Ebel C, Zaccai G (2000) Halophilic adaptation of enzymes. Extremophiles 4: 91–98. [DOI] [PubMed] [Google Scholar]
- 68. Madern D, Pfister C, Zaccai G (1995) Mutation at a single acidic amino acid enhances the halophilic behaviour of malate dehydrogenase from Haloarcula marismortui in physiological salts. Eur J Biochem 230: 1088–1095. [DOI] [PubMed] [Google Scholar]
- 69. Dong Y, Somero GN (2009) Temperature adaptation of cytosolic malate dehydrogenases of limpets (genus Lottia): differences in stability and function due to minor changes in sequence correlate with biogeographic and vertical distributions. J Exp Biol 212: 169–177. [DOI] [PubMed] [Google Scholar]
- 70. Kumar S, Tsai C (2000) Factors enhancing protein thermostability. Protein Eng 13: 179–191. [DOI] [PubMed] [Google Scholar]
- 71. Zeldovich BZ, Berezovsky IN, Shakhnovich EI (2007) Protein and DNS sequence determinants of thermophillic adaptation. PLoS Comput Biol 3: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu S-L, Baute GJ, Adams K (2011) Organ and cell type–specific complementary expression patterns and regulatory neofunctionalization between duplicated genes in Arabidopsis thaliana. . Genome Biol Evol 3: 1419–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Larkindale J, Hall JD, Knight MR, Vierling E (2005) Heat stress phenotypes of Arabidopsis thaliana mutants implicate multiple signalling pathways in the acquisition of thermotolerance. Plant Physiol 138: 882–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Burke JJ, O’Mahony PJ, Oliver MJ (2000) Isolation of Arabidopsis mutants lacking components of acquired thermotolerance. Plant Physiol 123: 575–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nguyen H, Brown RC, Lemmon B (2000) The specialized chalazal endosperm in Arabidopsis thaliana and Lepidium virginicum (Brassicaceae). Protoplasma 212: 99–110. [Google Scholar]
- 76. Ingram GC (2010) Family life at close quarters: communication and constraint in angiosperm seed development. Protoplasma 247: 195–214. [DOI] [PubMed] [Google Scholar]
- 77. Miyawaki K, Matsumoto-Kitano M, Kakimoto T (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J 37: 128–138. [DOI] [PubMed] [Google Scholar]
- 78. Sakakibara H (2006) Cytokinins: Activity, biosynthesis and translocation. Annu Rev Plant Biol 57: 431–449. [DOI] [PubMed] [Google Scholar]
- 79. Rasbery JM, Shan H, LeClair RJ, Norman M, Matsuda SPT, et al. (2007) Arabidopsis thaliana squalene epoxidase 1 is essential for root and seed development. J Biol Chem 282: 17002–17013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Predicted secondary structure of the region containing the AUG translation initiation codon of the FPS2::FPS1S, FPS2, and FPS2::FPS1S-mutdis mRNAs. Secondary structure models were generated by using the RNAfold web server (http://rna.tbi.univie.ac.at/). The AUG start codons are marked with an oval.
(TIF)
Normal embryo development in fps2-1 mutant seeds. Seeds with embryos at the indicated developmental stages were fixed and cleared for visualization as previously described [26].
(TIF)