Abstract
We present a methodology for the design, construction, and modification of synthetic gene networks. This method emphasizes post-assembly modification of constructs based on network behavior, thus facilitating iterative design strategies and rapid tuning and repurposing of gene networks. The ease of post-construction modifications afforded by this approach and the ever-increasing repository of components within the framework will help to fast-track the development of functional genetic circuits for synthetic biology.
Synthetic biology is focused on understanding biological design principles and programming new biological behaviors through the construction of artificial circuits from well-characterized genetic components. This engineering-driven approach was inspired by the development of two small genetic circuits: a bistable toggle switch1 and a self-sustaining oscillator2. Since these early demonstrations, the scale and complexity of synthetic gene networks have increased3–9.
The growing scale and complexity of synthetic gene networks requires new methods for network design, construction, and modification. Traditionally, ad hoc plasmid construction and modification schemes use DNA ligase to join restriction fragments. This becomes unwieldy when the number of components and the size of the construct limit the availability of unique restriction sites. To address this difficulty, several approaches have been recently developed10–12 that focus on standardizing the assembly of larger DNA fragments. Additive assembly, however, does not address other critical issues in the design and construction of synthetic gene networks, notably the need for post-assembly modifications and substitutions in response to the network’s observed performance.
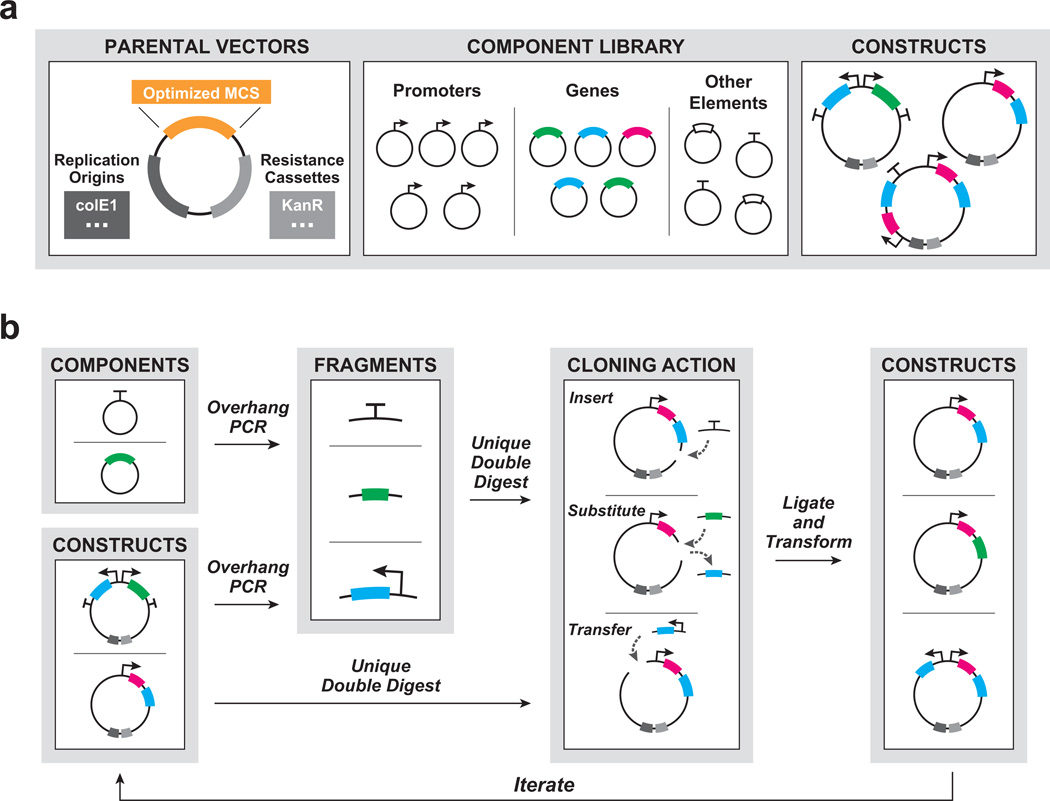
To address these needs, we developed a flexible plug-and-play approach for constructing and modifying synthetic gene networks. Drawing inspiration from the solderless breadboards used to develop electrical circuit prototypes, our platform provides for rapid and scalable assembly within the familiar molecular biology framework, while facilitating post-assembly modifications. The method features a set of optimal type IIp restriction enzymes whose respective restriction sites define the multiple cloning site (MCS) within the cloning vectors (Supplementary Table 1). The set of enzymes was chosen according to a specific set of parameters to ensure maximal compatibility during cloning (Online Methods). The method also features compatible genetic components, which have been optimized to exclude internal instances of the reserved sites. This permits post-assembly modifications by unique double digest (Fig. 1).
