Abstract
Background and Aims
The Glutathione S-transferase P1 (GSTP1) polymorphism have been considered a risk modifier for developing head and neck cancer (HNC) in many studies; however, the results of such studies are inconsistent. The aim of this study was to evaluate the possible association between the GSTP1 Ile105Val polymorphism and risk of HNC.
Method
We performed a search in the relevant electronic database and a meta-analysis based on 28 published case–control studies that included 6,404 cases and 6,523 controls. To take into account the possibility of heterogeneity across the studies, a Chi-square based I2-statistic test was performed. Crude pooled odds ratios (ORs) with 95% confidence intervals (CIs) were assessed using both fixed-effects and random-effects models.
Results
The results of this meta-analysis showed that the GSTP1 Ile105Val polymorphism was not significantly associated with risk of HNC in the overall study population (pooled OR 1.00, 95% CI 0.92–1.09) or in subgroup analyses stratified by ethnicity, sample size, tumor site or publication year. Moreover, substantial evidence of heterogeneity among the studies was observed. Publication year was identified as the main cause of heterogeneity.
Conclusion
This meta-analysis does not support a significant association between the GSTP1 Ile105Val polymorphism and risk of HNC.
Introduction
Head and neck cancer (HNC), including cancers of the oral cavity, pharynx, and larynx, is the sixth most common cancer worldwide, with an annual incidence of 500,000 cases [1]. The age-standardized incidence rates in developed and developing countries are 28.4 and 20.6 per 100,000 population, respectively [2]. The development of HNC is a multifactorial process associated with a variety of risk factors. Exposure to tobacco smoke and the consumption of alcohol are considered to be the most important etiological factors in the development of HNC [3]–[5]. However, not every smoker and/or alcohol consumer develops HNC, which suggests that genetic host factors might also contribute to its carcinogenesis.
Recent evidence indicates that carcinogen-metabolizing genes and DNA-repair genes play critical roles in determining individual susceptibility to HNC. Polymorphisms in such genes that encode enzymes may increase or decrease carcinogen activation/detoxification and modulate DNA repair capacity, possibly by altering their expression and function. One of the most important systems in detoxification is the glutathione S-transferase (GST) family of enzymes. GSTs are phase II xenobiotic metabolizing enzymes involved in catalyzing the conjugation reactions of reactive intermediates of electrophilic compounds with cytosolic glutathione (GSH). Based on sequence similarities, human cytosolic GSTs are mainly coded for at 5 loci: GSTA (a), GSTT1 (h), GSTM1 (l), GSTP1 (p), and GSTM3 (c). GSTP1 is a major GST isoform that catalyzes the conjugation of glutathione to toxic compounds, resulting in more water-soluble and less biologically active products that are easily excreted.
GSTP1 is located on chromosome 11q13. To date, three polymorphic alleles of GSTP1 are known–GSTP1*B, GSTP1*C, and GSTP1*D–in addition to the wild-type allele, GSTP1*A [6]. GSTP1*B alleles have an A-to-G transition at nucleotide 313 (codon 105, exon 5), causing an isoleucine-to-valine change, while GSTP1*D contains a C-to-T transition at nucleotide 341 (codon 113), resulting in an Ala114-Val114 (A114V) substitution. GSTP1*C contains both these transitions [6], [7]. Enzymes with the valine at amino-acid 105 have a sevenfold higher catalytic efficiency for the diol epoxides of polycyclic aromatic hydrocarbons (PAH) than the isoenzymes with the isoleucine at this position. In contrast, the Val105 enzyme is threefold less effective with 1-chloro-2,4-dinitrobenzene as a substrate [6], [8], [9]. There is still no evidence of a functional effect of the A114V substitution alone (GSTP1*D), although it has been suggested that it augments the increased PAH activity of the I105V substitution (GSTP1*C) [8]. The missense substitution Ile105Val results from an A/G base substitution at nucleotide 313. The Val105 form of the GSTP1 enzyme may be 2–3 times less stable than the canonical Ile105 form [10] and may be associated with a higher level of DNA adducts [11].
The association between GSTP1 polymorphism and risk of HNC has been investigated, but these studies yielded controversial results. Some suggested that genetic polymorphisms of GSTP1 genes could influence the balance between metabolic activation and detoxification of carcinogens and are therefore, related to individual susceptibility to HNC [12]–[16], other reports, however, did not support these findings [17]–[21]. Whether GSTP1 polymorphism modifies the risk of HNC remains uncertain.
Meta-analyses have been conducted on the association between HNC and polymorphisms of GSTM1 and GSTT1 [22]–[24]. Additionally, a meta-analysis review of the association between HNC and GSTM1, GSTT, and GSTP1 that included journal articles published between 1993 and 2003 was reported [25]. However, that paper included only a limited number of published studies on GSTP1, and results from new studies have been reported recently. Therefore, we performed the current meta-analysis, including journal articles published from 1997 to 2011, to more comprehensively investigate the association between GSTP1 Ile05Val1 polymorphism and the risk of HNC.
Materials and Methods
Identification of Eligible Studies
To identify all articles that examined the association between the GSTP1 Ile105Val polymorphism and risk of HNC, we conducted a literature search of PubMed using the following combination of keywords: Glutathione S-transferases P, polymorphism, and head and neck cancer, oral cancer/neoplasms, laryngeal cancer/neoplasms, pharyngeal cancer/neoplasms, or upper aerodigestive tract cancer/neoplasms. The language of publication was restricted to English.