Figure 1.
Plug-and-play methodology for synthetic gene networks (a) Elements comprising the framework: parental cloning vectors harboring a custom multiple cloning site (MCS) of optimal restriction enzyme sites; a library of commonly-used synthetic genetic components designed to exclude the restriction sites; a repository of assembled constructs that includes synthetic modules, intermediates, and circuits. (b) Generalized workflow for constructing and modifying synthetic gene networks, which prioritizes and streamlines the iterative process of arriving at functional networks and modules.
We selected an initial set of 26 well-characterized genetic components, including 12 genes and eight promoters (Fig. 1a, Supplementary Table 2), based on common usage in previously published Escherichia coli synthetic gene networks. We then optimized the sequences to exclude the MCS restriction sites without altering component function, by synonymous codon substitution for genes and annotation-guided or randomized mutagenesis for promoters and other regulatory elements (Supplementary Note). The components were then constructed either by synthesis (DNA2.0, Menlo Park, CA), PCR amplification, or site-directed mutagenesis of the source components. We confirmed library parts with optimized sequences for proper functionality and, when possible, compared with their non-optimized counterparts (Supplementary Figs. 1–3).
Constructing synthetic gene networks using this cloning process is straightforward (Fig. 1b). Components are each assigned to a directional “slot”, a pair of adjacent restriction sites within the MCS, and cloning is performed using classical molecular biology techniques. To demonstrate the approach, we recapitulated the original genetic toggle switch1 by designing, constructing, and tuning a bistable LacI-TetR genetic toggle switch from optimized vector and library components (Fig. 2a,b). The bistable toggle switch can maintain its respective genetic state upon removal of the chemical inducers. Induction with anhydrotetracycline (aTc) relieves TetR repression, allowing for high expression of LacI and GFP, while induction with isopropyl-β-D-1-thiogalactopyranoside (IPTG) relieves the LacI repression and produces the high TetR and mCherry state. We switched the toggle between the states via the addition of the respective chemical inputs and reliably maintained the states upon removal of the inducers (Fig. 2c, Supplementary Fig. 4).
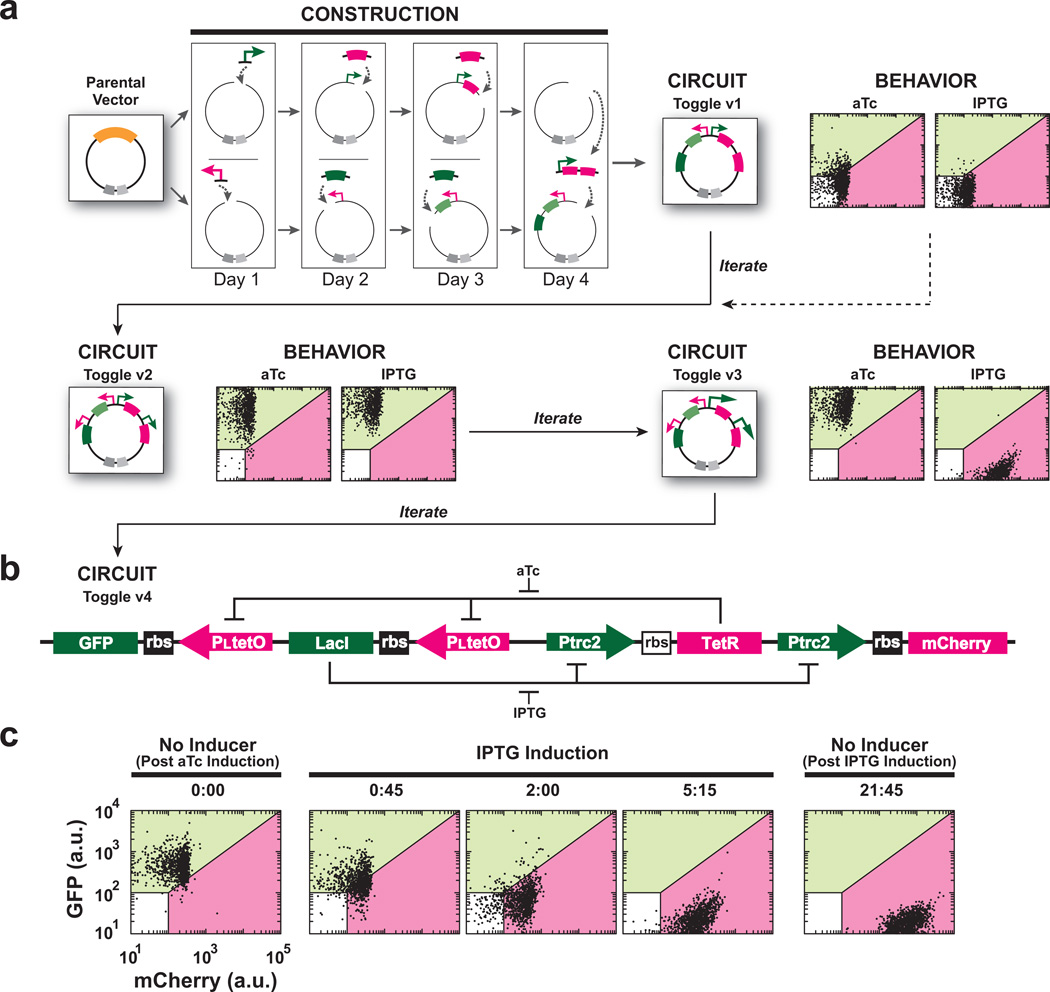
Figure 2.
Construction and tuning of a bistable genetic toggle switch. (a) Representation of the construction and characterization-driven tuning of a genetic toggle switch. Each of the intermediate toggle constructs was induced overnight with either aTc or IPTG, and cells were assayed for expression of fluorescent proteins (GFP and mCherry) by flow cytometry. The Parental Vector contains an antibiotic resistance gene (dark grey) and an origin of replication (light grey). (b) Schematic of the final bistable toggle switch. (c) IPTG-induced switching and subsequent maintenance of the genetic toggle switch. A time-course of cells that harbor the circuit switching from the GFP state (0 hrs) to the mCherry state (0–5:15 hrs) through IPTG induction, and then maintained in the mCherry state when diluted into the no-inducer condition and grown overnight (21:45 hrs). Data were obtained by flow cytometry at the indicated times; n = 10,000 events per experiment.
We found that multiple post-assembly modifications were required to arrive at a functional, bistable genetic toggle. Our approach accelerates characterization-driven iteration by permitting modification in lieu of complete reassembly. In this case, our initial bicistronic toggle construct (Toggle v1) did not activate in response to either of the inducers (Fig. 2a). In order to eliminate possible problems related to poor polycistronic expression and ensure transcription of the fluorescent reporters, we added an additional instance of both promoters to each node converting the toggle to a monocistronic design (Toggle v2). Toggle v2 exhibited aTc-induced GFP expression, but failed to produce mCherry expression in response to IPTG (Fig. 2a). This suggested poor promoter-driven expression of both mCherry and TetR. Our component library contained multiple LacI-controlled promoters, so we simply swapped both instances of the promoter PLlacO with Ptrc-2 (Fig. 2a), which we found to be a stronger promoter (Supplementary Fig. 1). The resulting toggle (Toggle v3) activated each state upon addition of the respective inducer. However, Toggle v3 was unable to maintain the high LacI (GFP) state in the absence of aTc, displaying a bias towards the high TetR (mCherry) state. We therefore sought to balance the circuit by performing random mutagenesis of the Shine-Dalgarno sequence in the ribosome-binding site (RBS) that controls the translation initiation rate of TetR. The fully functional, bistable toggle switch (Toggle v4) could be triggered to each state by addition of the respective inducer and successfully displayed memory of the states upon removal of the inducers (Fig. 2c). Of note, construction of the initial architecture of the toggle took five days. We completed the subsequent iterative tuning in separate three-day efforts, guided by the network’s performance (Fig. 2a).
Our method also enables large-scale reconstruction and design changes. For example, existing modules and circuits can be repurposed into usable cloning parts for novel synthetic gene networks that share common components and architectures. This eliminates the need for additional time-consuming de novo reconstruction of existing modules. To demonstrate this reconfigurability, we used the Toggle v4 as the starting point for the construction of three-node and four-node coherent feed-forward loops (FFLs)13 (Fig. 3a), which are functionally distinct from the bistable toggle switch.