Inclusion and Exclusion Criteria
The following inclusion criteria were used for the literature selection: (a) case–control study methodology; (b) association of HNCs (including oral cancer, laryngeal cancer, pharyngeal cancer, and upper aerodigestive cancer) with GSTP1 polymorphisms explored; (c) study sample size, odds ratios (ORs), and 95% confidence intervals (CIs) stated in the article; and (d) HNC cases confirmed using histopathology.
Major exclusion criteria were as follows: (a) aim and design of the study obviously different from our research objectives; (b) not case-control study; (c) control population included malignant tumor cases; and (d) article was a review or duplication of previous publication.
After performing the literature search, we reviewed all papers in accordance with the criteria defined above. In addition, the Hardy–Weinberg equilibrium (HWE) test was conducted to evaluate the genetic equilibrium of each study [26].
Data Extraction
Two investigators (Lang and Song) reviewed and extracted information independently from selected publications in accordance with the criteria for inclusion and exclusion. Data were then entered into a database. Any conflicts over study/data inclusion were settled by a discussion between the investigators.
Statistical Analysis
The crude odds ratios (ORs) and 95% confidence intervals (95% CIs) of GSTP1 Ile105Val polymorphism and risk of HNC were estimated for each study. For detection of any possible sample size biases, the OR and its 95% CI to each study were plotted respectively against the number of participants. A Chi-square based I2-statistic test was performed to assess the potential heterogeneity among the studies. An I2 value of less than 25% indicates low heterogeneity, 25% to 50% indicates moderate heterogeneity, and greater than 50% indicates high heterogeneity. If the result of the heterogeneity test was p>0.05, ORs were pooled according to the fixed-effect model [27]. Otherwise, the random-effect model was used [28]. The significance of the pooled ORs was determined by the Z-test. The HWE was assessed via Fisher’s exact test. Publication bias was assessed by visual inspection of Begg’s funnel plots and linear regression, respectively [29], [30]. All statistical analyses were undertaken using the Stata 10.0 software program (Stata Corporation, College station, TX).
Results
Literature Search and Studies Characteristics
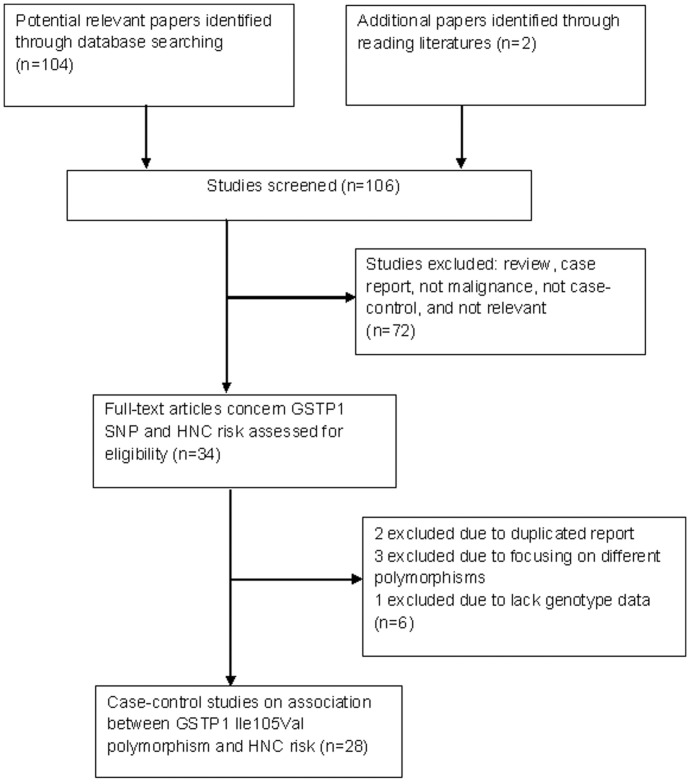
Our keyword search identified 104 papers and two additional relevant papers were adopted through reading literatures. Among them, 72 papers did not meet our criteria and were excluded after review of the abstracts. After reading the full texts of the remaining 34 papers, we eliminated an additional 6 papers, including 2 duplicated reports, 3 investigating different polymorphisms, and 1 lack of genotype data (Fig. 1). Therefore, a total of 28 case-control studies were identified, with 6404 cases and 6523 controls, of which 3136 cases and 3171 controls had the combined variant genotypes (Ile/Val and Val/Val) [12]–[21], [31]–[48]. The frequency of the GSTP1 valine genotype was 23.8–72.7% among controls and 24.9–72.3% among cases. Among these 28 studies, 11 studies were performed in Asian populations, 11 in Caucasians, and 4 in “Whites”, in 2 studies the population was unclear. Controls in 6 studies were population-based and the other 22 studies adopted hospital-based population as controls. Nine papers focused on the oral cavity or oropharyngeal cancer, 2 papers on laryngeal cancer, 1 paper on nasopharyngeal cancer, and the other 16 papers on unspecific HNCs (including 2 papers that also provided data on non-HNCs). The study characteristics are listed in Table 1.
Figure 1. The flow diagram of included/excluded studies.