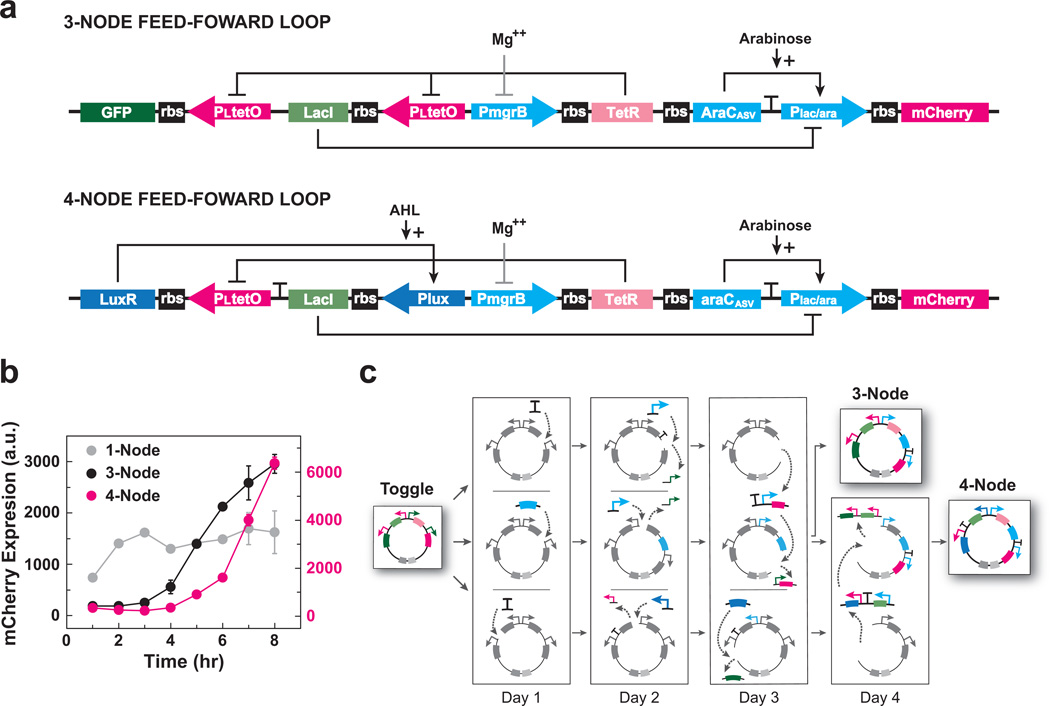
Figure 3.
Transformation of the genetic toggle switch into functional three- and four-node feed-forward loops. (a) Schematics of the three- and four-node feed-forward loop networks. Mg++ indirectly inhibits PmgrB via the PhoPQ two-component system. (b) Pulse response for one-, three- and four-node feed-forward loops. Cells containing the three- or four-node feed-forward loops were induced by the absence of MgCl2. After a 6 hour induction, MgCl2 was spiked into the media to stop induction. Flow cytometry measured mCherry fluorescence (a.u.) at one-hour intervals over an 8 hour time-course. Fluorescence data for one-node and three-node loops are plotted on the left (black) y-axis; fluorescence data for the four-node loop is plotted on the right (red) y-axis. Points represent mean values for three experiments ± s.d.; n = 10,000 events per experiment. (c) Representation of the five-day, parallel subcloning process used to construct plasmids encoding three- and four-node feed-forward loops starting from a genetic toggle switch.
Coherent feed-forward loops act as sign-sensitive delays in response to step stimuli13. Our FFL designs are comprised of a direct and indirect path to a final logical node controlling expression of a fluorescent reporter (mCherry) (Fig. 3a). Additional nodes added to the indirect path of the FFL increase the threshold duration. In the three-node FFL (Fig. 3a), which is operated in a background presence of arabinose, the promoter PmgrB triggers expression of the first node when induced by the absence of magnesium (Mg++) in the media. This activates both paths, producing AraC in the direct path and TetR in the indirect path. TetR subsequently represses the production of LacI in the next node of the indirect path. The hybrid promoter Plac/ara, which approximates an AND-like gate to produce mCherry, then integrates the two paths at the final node. The four-node FFL (Fig. 3a), which is operated in a background presence of arabinose and N-acyl homoserine lactone (AHL), has an additional transcriptional node involving LuxR in the indirect path.
We compared the outputs of the three- and four-node FFLs to each other and to a one-node control (Fig. 3b). In the control circuit, the promoter PmgrB drives the expression of mCherry directly. We induced the three circuits for 6 hours in the absence of Mg++ and made fluorescence measurements every hour during an 8 hour time-course. The circuit responses were markedly different. The one-node control responded immediately to the inducer, while the three- and four-node FFLs required longer induction times before producing outputs (Fig. 3b).
To arrive at these new networks, we designed a parallelized construction strategy originating from Toggle v4 (Fig. 3c). Transformation from the toggle to the three-node FFL required two insertions and two substitutions, while the four-node FFL required a further insertion and two additional substitutions. By making these modifications in parallel, we generated both feed-forward constructs in four rounds of subcloning over the course of five days (Fig. 3c).
Arriving at a functional synthetic gene network that behaves according to predicted phenotype is a two-part process. It requires physically constructing the gene network by assembling DNA encoding the network components, then tuning the network components and architecture to achieve desired functionality. Many new DNA assembly techniques, such as Golden Gate10 and Gibson11 cloning, are addressing the front end of this process. Our method provides a flexible platform that focuses on the back end of the development cycle, and is fully compatible with various assembly techniques that can be used at the front end, provided design considerations are made to incorporate the set of reserved restriction sites and components. Also, recycling existing circuits, intermediates, and components, as afforded by our approach, reduces the time required to re-synthesize or recreate validated parts. As the collection of well-characterized synthetic components, modules, and circuits grows, their reuse and repurposing will simplify the assembly of subsequent networks. Altogether, our platform accelerates the design-build-test cycle in synthetic biology by integrating assembly and debugging. Furthermore, our approach is well suited for integration with emerging computational platforms intended to implement rational design and automation in synthetic biology14–17, 18–20. Ultimately, our methodolgy, used with appropriate vectors and components, could be expanded to other bacterial systems, as well as yeast, plant and mammalian systems, facilitating the creation of complex synthetic gene networks in a wide range of organisms.
Online Methods
Restriction Enzyme Selection
The restriction enzymes used in our approach have recognition sites that are non-degenerate, palindromic, and six bp in length. To achieve consistent digest reaction conditions, the MCS restriction enzymes were also chosen to have at least 50 percent activity in the common buffer NEBuffer 4 (New England Biolabs) at 37 °C. The spacing of restriction sites in the MCS allows for double digests at adjacent pairs of restriction sites, enabling unique and directional insertions. Additionally, the inclusion of non-adjacent restriction sites that produce blunt or compatible cohesive ends facilitates large-scale reorganizations of the MCS. The set of high-copy cloning vectors that include this MCS was based on the pZE vector family21 (Fig. 1a). Instances of reserved restriction sites were removed from the vector backbones via codon substitution or randomized site-directed mutagenesis.
Cloning Details
The desired restriction sites are added to the 5’ and 3’ ends of each component using overhang-PCR. For each insertion, both component and vector are uniquely double-digested. During post-assembly modifications, the prior insert is removed by gel purification following vector digest. The fragments are then ligated and transformed, and the resulting clones are screened for the desired product. Since the restriction sites used throughout construction remain unique, assembly order is not constrained, which allows for a greater degree of flexibility in planning parallel assembly.
Component Characterization
Input/output functions were obtained for all of the optimized components and, when possible, compared to those for their unaltered counterparts. To obtain characterization data, custom test circuits were first constructed for each class of components, as detailed below. Then appropriate dose responses were generated with fluorescence measurements being performed with a BD FACSAriaII flow cytometer (BD Biosciences).
For promoters, cassettes were constructed in which the promoters drive the expression of a GFP reporter (Supplementary Fig. 1); these plasmids were transformed into MG1655 Pro cells. Dose response experiments were performed using the appropriate inducers for each promoter (Supplementary Fig. 1). The promoters were compared to their commonly-used, unmodified equivalents. In virtually all cases, the input/output functions matched well. Notably, the optimized PLlacO promoter exhibited a larger dynamic range than the original unaltered version, with other dose response dynamics matching closely. The PLtetO promoter was not optimized, since the original did not have any reserved restriction sites in its sequence.