Table 1. Characteristics of eligible studies.
| Author | Year | Country | Ethnicity | Genotyping Method | Tumor Site | Source of Control | Sample Size (case/control) | Matching | HWE | ||
| Ruwali M | 2011 | India | Asian | PCR-RFLP | Head & Neck | HB | 500/500 | Age/Gender/Race | Y | ||
| Karen-Ng LP | 2011 | Malaysia | Asian | PCR-RFLP | Oral Cavity | HB | 115/116 | None | UK | ||
| Chen MK | 2010 | China | Asian | PCR-RFLP | Oral Cavity | HB | 164/274 | Age/Gender/Race | Y | ||
| Ruwali M | 2009 | India | Asiana | PCR | Head & Neck | HB | 350/350 | Age/Gender/Race | Y | ||
| Singh M | 2008 | India | Asiana | PCR | Head & Neck | HB | 175/200 | Age/Gender | N | ||
| Datta S | 2007 | India | Asian | PCR-RFLP | Oral Cavity | HB | 307/209 | Smoke | Y | ||
| Cho CG | 2006 | Korea | Asian | PCR | Head & Neck | HB | 294/333 | UK | Y | ||
| Cheng YJ | 2003 | China | Asian | PCR-RFLP | Nasopharynx | PB | 264/323 | UK | Y | ||
| Katoh T | 1999 | Japan | Asian | PCR-RFLP | Oral Cavity | HB | 83/122 | UK | N | ||
| Morita S | 1999 | Japan | Asian | PCR-RFLP | Head & Neck | PB | 145/164 | None | Y | ||
| Soya SS | 2007 | India | Asian | PCR-RFLP | Head & Neck | HB | 408/222 | Age/Gender/Race | Y | ||
| Oude Ophuis MB | 2003 | Netherland | Caucasian | PCR-RFLP | Head & Neck | HB | 235/285 | Age | Y | ||
| Kelders WP | 2002 | Netherland | UKb | PCR-RFLP | Head & Neck | HB | 85/51 | UK | Y | ||
| McWilliams JE | 2000 | USA | Caucasian | PCR-RFLP | Head & Neck | HB | 146/124 | Race/Alcohol | Y | ||
| Park JY | 1999 | USA | Caucasian + American | PCR-RFLP | Oral Cavity | HB | 157/260 | UK | N | ||
| Jourenkova-MN | 1999 | Europe | Caucasian | PCR-RFLP | Oral Cavity | HB | 121/172 | Age/Gender/Race/Smoke | Y | ||
| Jourenkova-MN | 1999 | France | Caucasian | PCR-RFLP | Larynx | HB | 129/172 | UK | Y | ||
| Matthias C | 1998 | Germany | Caucasian | PCR-RFLP | Head & Neck | HB | 380/180 | Race | N | ||
| To-Figueras J | 2002 | Spain | Caucasian | PCR-RFLP | Larynx | HB | 204/203 | None | Y | ||
| Soucek P | 2010 | Czech and Poland | Caucasian | PCR-RFLP | Head & Neck | PB | 116/122 | Age/Gender/Alcohol | Y | ||
| Reszka E | 2008 | Poland | Caucasian | PCR-RFLP | Head & Neck | PB | 127/151 | Gender/Alcohol/Occupation | UK | ||
| Harth V | 2008 | Germany | Caucasian | RT-PCR | Head & Neck | HB | 312/300 | UK | Y | ||
| Evans AJ | 2004 | USA | Caucasian | PCR-RFLP | Head & Neck | PB | 283/208 | None | Y | ||
| Leichsenring A | 2006 | Brazil | UKb | PCR-RFLP | Oral Cavity | HB | 72/60 | Gender | Y | ||
| Hataqima A | 2008 | Brazil | White+Black | PCR-RFLP | Oral Cavity | HB | 231/212 | Age/Gender/Race | Y | ||
| Peters ES | 2006 | USA | Whitec | PCR-RFLP | Head & Neck | PB | 690/748 | Age/Gender | Y | ||
| Amador AG | 2002 | USA | Whitec | PCR-RFLP | Head & Neck | HB | 137/99 | UK | N | ||
| Olshan AF | 2006 | USA | Whitec | PCR-RFLP | Head & Neck | HB | 172/193 | UK | Y | ||
male only;
Brazilian;
majority is White;
HWE, Hardy–Weinberg equilibrium; PCR-RFLP, polymerase chain reaction–restriction fragment length polymorphism; HB: Hospital based; PB: Population based; UK: Unknown or Unstated; Smoke: Tobacco consumption; Alcohol: Alcohol consumption. RT-PCR, Real-time–polymerase chain reaction.
Test of Heterogeneity
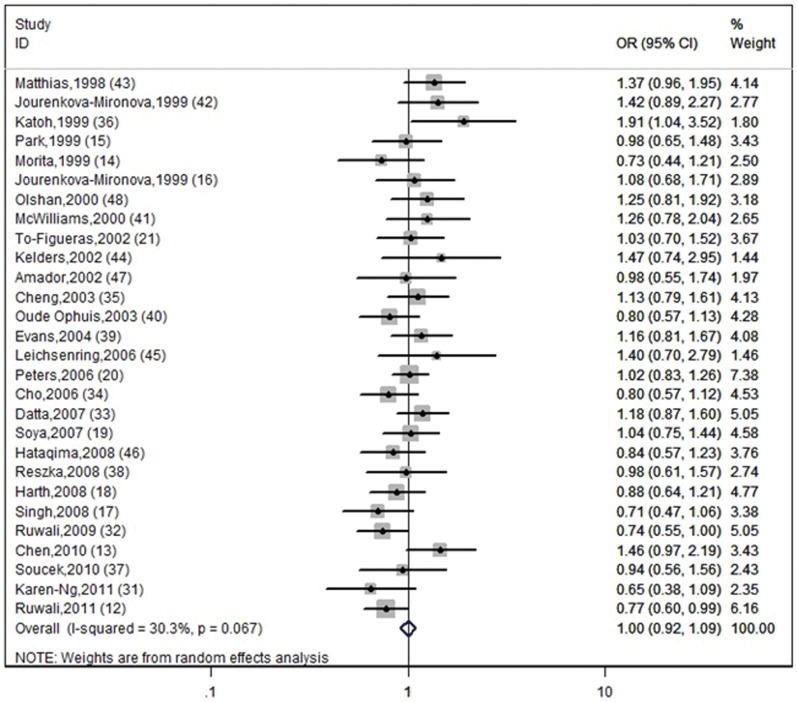
Figure 2 shows the association between the GSTP1 Ile105Val polymorphism and risk of HNC. We analyzed the heterogeneity for all 28 studies and the test value of Chi-square was 38.62, with 27 degrees of freedom (d.f.) and 0.05<P<0.1 (p = 0.069). This result shows there is heterogeneity among the studies. Additionally, I2 value is calculated as another index for the heterogeneity test. As shown in Figure 2, the I2 value was 30.1% (between 25% to 50%), which suggests slight to moderate heterogeneity. Thus, the random-effect model was utilized for evaluation. In Figure 2, one can observe that 5 studies [13], [31], [36], [44], [45] may attribute to the major sources of heterogeneity. Further stratified meta-analysis is needed to perform.