For transcription factors (TFs), cascade circuits were constructed in which the magnesium-responsive PmgbrB drives the inducible expression of the TF, which subsequently operates on a specific promoter to drive the expression of a GFP reporter (Supplementary Fig. 2). These circuits were transformed into the respective MG1655-derived knockout strains (i.e., strains that have the native TF gene knocked out). Dose responses were performed on the TF-sensitive promoters in the absence (with magnesium) and presence (without magnesium) of the TF. In the case of the cI protein, the dose response was performed with magnesium as the inducer, since cI is not regulated by a chemical inducer. All of our optimized TFs were able to be expressed and predictably regulated their specific promoters in a dose-dependent fashion.
For our optimized terminator, two simple circuits were constructed in which PBAD drove the expression of two bicistronic reporters, GFP and mCherry, with and without the terminator between the two reporters (Supplementary Fig. 3). These circuits were transformed into MG1655 cells. The expression of the two reporters was measured in response to induction. For the circuit with the terminator present, the downstream reporter, mCherry, produced no expression confirming proper function.
Two optimized reporters were also tested for fluorescence output. The test circuits consisted of PLtetO driving the expression of the respective fluorescent reporter (Supplementary Fig. 3). The circuits were transformed into MG1655 cells. The tested reporters all exhibited dose-dependent expression.
Primer Design
The PCR primers used for plasmid constriction and modification were designed according to the following algorithm. Each primer began (5’) with six bases arbitrarily selected in a manner to create primers with similar Tm, calculated using OligoCalc (http://www.basic.northwestern.edu/biotools/oligocalc.html). The next six bases (5’-3’) comprised the desired restriction enzyme recognition site. The remainder of the primer, the 3’ end, consisted of either the “Fw. Primer Homology” or the “Rev. Primer Homology” sequences, annotated in the component sequence entries. Thus, the final primer design was:
5’ - six arbitrary bases + six bases for recognition site + Fw./Rev. Primer Homology – 3’
The primers were ordered from IDT.
Induction Conditions
All toggle experiments were performed in M9 minimal media supplemented with 0.2 % glucose and 0.02 % Casamino acids. We induced the Ptrc2 promoter with IPTG at a concentration of 1 mM and the PLtetO promoter with 100ng/mL aTc.
All feed forward loop experiments were performed in M9 minimal media supplemented with 0.2 % glucose and 0.02 % Casamino acids and a background of 0.01 % Arabinose. Experiments involving the four-node feed forward loop were also performed in the presence of 10 µM AHL. We performed experiments on the feed-forward loops by repressing the PmgrB promoter with 50 mM MgCl2.
Editorial Summary:
A unique multiple cloning site (MCS) and defined genetic components without MCS restriction sites allow for the rapid construction and iterative tuning of synthetic genetic circuits.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the Howard Hughes Medical Institute and the Office of Naval Research MURI program.
Footnotes
AUTHOR CONTRIBUTIONS
K.D.L., R.B.A., A.S.K., and J.J.C. designed the study. K.D.L., R.B.A., R.J.K., and A.S.K. performed experiments. K.D.L., R.B.A., R.J.K., A.S.K., and J.J.C. wrote the manuscript. A.S.K. and J.J.C. supervised the project.
References
- 1.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 2.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 3.Culler SJ, Hoff KG, Smolke CD. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science. 2010;330:1251–1255. doi: 10.1126/science.1192128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danino T, Mondragon-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khalil AS, et al. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 8.Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded NOR gates and chemical 'wires'. Nature. 2011;469:212–215. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- 10.Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 2009;4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 12.Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 13.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cambray G, Mutalik VK, Arkin AP. Toward rational design of bacterial genomes. Curr Opin Microbiol. 2011;14:624–630. doi: 10.1016/j.mib.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Carothers JM, Goler JA, Juminaga D, Keasling JD. Model-driven engineering of RNA devices to quantitatively program gene expression. Science. 2011;334:1716–1719. doi: 10.1126/science.1212209. [DOI] [PubMed] [Google Scholar]
- 16.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beal J, et al. An End-to-End Workflow for Engineering of Biological Networks from High-Level Specifications. ACS Synthetic Biology. 2012;1:317–331. doi: 10.1021/sb300030d. [DOI] [PubMed] [Google Scholar]
- 19.Lux MW, Bramlett BW, Ball DA, Peccoud J. Genetic design automation: engineering fantasy or scientific renewal? Trends Biotechnol. 2012;30:120–126. doi: 10.1016/j.tibtech.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandran D, Bergmann F, Sauro H. TinkerCell: modular CAD tool for synthetic biology. J Biol Eng. 2009;3:19. doi: 10.1186/1754-1611-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.