Figure 2. Forest plot with a random-effect model for GSTP1 Ile105Val polymorphism and risk for HNC.
The center of each square represents the OR value, the area of each square is proportional to the sample size and thus the weight of the corresponding study, and the horizontal short line indicates the 95% confidence interval. The pooled OR is represented by the diamond. (Test for heterogeneity: chi2 = 38.75, df = 27, p = 0.067. Test for overall effect: z = 0.02, p = 0.984).
Meta-analysis Results
The summary OR for the GSTP1 Ile105Val genotype was 1.00 (OR = 1.00, 95% CI = 0.92–1.09) and the test for overall effect Z value was 0.02 (p = 0.984). The overall meta-analysis showed that there was no significant association between the risk of HNC and GSTP1 Ile105Val polymorphism (p>0.05). Figure 2 shows the pooled OR with 95% CI of association between the GSTP1 Ile105Val polymorphism and risk of HNC.
To determine the cause of the moderate heterogeneity among the studies and to obtain more accurate results, we conducted further meta-analyses stratified according to tumor site, study sample size, ethnic group, publication year, source of controls, and consistency of frequency with HWE. In four unspecific “Head and Neck” tumor site studies, the sample size of subtype cancers were also available: thus, a total of 12 studies on oral and oropharyngeal cancers, 6 studies on laryngeal cancer, 1 on nasopharyngeal cancer, and 13 on mixed HNCs were examined in a stratified meta-analysis (Table 2). Meta-regression was employed to calculate the between-study variance. Publication year was identified as the main cause of heterogeneity. Only 8.22% of residual variation heterogeneity was left if we excluded the study year from the meta-analysis, and the estimate of between-study variance was tau = 0.000561, p = 0.005. The pooled OR of studies published before 2005 appeared to indicate an association between the GSTP1 Ile105Val polymorphism and risk of HNC, although p>0.05. Meta-analyses stratified according to other factors, such as tumor site, source of controls, ethnicity, sample size, and consistency of HWE, did not show a significant association between the GSTP1 Ile105Val polymorphism and risk of HNC (Table 2). The residual variation I2 values (heterogeneities) of stratified meta-regression were 26.1% of tumor site, 28.4% of ethnicity, 32.9% of source of control, 31.3% of sample size, 32.8% of HWE consistency respectively. Compared with the overall I2 value of 30.1%, none of these factors predominantly contribute to the overall heterogeneity, except publication year.
Table 2. Stratified Meta-analysis and Meta-regression Analysis of Heterogeneity.
| Stratification | N | Meta-regression* | OR | 95% CI | P value |
| Overall | 28 | I2=30.3% | 1.00 | 0.921.10 | 0.984 |
| Tumor site | P=0.157 (I2 res=26.1%) | ||||
| Mixed HNC | 13 | 0.94 | 0.851.05 | 0.267 | |
| Oral/Oropharynx | 12 | 1.10 | 0.901.33 | 0.355 | |
| Larynx | 6 | 0.87 | 0.691.11 | 0.260 | |
| Nasopharynx | 1 | 1.13 | 0.791.61 | 0.742 | |
| Ethnicity | P=0.208 (I2 res=28.4%) | ||||
| Asian | 11 | 0.94 | 0.791.11 | 0.451 | |
| Caucasian | 11 | 1.05 | 0.931.18 | 0.441 | |
| Other | 6 | 1.05 | 0.901.22 | 0.549 | |
| Source of control | P=0.894 (I2 res=32.9%) | ||||
| Hospital-based | 22 | 1.00 | 0.901.12 | 0.941 | |
| Population-based | 6 | 1.02 | 0.891.18 | 0.762 | |
| Publication year | P=0.005 (I2 res=8.2%) | ||||
| Before 2005 | 14 | 1.12 | 0.991.26 | 0.066 | |
| 2005 and after | 14 | 0.92 | 0.821.03 | 0.142 | |
| Sample Size | P=0.363 (I2 res=31.3%) | ||||
| 300 | 21 | 1.02 | 0.911.14 | 0.714 | |
| 300 | 7 | 0.97 | 0.831.13 | 0.686 | |
| HWE | P=0.974 (I2 res=32.8%) | ||||
| Yes | 21 | 1.00 | 0.911.09 | 0.933 | |
| No | 7 | 1.01 | 0.781.30 | 0.972 |
Meta-regression indicates the between-study variance.
Includes studies without enough data to calculate HWE.
N, number; OR, odds ratio; 95%CI, confidence interval; “I2” indicates variation in OR attributable to heterogeneity; “I2 res” in the parentheses indicates residual variation due to heterogeneity of each factor; HWE, Hardy–Weinberg equilibrium.
Sensitivity Analysis
In order to compare the differences between the meta-analyses and evaluate their sensitivity, we also reported the results of the fixed-effect model for GSTP1, as follows: the combined OR was 0.99 with 95% CI from 0.92 to 1.06 (z = 0.27, p = 0.790), similar to the results of the random-effect models (test of heterogeneity χ2 = 38.75, df = 27, p = 0.067).
Bias Diagnostics
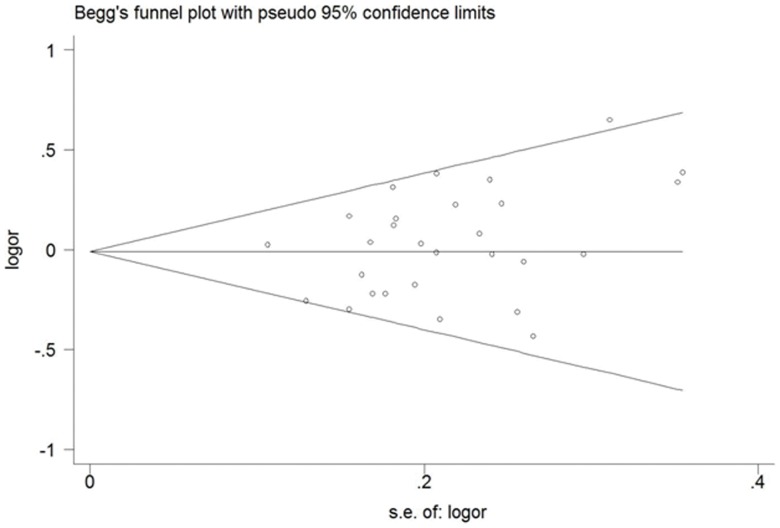
A Begg’s funnel plot created to assess possible publication biases showed nearly symmetrical pattern, indicating that there was no publication bias (Fig. 3). In addition, Egger’s test used to quantitatively evaluate the publication bias, found no evidence of bias (p = 0.128).
Figure 3. Begg’s funnel plot for publication bias assessment.
Each hollow cycle represents a separate study for the indicated association.
Discussion
GSTP1 polymorphisms have been evaluated as risk factors for cancers in a number of studies. Extensive molecular epidemiological studies indicate that the GSTP1 variant is more likely to lead to development of cancer than its wild type. A series of studies demonstrated that the GSTP1 codon 105 polymorphism is associated with various types of cancer, including breast, prostate, and lung cancer [49]–[51]. However, in this meta-analysis of 28 case–control studies, there was no evidence supporting the hypothesis that the GSTP1 Ile105Val polymorphism is significantly associated with risk of HNC in the general population.
One possible explanation for this lack of association may be suboptimal study design. Considering the role of GSTP1 as a carcinogen-metabolizing gene, potential effect of tobacco and alcohol to HNC should be taken into consideration of the study design. As shown in Table 1, the rate of matched tobacco or alcohol consumption between case and control was low. There were only 2 studies [33], [42] with smoking matching and 3 with alcohol consumption matching [37], [38], [41]. Although adjustment according to smoking and alcohol has been done in most studies, this may still cause inevitable heterogeneity among studies.
There is also evidence of heterogeneity on other aspects among the studies in this systematic review and meta-analysis. Potential sources of heterogeneity include the publication year, case-control matching, and sample size. The pooled subgroup analysis of a subset of studies published before 2005 suggested a weak association, although it was not statistically significant (p = 0.066). The reason for this is not clear. It might be due to uncontrolled confounding factors or to inherent bias in the study design. It is clear from this meta-analysis that the design of some of the case–control studies was suboptimal. From the forest plot (Fig. 2), one can observe that 5 studies are the major sources of heterogeneity [13], [31], [36], [44], [45]. In some papers, the study design included important oversights, for example, some studies used small sample sizes [36], [44], [45]. Selection bias may be another source of heterogeneity. Some studies used samples with highly heterogeneous ethnic origins [45], [47] or the composition of ethnicity was not clearly stated [44]. Other studies recruited control subjects from hospital-based population. Since it is conceivable that the GSTP1 gene might confer susceptibility to non-cancer disease, the genotype frequencies might be different between population-based and hospital-based controls, and this might introduce heterogeneity among studies. The use of population-based controls is, therefore, more appropriate in association studies.
Due to the fact that deviation from HWE may point to methodological weaknesses, such as biased selection of subjects, genotyping errors, or population stratification, we performed further stratified analyses. The meta-analyses that excluded studies whose genotype frequencies in controls significantly departed from HWE did not result in any substantial modification of crude OR results pertaining to the GSTP1 Ile105Val (Table 2). Although studies with heterogeneity do not significantly alter the estimate of the overall OR and result in a type I error, more optimal and well-designed studies are needed to investigate this association more closely and systematically.
Studies from Asian countries tended to support the association between the GSTP1 Ile105Val polymorphism and risk of HNC, whereas most studies from European countries failed to demonstrate this association. According to our results, after subgroup analysis by ethnicity, cancer sites, and source of control group, no significant associations were observed. However, ethnicity is definitely an important factor when investigating the association of genetic polymorphisms with cancer risk. Further large-scale investigation might be needed to validate our results.
Our results showed no association between the GSTP1 Ile105Val polymorphism and the risk of HNC in general, as well as between this polymorphism and the risk of oral cancer or laryngeal cancer when we stratified HNC according to subtypes of tumor sites. Our results are consistent with those of Hashibe and colleagues, except the finding of the higher risk of oral cancer than the risk of laryngeal cancer for the GSTP1 any valine genotype [25]. It was reported that the metabolic action of GST enzymes may differ by cancer site; the highest concentrations of GSTP1 have been observed in oral and pharyngeal tissues, and the highest concentrations of GSTM1 have been observed in laryngeal tissue, relative to the other GSTs [52]. Studies on GSTP1 polymorphism and the risk of oral cavity cancer reached controversial conclusions [13], [15], [31], [36], [45]. In this study, no positive association was found between the GSTP1 polymorphism and the risk of oral or oropharyngeal cancer. Since the data on subsets of oral cavity cancer and oropharyngeal cancer were not available, further stratified meta-analysis was not able to be performed. If there is association between the GSTP1 Ile105Val polymorphism and the risk of oral cavity cancer or oropharyngeal cancer still remains unclear, although it is negative as combined subset according to our results.
Although considerable effort was made to test for the possible association between the GSTP1 Ile105Val polymorphism and risk of HNC, there are still some limitations inherited from the published studies. First, due to limited detailed data presented in the published studies, the potential effect of important risk factors to HNC was not examined, such as smoking (data of sample size associated with smoking was available in only 7 studies) and alcohol consumption. Second, the results are only based on single-factor estimates, without adjustment for other risk factors such as age, ethnicity, family history, and environmental factors. Third, GSTP1 may influence susceptibility to head and neck cancer independently or with other genes. However, due to lack of individual data in the present review, we did not perform more detailed analyses, such as analyses of joint effects with other risk factors or gene-gene or gene-environment interactions.
In conclusion, this meta-analysis demonstrates that the GSTP1 Ile105Val polymorphism appears to not be associated with risk of HNC. To confirm our findings, well-designed studies with large sample sizes in diverse ethnic populations are warranted.
Supporting Information
Flowchart for selection of studies.
(PDF)
PRISMA Checklist.
(PDF)
Funding Statement
The authors have no funding or support to report.
References
- 1. Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, et al. (2004) Cancer statistics, 2004. CA Cancer J Clin 54: 8–29. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F (2011) Center MM, Ferlay J, Ward E, et al (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 3. Maier H, Dietz A, Gewelke U, Heller WD, Weidauer H (1992) Tobacco and alcohol and the risk of head and neck cancer. Clin Investig 70: 320–327. [DOI] [PubMed] [Google Scholar]
- 4. Day GL, Blot WJ, Austin DF, Bernstein L, Greenberg RS, et al. (1993) Racial differences in risk of oral and pharyngeal cancer: alcohol, tobacco, and other determinants. J Natl Cancer Inst 85: 465–473. [DOI] [PubMed] [Google Scholar]
- 5. Hecht SS (2003) Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer 3: 733–744. [DOI] [PubMed] [Google Scholar]
- 6. Ali-Osman F, Akande O, Antoun G, Mao JX, Buolamwini J (1997) Molecular cloning, characterization, and expression in Escherichia coli of full-length cDNAs of three human glutathione S-transferase Pi gene variants. Evidence for differential catalytic activity of the encoded proteins. J Biol Chem 272: 10004–10012. [DOI] [PubMed] [Google Scholar]
- 7. Zimniak P, Nanduri B, Pikula S, Bandorowicz-Pikula J, Singhal SS, et al. (1994) Naturally occurring human glutathione S-transferase GSTP1-1 isoforms with isoleucine and valine in position 104 differ in enzymic properties. Eur J Biochem 224: 893–899. [DOI] [PubMed] [Google Scholar]
- 8. Hu X, Xia H, Srivastava SK, Herzog C, Awasthi YC, et al. (1997) Activity of four allelic forms of glutathione S-transferase hGSTP1-1 for diol epoxides of polycyclic aromatic hydrocarbons. Biochem Biophys Res Commun 238: 397–402. [DOI] [PubMed] [Google Scholar]
- 9. Sundberg K, Johansson AS, Stenberg G, Widersten M, Seidel A, et al. (1998) Differences in the catalytic efficiencies of allelic variants of glutathione transferase P1-1 towards carcinogenic diol epoxides of polycyclic aromatic hydrocarbons. Carcinogenesis 19: 433–436. [DOI] [PubMed] [Google Scholar]
- 10. Johansson AS, Stenberg G, Widersten M, Mannervik B (1998) Structure-activity relationships and thermal stability of human glutathione transferase P1-1 governed by the H-site residue 105. J Mol Biol 278: 687–698. [DOI] [PubMed] [Google Scholar]
- 11. Ryberg D, Skaug V, Hewer A, Phillips DH, Harries LW, et al. (1997) Genotypes of glutathione transferase M1 and P1 and their significance for lung DNA adduct levels and cancer risk. Carcinogenesis 18: 1285–1289. [DOI] [PubMed] [Google Scholar]
- 12.Ruwali M, Singh M, Pant MC, Parmar D (2011) Polymorphism in glutathione S-transferases: susceptibility and treatment outcome for head and neck cancer. Xenobiotica. 2011/10/01 ed. 1122–1130. [DOI] [PubMed]
- 13. Chen MK, Tsai HT, Chung TT, Su SC, Kao TY, et al. (2010) Glutathione S-transferase P1 and alpha gene variants; role in susceptibility and tumor size development of oral cancer. Head Neck 32: 1079–1087. [DOI] [PubMed] [Google Scholar]
- 14. Morita S, Yano M, Tsujinaka T, Akiyama Y, Taniguchi M, et al. (1999) Genetic polymorphisms of drug-metabolizing enzymes and susceptibility to head-and-neck squamous-cell carcinoma. Int J Cancer 80: 685–688. [DOI] [PubMed] [Google Scholar]
- 15. Park JY, Schantz SP, Stern JC, Kaur T, Lazarus P (1999) Association between glutathione S-transferase pi genetic polymorphisms and oral cancer risk. Pharmacogenetics 9: 497–504. [PubMed] [Google Scholar]
- 16. Jourenkova-Mironova N, Voho A, Bouchardy C, Wikman H, Dayer P, et al. (1999) Glutathione S-transferase GSTM3 and GSTP1 genotypes and larynx cancer risk. Cancer Epidemiol Biomarkers Prev 8: 185–188. [PubMed] [Google Scholar]
- 17. Singh M, Shah PP, Singh AP, Ruwali M, Mathur N, et al. (2008) Association of genetic polymorphisms in glutathione S-transferases and susceptibility to head and neck cancer. Mutat Res 638: 184–194. [DOI] [PubMed] [Google Scholar]
- 18. Harth V, Schafer M, Abel J, Maintz L, Neuhaus T, et al. (2008) Head and neck squamous-cell cancer and its association with polymorphic enzymes of xenobiotic metabolism and repair. J Toxicol Environ Health A 71: 887–897. [DOI] [PubMed] [Google Scholar]
- 19. Soya SS, Vinod T, Reddy KS, Gopalakrishnan S, Adithan C (2007) Genetic polymorphisms of glutathione-S-transferase genes (GSTM1, GSTT1 and GSTP1) and upper aerodigestive tract cancer risk among smokers, tobacco chewers and alcoholics in an Indian population. Eur J Cancer 43: 2698–2706. [DOI] [PubMed] [Google Scholar]
- 20. Peters ES, McClean MD, Marsit CJ, Luckett B, Kelsey KT (2006) Glutathione S-transferase polymorphisms and the synergy of alcohol and tobacco in oral, pharyngeal, and laryngeal carcinoma. Cancer Epidemiol Biomarkers Prev 15: 2196–2202. [DOI] [PubMed] [Google Scholar]
- 21. To-Figueras J, Gene M, Gomez-Catalan J, Pique E, Borrego N, et al. (2002) Microsomal epoxide hydrolase and glutathione S-transferase polymorphisms in relation to laryngeal carcinoma risk. Cancer Lett 187: 95–101. [DOI] [PubMed] [Google Scholar]
- 22. Zhang ZJ, Hao K, Shi R, Zhao G, Jiang GX, et al. (2011) Glutathione S-transferase M1 (GSTM1) and glutathione S-transferase T1 (GSTT1) null polymorphisms, smoking, and their interaction in oral cancer: a HuGE review and meta-analysis. Am J Epidemiol 173: 847–857. [DOI] [PubMed] [Google Scholar]
- 23. Zhuo X, Cai L, Xiang Z, Li Q, Zhang X (2009) GSTM1 and GSTT1 polymorphisms and nasopharyngeal cancer risk: an evidence-based meta-analysis. J Exp Clin Cancer Res 28: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhuo WL, Wang Y, Zhuo XL, Zhu B, Zhu Y, et al. (2009) Polymorphisms of CYP1A1 and GSTM1 and laryngeal cancer risk: evidence-based meta-analyses. J Cancer Res Clin Oncol 135: 1081–1090. [DOI] [PubMed] [Google Scholar]
- 25. Hashibe M, Brennan P, Strange RC, Bhisey R, Cascorbi I, et al. (2003) Meta- and pooled analyses of GSTM1, GSTT1, GSTP1, and CYP1A1 genotypes and risk of head and neck cancer. Cancer Epidemiol Biomarkers Prev 12: 1509–1517. [PubMed] [Google Scholar]
- 26. Wittke-Thompson JK, Pluzhnikov A, Cox NJ (2005) Rational inferences about departures from Hardy-Weinberg equilibrium. Am J Hum Genet 76: 967–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 28. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 29. Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50: 1088–1101. [PubMed] [Google Scholar]
- 30. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karen-Ng LP, Marhazlinda J, Rahman ZA, Yang YH, Jalil N, et al. (2011) Combined effects of isothiocyanate intake, glutathione S-transferase polymorphisms and risk habits for age of oral squamous cell carcinoma development. Asian Pac J Cancer Prev 12: 1161–1166. [PubMed] [Google Scholar]
- 32. Ruwali M, Pant MC, Shah PP, Mishra BN, Parmar D (2009) Polymorphism in cytochrome P450 2A6 and glutathione S-transferase P1 modifies head and neck cancer risk and treatment outcome. Mutat Res 669: 36–41. [DOI] [PubMed] [Google Scholar]
- 33. Datta S, Majumder M, Biswas NK, Sikdar N, Roy B (2007) Increased risk of oral cancer in relation to common Indian mitochondrial polymorphisms and Autosomal GSTP1 locus. Cancer 110: 1991–1999. [DOI] [PubMed] [Google Scholar]
- 34. Cho CG, Lee SK, Nam SY, Lee MS, Lee SW, et al. (2006) Association of the GSTP1 and NQO1 polymorphisms and head and neck squamous cell carcinoma risk. J Korean Med Sci 21: 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng YJ, Chien YC, Hildesheim A, Hsu MM, Chen IH, et al. (2003) No association between genetic polymorphisms of CYP1A1, GSTM1, GSTT1, GSTP1, NAT2, and nasopharyngeal carcinoma in Taiwan. Cancer Epidemiol Biomarkers Prev 12: 179–180. [PubMed] [Google Scholar]
- 36. Katoh T, Kaneko S, Takasawa S, Nagata N, Inatomi H, et al. (1999) Human glutathione S-transferase P1 polymorphism and susceptibility to smoking related epithelial cancer; oral, lung, gastric, colorectal and urothelial cancer. Pharmacogenetics 9: 165–169. [PubMed] [Google Scholar]
- 37. Soucek P, Susova S, Mohelnikova-Duchonova B, Gromadzinska J, Moraviec-Sztandera A, et al. (2010) Polymorphisms in metabolizing enzymes and the risk of head and neck squamous cell carcinoma in the Slavic population of the central Europe. Neoplasma 57: 415–421. [DOI] [PubMed] [Google Scholar]
- 38. Reszka E, Czekaj P, Adamska J, Wasowicz W (2008) Relevance of glutathione S-transferase M1 and cytochrome P450 1A1 genetic polymorphisms to the development of head and neck cancers. Clin Chem Lab Med 46: 1090–1096. [DOI] [PubMed] [Google Scholar]
- 39. Evans AJ, Henner WD, Eilers KM, Montalto MA, Wersinger EM, et al. (2004) Polymorphisms of GSTT1 and related genes in head and neck cancer risk. Head Neck 26: 63–70. [DOI] [PubMed] [Google Scholar]
- 40. Oude Ophuis MB, Roelofs HM, van den Brandt PA, Peters WH, Manni JJ (2003) Polymorphisms of the glutathione S-transferase P1 gene and head and neck cancer susceptibility. Head Neck 25: 37–43. [DOI] [PubMed] [Google Scholar]
- 41. McWilliams JE, Evans AJ, Beer TM, Andersen PE, Cohen JI, et al. (2000) Genetic polymorphisms in head and neck cancer risk. Head Neck 22: 609–617. [DOI] [PubMed] [Google Scholar]
- 42. Jourenkova-Mironova N, Voho A, Bouchardy C, Wikman H, Dayer P, et al. (1999) Glutathione S-transferase GSTM1, GSTM3, GSTP1 and GSTT1 genotypes and the risk of smoking-related oral and pharyngeal cancers. Int J Cancer 81: 44–48. [DOI] [PubMed] [Google Scholar]
- 43. Matthias C, Bockmuhl U, Jahnke V, Harries LW, Wolf CR, et al. (1998) The glutathione S-transferase GSTP1 polymorphism: effects on susceptibility to oral/pharyngeal and laryngeal carcinomas. Pharmacogenetics 8: 1–6. [DOI] [PubMed] [Google Scholar]
- 44. Kelders WP, Oude Ophuis MB, Roelofs HM, Peters WH, Manni JJ (2002) The association between glutathione S-transferase P1 genotype and plasma level in head and neck cancer. Laryngoscope 112: 462–466. [DOI] [PubMed] [Google Scholar]
- 45. Leichsenring A, Losi-Guembarovski R, Maciel ME, Losi-Guembarovski A, Oliveira BW, et al. (2006) CYP1A1 and GSTP1 polymorphisms in an oral cancer case-control study. Braz J Med Biol Res 39: 1569–1574. [DOI] [PubMed] [Google Scholar]
- 46. Hatagima A, Costa EC, Marques CF, Koifman RJ, Boffetta P, et al. (2008) Glutathione S-transferase polymorphisms and oral cancer: a case-control study in Rio de Janeiro, Brazil. Oral Oncol 44: 200–207. [DOI] [PubMed] [Google Scholar]
- 47. Amador AG, Righi PD, Radpour S, Everett ET, Weisberger E, et al. (2002) Polymorphisms of xenobiotic metabolizing genes in oropharyngeal carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 93: 440–445. [DOI] [PubMed] [Google Scholar]
- 48. Olshan AF, Weissler MC, Watson MA, Bell DA (2000) GSTM1, GSTT1, GSTP1, CYP1A1, and NAT1 polymorphisms, tobacco use, and the risk of head and neck cancer. Cancer Epidemiol Biomarkers Prev 9: 185–191. [PubMed] [Google Scholar]
- 49. Rybicki BA, Neslund-Dudas C, Nock NL, Schultz LR, Eklund L, et al. (2006) Prostate cancer risk from occupational exposure to polycyclic aromatic hydrocarbons interacting with the GSTP1 Ile105Val polymorphism. Cancer Detect Prev 30: 412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee SA, Fowke JH, Lu W, Ye C, Zheng Y, et al. (2008) Cruciferous vegetables, the GSTP1 Ile105Val genetic polymorphism, and breast cancer risk. Am J Clin Nutr 87: 753–760. [DOI] [PubMed] [Google Scholar]
- 51. Miller DP, De Vivo I, Neuberg D, Wain JC, Lynch TJ, et al. (2003) Association between self-reported environmental tobacco smoke exposure and lung cancer: modification by GSTP1 polymorphism. Int J Cancer 104: 758–763. [DOI] [PubMed] [Google Scholar]
- 52. Geisler SA, Olshan AF (2001) GSTM1, GSTT1, and the risk of squamous cell carcinoma of the head and neck: a mini-HuGE review. Am J Epidemiol 154: 95–105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flowchart for selection of studies.
(PDF)
PRISMA Checklist.
(PDF)